Abstract
DNA methylation, an essential epigenetic feature of DNA that modulates gene expression and genomic integrity, is catalyzed by methyltransferases that use the universal methyl donor S-adenosyl-l-methionine. Methylenetetrahydrofolate reductase (MTHFR) catalyzes the synthesis of 5-methyltetrahydrofolate (5-methylTHF), the methyl donor for synthesis of methionine from homocysteine and precursor of S-adenosyl-l-methionine. In the present study we sought to determine the effect of folate status on genomic DNA methylation with an emphasis on the interaction with the common C677T mutation in the MTHFR gene. A liquid chromatography/MS method for the analysis of nucleotide bases was used to assess genomic DNA methylation in peripheral blood mononuclear cell DNA from 105 subjects homozygous for this mutation (T/T) and 187 homozygous for the wild-type (C/C) MTHFR genotype. The results show that genomic DNA methylation directly correlates with folate status and inversely with plasma homocysteine (tHcy) levels (P < 0.01). T/T genotypes had a diminished level of DNA methylation compared with those with the C/C wild-type (32.23 vs.62.24 ng 5-methylcytosine/μg DNA, P < 0.0001). When analyzed according to folate status, however, only the T/T subjects with low levels of folate accounted for the diminished DNA methylation (P < 0.0001). Moreover, in T/T subjects DNA methylation status correlated with the methylated proportion of red blood cell folate and was inversely related to the formylated proportion of red blood cell folates (P < 0.03) that is known to be solely represented in those individuals. These results indicate that the MTHFR C677T polymorphism influences DNA methylation status through an interaction with folate status.
DNA methylation is an important epigenetic feature of DNA that plays critical mechanistic roles in gene regulation (1–3) and cellular differentiation (4–6). In eukaryote cells DNA methylation is primarily comprised of methyl groups at the 5′ position of cytosine, a feature that is especially prevalent in CpG-rich sequences known as CpG islands (7–10), most of which reside within gene promoter regions (11). Aberrant genomic DNA methylation is widely recognized to be associated with different diseases, and is implicated in the genesis of cancer (12, 13) and neurodevelopmental disorders (2).
DNA methylation patterns in the cell are established and maintained by DNA methyltransferases (14–17) which, as do many other cellular methylation reactions, use S-adenosyl-L-methionine (SAM) as the donor of methyl groups (14). A major source of methyl groups used by SAM for methylation reactions is the folate-dependent de novo synthesis of one-carbon units (18).
Two specific enzymes are involved in this methyl group synthesis process: the B12-dependent methionine synthase that uses 5-methylTHF for the methylation of homocysteine to methionine, and the methylenetetrahydrofolate reductase (MTHFR) [5-methylTHF:(acceptor) oxidoreductase, EC 1.7.99.5]. MTHFR is considered a key enzyme in the one-carbon metabolism, because it is responsible for the irreversible conversion of 5,10-methylenetetrahydrofolate to 5-methylTHF, which serves as methyl donor for methionine, precursor of SAM.
Not only is this pathway important for methyl group synthesis but the lack of methyl group production results in an intracellular accumulation of S-adenosylhomocysteine (SAH), as well as an increase in plasma of total homocysteine levels (tHcy) (18).
Soon after the isolation of cDNA and gene mapping of the human MTHFR (19), a common missense mutation was identified in the MTHFR gene, the C to T substitution at position 677, that encodes for a thermolabile variant with reduced enzymatic function (20). By determination of tHcy, a strong gene–nutrient interaction has been demonstrated in the phenotypic expression of this genetic mutation in the MTHFR enzyme (21, 22). Under conditions of low folate plasma levels, the mutant T allele has been shown to be associated with higher levels of tHcy. This relationship between the MTHFR polymorphism and plasma folate values has been implicated as the likely link between the C677T polymorphism and cardiovascular disease (23, 24), as well as neural tube defects (25, 26).
Consistent with the concept that T/T homozygosity reduces the in vivo availability of 5-methylTHF is our recent observation that the distribution of different coenzymatic forms of folate is altered in T/T homozygotes (27). The red blood cells (RBCs) of the T/T homozygous mutants have diminished proportions of methylated tetrahydrofolates, as well as variable amounts of formylated folate. In contrast, cells from the wild-type individuals contain exclusively methylated folate derivatives (27). However, whether the different availability of methylated forms of folate may affect biological methylation reactions, such as DNA methylation, has not been demonstrated before.
We recently observed in a preliminary fashion that homozygous individuals for the MTHFR C677T mutation possess a lower degree of DNA methylation (28). However, the small number of subjects and the indirect method used to assess genomic DNA methylation (29, 30) limited the conclusions that could be drawn.
We designed the present study to evaluate, in a large cohort of subjects, the effect of folate status on genomic DNA methylation. To directly measure genomic DNA methylation we developed a quantitative, highly specific assay using liquid chromatography (LC)/MS techniques. This method allows for the direct measurement of 5-methylcytosine (mCyt) in DNA digests and differs from indirect enzymatic methods, which are semiquantitative and known to suffer from considerable interassay variability (31).
