Abstract
The emergence of antibiotic resistance among nosocomial pathogens has reemphasized the need for effective infection control strategies. The spread of resistant pathogens within hospital settings proceeds along various routes of transmission and is characterized by large fluctuations in prevalence, which are typical for small populations. Identification of the most important route of colonization (exogenous by cross-transmission or endogenous caused by the selective pressure of antibiotics) is important for the design of optimal infection control strategies. Such identification can be based on a combination of epidemiological surveillance and costly and laborious as well as time-consuming methods of genotyping. Furthermore, analysis of the effects of interventions is hampered by the natural fluctuations in prevalence. To overcome these problems, we introduce a mathematical algorithm based on a Markov chain description. The input is longitudinal prevalence data only. The output is estimates of the key parameters characterizing the two colonization routes. The algorithm is tested on two longitudinal surveillance data sets of intensive care patients. The quality of the estimates is determined by comparing them to accurate estimates based on additional information obtained by genotyping. The results warrant optimism that this algorithm may help to quantify transmission dynamics and can be used to evaluate the effects of infection control interventions more carefully.
Infections with antibiotic-resistant bacteria are emerging in hospitalized patients, especially those treated in intensive care units (ICUs) (1). Nosocomial pathogens with resistance to almost all commercially available antibiotics [e.g., vancomycin-resistant enterococci (VRE) and vancomycin-intermediate Staphylococcus aureus] have confronted physicians with the possibility of a postantibiotic era (2, 3). Therefore, prevention of transmission of such microorganisms has become even more important (4).
Patient colonization with antibiotic-resistant microorganisms within hospital settings is determined by admission and discharge rates of colonized and noncolonized patients and on the likelihood that noncolonized patients acquire colonization. Several distinct routes of colonization can be distinguished. Colonization may result from exogenous acquisition—i.e., from the environment or from other patients treated in the ward, usually via temporarily contaminated hands of health care workers (5). Alternatively, colonization may be undetectable until, because of the selective growth advantage provided by antimicrobial treatment, bacterial growth of resistant pathogens is such that detection limits of culture methods are exceeded. In these cases, colonization is regarded to be from endogenous sources. Because of the small patient populations involved (typically 10–20 patients in one ICU) large fluctuations in prevalence frequently occur just by chance. In longitudinal studies, fluctuations in point prevalences of colonization with antibiotic-resistant microorganisms in ICUs from 0% to 80% have been observed, in periods without changes in infection control or antibiotic prescription policies (6, 7).
The prevalence of antibiotic-resistant microorganisms in hospital settings is the resultant of the “traffic” along the various routes. Prevention of colonization and infection should be tailored on the relative importance of the transmission routes. Unfortunately, assessing the importance seems to require the use of microbiological surveillance in combination with genotyping of microorganisms. Microbiological surveillance is necessary to determine prevalence, and genotyping is essential to demonstrate that acquired colonization is a result of either cross-transmission or endogenous colonization. As genotyping techniques are costly and laborious, wide-scaled use is precluded. As a result, the relative importance of the different routes of colonization in ICUs is generally not known. Such knowledge is also necessary to evaluate the effects of interventions reliably. Because of the natural occurrence of large fluctuations in prevalence, it is difficult to ascribe a change in prevalence to a specific intervention. Separate quantification of exogenous and endogenous incidences will provide more reliable information.
In the present study, therefore, we develop a method for estimating the relative importance on the basis of readily available longitudinal surveillance data. We do so within the framework of a Markov chain model, which involves assumptions concerning the admission and discharge of patients as well as pathogen transmission.
Recently, several mathematical models have been used to investigate transmission dynamics of antibiotic-resistant pathogens within hospitals and hospital wards (6, 8–10). For example, Austin et al. (6) used the Ross model of vector-borne diseases to describe the dynamics of VRE in ICUs and validated the model on epidemiological data obtained in an ICU where colonization with VRE was endemic. Special emphasis was put on the quantitative effects of classical infection control measures, such as hand disinfection and staff cohorting (6). Independently, Lipsitch et al. (8) constructed a model of the spread of antibiotic resistance in hospitals, and qualitatively predicted the effects of different antibiotic strategies. Both models were basically deterministic, using differential equations to describe bacterial transmission. Such deterministic models may not be reliable when populations are small, such as those in hospital settings, as opposed to general populations. In some models the conclusions were, therefore, further tested against Monte Carlo simulations (6, 10).
We will show that our algorithm for data analysis, which is based on a stochastic (Markov chain) model, provides a useful tool to analyze the dynamics of transmission and to determine the relative importance of different routes of colonization within hospital settings by using only longitudinal data on the number of patients being colonized. Our assumptions concerning discharge and colonization formulated below are identical to those of Austin et al. (6). The difference is that we explicitly represent the patients in the actual number of beds in a ward, rather than concentrating on the mean and the variance of the number of colonized patients.
To test the method we have applied it to two data sets, both containing detailed information obtained by genotyping (6, 7). For this purpose we have initially ignored the information based on genotyping and have used only the time series of colonized patients. The accuracy of the model predictions, however, was assessed by comparison with the conclusions that were derived from the full data set, including genotyping results. In other words, the results of genotyping were used as a gold standard, to check how accurately we can assess the situation when such information is not available.
Formulation of the Model
We consider the spread of one species of bacteria in an ICU composed of N beds, which, reflecting the resources needed to maintain them, are assumed to be occupied to full capacity at all times. The patients are either colonized or noncolonized. A patient is considered colonized when colonization has been demonstrated by standard microbiological culture techniques. Rates of discharge are denoted as 1/d (in days) for noncolonized patients and 1/d′ for colonized patients. A change in the number of colonized patients in ICU, say i, can occur in the following ways:
(i) A noncolonized patient is replaced by a colonized patient. If admitted patients are colonized with probability q, this replacement occurs with per bed rate q/d.
(ii) A colonized patient is replaced by a noncolonized patient. This replacement occurs with per bed rate (1 − q)/d′.
(iii) A noncolonized patient acquires colonization spontaneously—i.e., endogenous colonization occurs. The chance for this event per unit time per patient is denoted by α. Explosive growth under antibiotic treatment of preexisting small numbers of resistant microorganisms is the most likely explanation, but the development of resistance de novo is an alternative or additional possibility. We assume that every patient may acquire colonization spontaneously and disappearance of evident colonization will not occur (true for typical lengths of stay).
(iv) A noncolonized patient acquires colonization from another patient [cross-colonization, usually via contaminated hands of health care workers (HCWs)]. The rate of transmission is assumed to be equal to θi/N, with θ a constant depending on the details of patient–HCW interactions, but independent of the number of colonized patients, i, whenever the duration of contamination for HCWs is small.
We can, and do, ignore the replacement of a patient by another patient with the same colonization status. With these transition rates specified we can now formulate a bookkeeping equation (the so-called master equation) for this continuous-time Markov process:
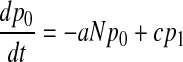 |
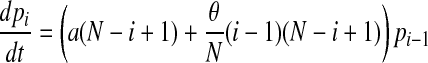 |
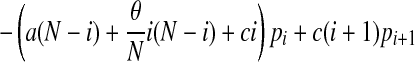 |
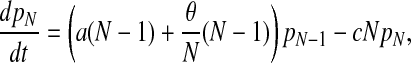 |
1 |
with pi(t) denoting the probability of having i colonized patients at time t and the parameters a and c defined in terms of q, d, d′, and α introduced above, by
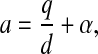 |
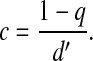 |
2 |
The compound parameter a is the spontaneous colonization rate, including replacement of noncolonized by colonized patients; c is the “decolonization” rate (i.e., replacement of colonized by noncolonized patients), and θ is the transmission rate. The changes in the course of time of the number of colonized patients in the model are completely determined by these three parameters.
The Stationary Distribution
The distribution of colonized patients will, in the long run, converge to the stationary distribution ps, which is determined by the condition dPi/dt = 0 and given explicitly by the formula
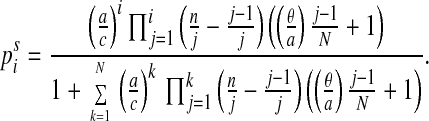 |
3 |
Note that the stationary distribution depends only on the ratio
a/c between the spontaneous colonization rate
(a) and the “decolonization” rate (c), and
the ratio θ/a between the transmission rate (θ) and
the spontaneous colonization rate (a). The key point to note
about the meaning of the stationary distribution is that
p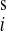 gives the probability to find
precisely i colonized patients in the ICU at any given
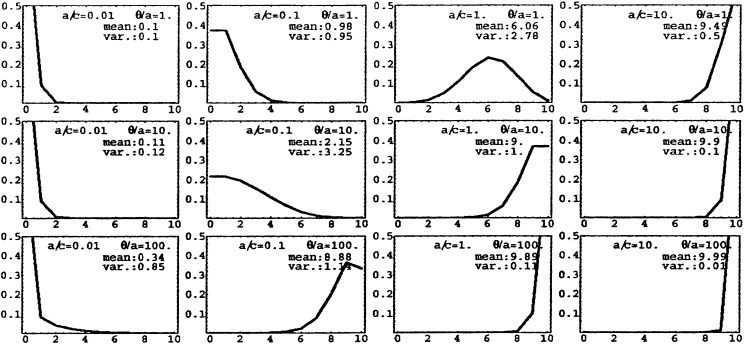
moment of time. The stationary distribution for n = 10
and a variety of parameter values a/c and
a/θ are depicted in Fig.
1. An almost normal distribution occurs
when a/c = 1 and θ/a =
1, but for other parameter values the shape of the distribution is very
different. Note that the distributions are not very sharply peaked,
meaning that in the course of time nonnegligible fluctuations in the
actual number of colonized patients are to be expected. Therefore, an
observed increase in the number of colonized patients does not
necessarily indicate a deterioration of hygienic standards, as it may
be a chance fluctuation. Neither should low colonization rates be a
reason for complacency. Note that large θ/a values give
large “tails” to the distribution corresponding to outbreak
situations, but increases in a also shift the distribution
rightwards. Incidentally, we remark that for q tending to
zero the stationary distribution should converge to the quasistationary
distribution as studied by Nasell (11), but we did not explore this
connection.
gives the probability to find
precisely i colonized patients in the ICU at any given
moment of time. The stationary distribution for n = 10
and a variety of parameter values a/c and
a/θ are depicted in Fig.
1. An almost normal distribution occurs
when a/c = 1 and θ/a =
1, but for other parameter values the shape of the distribution is very
different. Note that the distributions are not very sharply peaked,
meaning that in the course of time nonnegligible fluctuations in the
actual number of colonized patients are to be expected. Therefore, an
observed increase in the number of colonized patients does not
necessarily indicate a deterioration of hygienic standards, as it may
be a chance fluctuation. Neither should low colonization rates be a
reason for complacency. Note that large θ/a values give
large “tails” to the distribution corresponding to outbreak
situations, but increases in a also shift the distribution
rightwards. Incidentally, we remark that for q tending to
zero the stationary distribution should converge to the quasistationary
distribution as studied by Nasell (11), but we did not explore this
connection.
Figure 1.
The stationary distribution of colonized patients (Eq. 2) for various values of the parameters a/c and θ/a (n = 10). On the horizontal axis is the number of colonized patients i, and on the vertical axis is the probability of encountering i colonized patients (or, equivalently, the fraction of the time i patients are colonized in the long run).
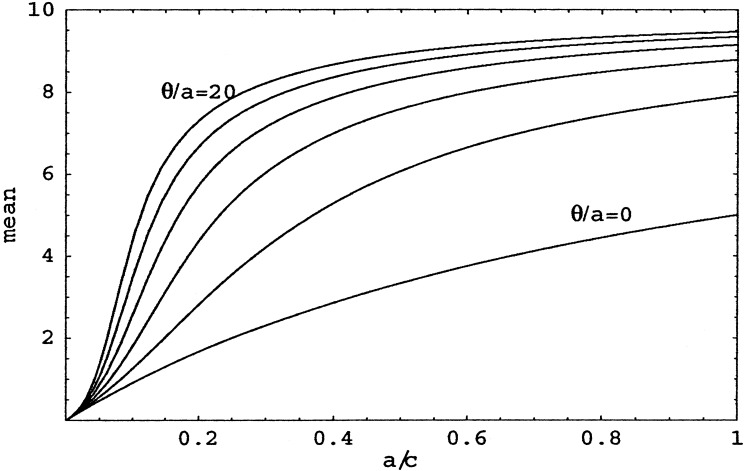
Concentrating for the moment on the mean number of colonized patients, we see (Fig. 2) that it goes to 0 when a/c decreases (i.e., when decolonization occurs much more frequently than spontaneous colonization), but rapidly increases to N when a/c increases. Even for large θ/a, when transmission is relatively important, the number of colonized patients increases only linearly with slope N, for small a/c. This observation confirms the importance of infection control strategies that separate colonized patients from noncolonized patients as quickly as possible (leading to a small length of stay d′ and thus to a small a/c).
Figure 2.
Mean number of colonized patients for various θ/a as a function of a/c. From the top, the curves are for θ/a = 20, 16, 12, 8, 4, and 0.
Estimating Parameters from Observational Data
Two sets of data on the epidemiology of, respectively, VRE and Pseudomonas aeruginosa were obtained from different ICUs (7, 12). In both studies extensive surveillance of colonization and genotyping were combined to determine prevalences of colonization and the contributions of endogenous and exogenous colonization to the incidence. Pulsed-field gel electrophoresis (PFGE) techniques were used for genotyping (13). Cross-transmission was defined as a case of acquired colonization with a pathogen with a genotype identical to that of an isolate of another patient treated in the same ward and in the same time period. Endogenous colonization was defined as acquired colonization with a genotype that was not found to be associated with colonization in any other patient. With these definitions, cross-transmission was the responsible route of colonization in 85% of the patients acquiring colonization with VRE and in 13% of the patients acquiring colonization with P. aeruginosa (7, 12). Table 1 summarizes the main epidemiological determinants for these studies. The parameters c, a, and θ of the model can be computed directly from the information provided in Table 1, and are represented under “Observed” in Table 2.
Table 1.
Epidemiological data concerning studies of VRE and Pseudomonas aeruginosa
| Data | Value
|
|
|---|---|---|
| VRE | P. aeruginosa | |
| Number of beds in ICU (mean occupancy) | 16 (14) | 9 (7) |
| Duration of study period, days | 133 | 299 |
| Number of patients admitted | 189 | 289 |
| Number of patients colonized on admission | 28 | 12 |
| Number of patients with acquired colonization | 45 | 23 |
| Fraction of incoming patients colonized | 0.148 | 0.04 |
| Spontaneous colonization fraction, % | 15 | 87 |
| Spontaneous colonization rate, per day | 0.05 | 0.07 |
| Cross-transmission fraction, % | 85 | 13 |
| Cross-transmission rate, per day | 0.29 | 0.01 |
| Mean endemic prevalence of colonization (X1), % | 36 | 34 |
| Observed mean length of stay of colonized patients (LOS′),* days | 15 | 24 |
| Discharge rate for colonized patients (1/d′) | 1/8 | 1/17 |
| Observed mean length of stay of noncolonized patients (LOS),* days | 7 | 7 |
| Discharge rate for noncolonized patients (1/d) | 1/10 | 1/10 |
Calculation of d and d′:d =LOS/(1 − X1); d′ = LOS′ − LOS.
Table 2.
Estimates of c, a, and θ
| VRE
|
P. aeruginosa
|
|||
|---|---|---|---|---|
| Observed | MLE fit to model | Observed | MLE fit to model | |
| c | 0.09 (0.01) | 0.11 (0.05) | 0.065 (0.015) | 0.069 (0.03) |
| a | 0.03 (0.015) | 0.03 (0.02) | 0.02 (0.01) | 0.021 (0.01) |
| θ | 0.08 (0.03) | 0.09 (0.05) | 0.01 (0.01) | 0.00 (0.02) |
Errors are given in parentheses. MLE, maximum likelihood estimate.
Independently, we determined the parameters c, a, and θ by maximizing the a priori probability of occurrence of the observed time series of the number of colonized patients. This probability follows from the model as follows: if we write Eq. 1 in the form dp/dt = Ap, then the chance of finding j colonized patients at time t + Δt, given the fact that i patients are colonized at time t, is given by (eΔtA)ij; so, given a list {c1, … , cn} of the number of colonized patients at times {t1, … , tn} it is easy to calculate the above-mentioned probability as
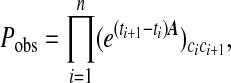 |
4 |
where A, and thereby Pobs, depends on a, c, and θ. Using standard numerical techniques, we then determined the parameters a, c, and θ that maximize Pobs. The outcome is listed in Table 2 under “Maximum likelihood estimate (MLE) fit to model.” As shown, the two methods to estimate a, c, and θ (genotyping and MLE) yield comparable results. The key point is that the MLE yields the relevant information with a minimal input of data. The only data that should be available for this analysis are the surveillance data. There is no need for bacterial genotyping. On the other hand, the data must encompass a sufficiently long period (how long is sufficient depends on the time scales in the ICU, but typically months) to reduce the uncertainty in the determination of the parameters.
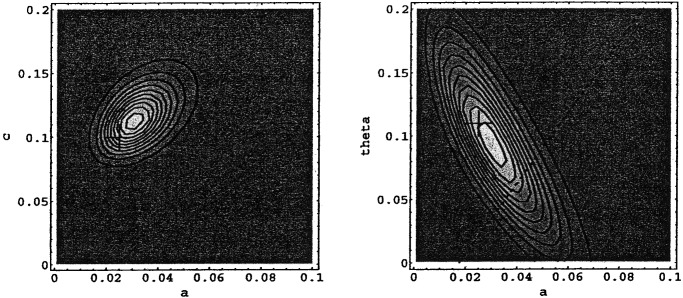
Fig. 3 shows contours of equal Pobs for VRE. Given the observed longitudinal data on the numbers of patients colonized with VRE, the combination of parameters a, c, and θ that maximizes Pobs is depicted by the central contour (and listed as MLE in Table 2). Indicated with crosses are the parameter values obtained by using the complete data set (i.e., those listed under “Observed” in Table 2). As noted before, the “directly observed parameters” (crosses) and the maximum likelihood parameter estimates derived from the Markov chain model are consistent. The contours of equal probability in Fig. 3 Right are stretched in one direction along the line of constant colonization pressure, defined by α + X1⋅θ (with X1 the mean prevalence of colonized patients). The shape of the contours reflects the inherent difficulty of distinguishing between two different routes of colonization (spontaneous and cross-transmission), because both increase the number of colonized patients. We can, however, quantify the relative importance of both colonization routes: Plotted in Fig. 3 Right is the line of equal importance of endogenous colonization and cross-transmission (approximately θ/α⋅X1 = 1). For VRE the probability of the observations is greater above this line [P(θ/α⋅X1 > 1) = 0.75], indicating that cross-transmission was at least as important as, but probably more important than endogenous colonization, in agreement with the conclusions based on bacterial genotyping (θ⋅X1/α ≈ 5 ± 1.5).
Figure 3.
Contour plots of the likelihood Pobs of a–c and a–θ slices of parameter space at θ = 0.91 and c = 0.113, respectively, for VRE. Contours are linear in Pobs. The dotted line indicates equality between spontaneous colonization and cross-transmission.
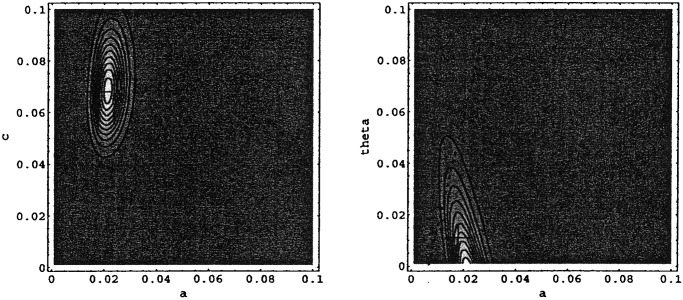
Similarly, the values of Pobs are depicted for P. aeruginosa in Fig. 4. Again, there is good agreement between the parameter estimates and the observations. The results show a much lower value for θ, which is in agreement with the direct observations. The probability of the observations above the equality line is low [P(θ/α⋅X1 > 1) = 0.08], which is in accordance with the fact that cross-transmission was relatively unimportant for P. aeruginosa in this ICU (θ⋅X1/α ≈ 0.1 ± 0.02).
Figure 4.
Contour plots of the likelihood Pobs of a–c slice at θ = 0.001 and of an a–θ slice at c = 0.069 for P. aeruginosa. Contours are linear in Pobs. The dotted line indicates equality between spontaneous colonization and cross-transmission.
Discussion
We have presented an algorithm to determine the relative importance of different routes of bacterial colonization within the context of a simple Markov chain model for small closed populations, such as ICUs or other hospital wards. It uses only readily available longitudinal data on prevalences of colonization in the ICU. We have demonstrated that it is possible to determine the relative importance of endogenous colonization and cross-transmission without resorting to costly and time-consuming methods of genotyping and despite the inherent intricacies of disentangling mechanisms that have the same effect.
The emergence of antibiotic resistance and the possibility of a “postantibiotic” era have reemphasized the need for effective measures to limit the spread of these pathogens. Classic infection control practices have focused on improving antibiotic policies and compliance with infection control measures, such as hand disinfection. Although these measures remain cornerstones of infection control, they have been insufficient to prevent the emergence of antibiotic resistance. A better understanding of the dynamics of colonization and infection may contribute to more successful infection control strategies. For example, the mean endemic prevalences of VRE and P. aeruginosa in the two different ICUs were almost identical. Yet, both fingerprinting of multiple isolates and our estimates from the longitudinal data clearly demonstrated that cross-transmission was the most important transmission route for VRE, whereas endogenous colonization was much more important for P. aeruginosa. Such insights are crucial when designing infection control measures.
Most of the models of nosocomial spread of pathogens are deterministic (6, 8–10)—i.e., the dynamic behavior of the system is completely determined by the initial conditions. However, because the populations within closed hospital settings are usually small, chance effects cannot be neglected (and that is why some authors supplement the analysis of a deterministic model by simulations of a stochastic variant—e.g., refs. 6 and 10). A Markov chain description has at least two advantages over more conventional deterministic models, even when incorporating Gaussian correction terms extends the latter. Markov chain descriptions capture much of the chance fluctuations that are due to the fact that we describe a finite, but often small, population of discrete entities—i.e., the individuals. In addition, there is a direct correspondence between observable quantities and model variables, which allows extracting maximal information about the model parameters from the available data. In typical pre- and postintervention studies, outcomes (such as numbers of patients colonized with a resistant microorganism) are usually compared by standard statistical tests, such as χ2 tests or logistic regression analyses. However, in doing so it is assumed that patients are affected independently, which is not true for transmitted pathogens. The proportion of other patients being colonized, also called colonization pressure, amplifies the risk for noncolonized patients to acquire colonization (14–17). Therefore, statistical tests may yield significant differences between two periods when these are in fact caused by chance. Estimation of the parameters of the Markov chain model before and after an intervention would avoid such problems. The estimates also show in what way the intervention was effective: Was transmission reduced or did the occurrence of spontaneous cases decline?
A central concept in infectious disease dynamics is the basic reproduction number, R0, which corresponds to the average number of secondary infected cases (18, 19). When applied to the dynamics of nosocomial pathogens, R0 represents the number of secondary colonized patients because of cross-transmission generated by a primary case in a pathogen-free ward. Infection prevention aims to decrease the effective R0 below unity [it is helpful to speak about the effective reproduction number when infection control measures have been applied (6)]. In our model the effects of infection control measures are captured in θ, and the effective R is equal to d′⋅θ, so to 0.7 ± 0.2 for VRE and 0.14 ± 0.04 for P. aeruginosa. Previously we found an effective R for VRE of 0.7, using the same data set but a different method (6). This finding implies that neither of the pathogens can persist in the ICU by transmission alone, and that the continuous admission of already colonized patients is needed for endemicity. R0 for VRE was estimated to be ≈3.8 In the absence of infection prevention, however (6).
Our model encompasses a number of simplifications, which may limit its practical applicability. We assumed that the patient population was homogeneous: all patients were considered to be identical with regard to the probability of their length of stay and susceptibility for colonization. Moreover, we have assumed that the various processes occur with certain constant probabilities in each time interval and that the transmission of bacteria from patient to patient occurred instantaneously. In reality some time will probably be needed for bacterial multiplication after an event of colonization before pathogens can be transmitted further. Moreover, some patients may be more likely to act as sources for bacterial spread, because they receive more patient contacts or such contacts are more likely to result in cross-transmission. In addition, our model does not allow for differences in the severity of colonization, as our patients are either considered noncolonized and thus not contagious or colonized and fully contagious. However, a more detailed model capturing all these uncertainties would have more parameters, and so would not necessarily be more reliable. It is also obvious that rapid changes in q (i.e., admission prevalences), for example during epidemics, will influence, and probably decrease, the reliability of the predictions. We assumed q to be constant, which we think is reasonable for the combination of time periods and the kind of nosocomial events we consider. Finally, some important general aspects of data analysis were neglected. For example, we just gave point estimates of the parameters without trying to obtain more information about the probability distribution of the parameters, given the data (20, 21).
In addition, all surveillance techniques will have a lower detection limit, which we have ignored even though it might have an impact on the conclusions. However, we think that the different routes of colonization, and thus relative impacts, would be equally affected. The use of more sensitive tests may identify more patients with presumed endogenous colonization, thereby diminishing the relative impact of exogenous colonization. On the other hand, such a test may identify more patients already colonized on admission, thereby diminishing the relative impact of endogenous colonization. And finally, a more sensitive test may identify more cases of cross-transmission, as patients may initially be colonized with very low numbers of microorganisms. Furthermore, more specific methods of microbial genotyping may diminish incidences of cross-transmission. For example, phenotypes of antimicrobial resistance profiles may not have sufficient discriminatory power to distinguish clonal difference (13). More work is needed to quantify the definition of colonization and its relation to transmissibility. Here, we have assumed that all colonized patients could be a source of transmission, which, we think, is a fair assumption.
In conclusion, we described a method that can identify the relative importance of various transmission routes of microorganisms on the basis of a sufficiently long time series of the number of colonized patients in hospital settings with, typically, a small total number of patients. Such information may both guide infection control strategies and help to carefully evaluate the outcome of intervention studies.
Acknowledgments
We thank M. Lipsitch and M. C. M. de Jong for stimulating discussions and helpful comments.
Abbreviations
- ICU
intensive care unit
- VRE
vancomycin-resistant enterococci
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Vincent J L, Bihari D J, Suter P M, Bruining H A, White J, Nicolas-Chanoin M H, Wolff M, Spencer R C, Hemmer M. J Am Med Assoc. 1995;274:639–644. [PubMed] [Google Scholar]
- 2.Murray B E. N Engl J Med. 2000;342:710–721. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- 3.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 4.Cohen M L. Science. 1992;257:1050–1055. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- 5.Bonten M J M, Weinstein R A. Semin Respir Infect. 2000;15:327–335. doi: 10.1053/srin.2000.20936. [DOI] [PubMed] [Google Scholar]
- 6.Austin D A, Bonten M J M, Slaughter S, Weinstein R A, Anderson R M. Proc Natl Acad Sci USA. 1999;96:6908–6913. doi: 10.1073/pnas.96.12.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonten M J M, Bergmans D C J J, Speijer H, Stobberingh E E. Am J Respir Crit Care Med. 1999;160:1212–1219. doi: 10.1164/ajrccm.160.4.9809031. [DOI] [PubMed] [Google Scholar]
- 8.Lipsitch M, Bergstrom C T, Levin B R. Proc Natl Acad Sci USA. 2000;97:1938–1943. doi: 10.1073/pnas.97.4.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sébille V, Chevret S, Valleron A-J. Infect Control Hosp Epidemiol. 1997;18:84–92. doi: 10.1086/647560. [DOI] [PubMed] [Google Scholar]
- 10.Cooper B S, Medley G F, Scott G M. J Hosp Infect. 1999;43:131–147. doi: 10.1053/jhin.1998.0647. [DOI] [PubMed] [Google Scholar]
- 11.Nasell I. Math Biosci. 1999;156:21–40. doi: 10.1016/s0025-5564(98)10059-7. [DOI] [PubMed] [Google Scholar]
- 12.Slaughter S, Hayden M K, Nathan C, Hu T C, Rice T, Van Voorhis J, Matushek M, Franklin C, Weinstein R A. Ann Intern Med. 1996;125:448–456. doi: 10.7326/0003-4819-125-6-199609150-00004. [DOI] [PubMed] [Google Scholar]
- 13.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonten M J M, Slaughter S, Ambergen A W, Hayden M K, van Voorhis J, Nathan C, Weinstein R A. Arch Intern Med. 1998;158:1127–1132. doi: 10.1001/archinte.158.10.1127. [DOI] [PubMed] [Google Scholar]
- 15.Merrer J, Santoli F, Appéré-De Vecchi C, Tran B, de Jonghe B, Outin H. Infect Control Hosp Epidemiol. 2000;21:718–723. doi: 10.1086/501721. [DOI] [PubMed] [Google Scholar]
- 16.de Man P, van der Veeke E, Leemreijze M, van Leeuwen W, Vos G, van Den Anker J, Verbrugh H, van Belkum A. J Infect Dis. 2001;184:211–214. doi: 10.1086/322014. [DOI] [PubMed] [Google Scholar]
- 17.Puzniak L A, Mayfield J, Leet T, Kollef M, Mundy L M. Clin Infect Dis. 2001;33:151–157. doi: 10.1086/321807. [DOI] [PubMed] [Google Scholar]
- 18.Anderson R M, May R M. Infectious Diseases of Humans: Dynamics and Control. Oxford: Oxford Univ. Press; 1991. [Google Scholar]
- 19.Diekmann O, Heesterbeek H. Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation. Chichester, U.K.: Wiley; 2000. [Google Scholar]
- 20.Becker N G. Analysis of Infectious Diseases Data. London: Chapman and Hall; 1989. [Google Scholar]
- 21.Andersson H, Britton T. Springer Lecture Notes in Statistics. New York: Springer; 2000. Stochastic Epidemic Models and Their Statistical Analysis. [Google Scholar]