We sought to emphasize: (i) the effect of folate status on DNA methylation as determined by direct measurement of mCyt; (ii) the impact of the interaction between the MTHFRC677T mutation and folate status on DNA methylation; and (iii) the relationship between different coenzymatic forms of folate in the T/T homozygous mutants and DNA methylation.
Materials and Methods
Study Population.
We studied a total of 292 unrelated, age- and sex-matched subjects recruited from a single geographical area (northern Italy). Characteristics of the study population including MTHFR genotype distribution are described in detail elsewhere (22, 32). Of these 292 subjects, 193 had angiographically documented coronary atherosclerosis. Ninety-nine had normal coronary arteries as documented by angiography and had neither clinical nor laboratory evidence of atherosclerosis. They were examined for reasons other than coronary artery disease (in most cases valvular heart disease; refs. 22 and 32). Informed consent was obtained from all subjects after a full explanation of the study. The University Hospital of Verona Review Board approved the study.
Biochemical Analysis.
Samples of venous blood were drawn from each subject after an overnight fast. DNA was extracted from whole blood by using a phenol:chloroform:isoamyl alcohol protocol. All of the laboratory methods for biochemical analyses have been performed as described (22). tHcy levels were measured by high-performance liquid chromatography (HPLC) with fluorescent detection according to Araki and Sako (33). Plasma folate and vitamin B12 concentrations were measured by an automated chemiluminescence method (Chiron Diagnostics, East Walpole, MA). Vitamin B6 levels were measured by HPLC method according to Kimura et al. (34). RBC folate form distribution was detected by affinity followed by reverse-phase chromatography with electrochemical detection as described (27, 35).
MTHFR Genotyping.
The analysis of the MTHFR C677T mutation was performed by PCR followed by Hinf I digestion according to Frosst et al. (20).
DNA Methylation Measurement.
To measure genomic DNA methylation we developed a method using LC/MS. The instrument was a HP-Bruker Esquire, LC Ion Trap LC/MS (Billerica, MA) equipped with an electrospray ionization source. One microgram of DNA was enzymatically hydrolyzed by sequential digestion with 2 units of nuclease P1, 0.002 units of venom phosphodiesterase I, and 0.5 units of alkaline phosphatase as described by Crain (36). Twenty microliters of the hydrolyzed-DNA solution was injected onto a Suplex pKb 100 analytical column (250 × 2.1 mm i.d.) protected by a 5-μm Suplex pKb 100 precolumn (Supelco, Bellefonte, PA) (37). The isocratic mobile phase consisted of 7 mM ammonium acetate pH 6.7/methanol (5% vol/vol) in water delivered at a flow rate of 0.3 ml/min for 13 min. This allowed the separation of the four DNA bases, as well as the identification of 5-methylcytosine. Mass spectrometry operating conditions relating to the electrospray source were as follows: capillary 30 nA, nebulizer 40.0 psi, N2 drying gas 9 l/min, and drying temperature 350°C. The ion trap was scanned from m/z 100–150. 2′-Deoxy[U-15N3]cytidine (Cambridge Isotopes Laboratories, Andover, MA) was used as an internal standard and mixtures of 5-methyl-2′-deoxycytidine and 2′-deoxycytidine (Sigma) were used as external standards. Identification of cytosine (Cyt) and mCyt was obtained by combined UV detection at 254 nm and MS analysis of the chromatographic peaks eluting after 4.5 ± 0.5 min and 6.5 ± 0.5 min, respectively. The ion peak at m/z = 112 corresponded to Cyt and the ion peak at m/z = 126 corresponded to mCyt, confirming what was described by others (38). The ion peak at m/z = 115 corresponded to the stable isotope used as internal standard. Quantification of Cyt was done by comparison to its internal standard, and mCyt quantification was obtained by comparison with external standards of known concentration because a stable isotope standard for mCyt was not commercially available. The linear working range of the method was from 100 pg to 10 μg. The limit of detection for both Cyt and mCyt was 100 pg. DNA methylation status was defined as the amount of mCyt per μg of DNA. Coefficients of variance (n = 5) were 1.6% (within day) and 5.7% (between days).
Statistical Analysis.
All computations were performed by using SYSTAT software (Version 10.0) for Windows 2000 (SPSS, Chicago). The distribution of continuous variables in groups are expressed as mean ± SD. Analysis was performed with log-transformed data for all skewed variables including DNA methylation. Therefore, geometric means (antilogarithms of the transformed means) are presented and 95% confidence intervals (CIs) are calculated using the transformed values and displayed as the antilogarithms of the transformed data. Statistical significance for differences in continuous variables was tested by Student's unpaired t test. Categorical variables were analyzed using a χ2 test or linear regression analysis when appropriate. Adjustment for confounding variables (i.e., age, sex, smoking, creatinine, and vitamin status when appropriate) was performed by general linear model analysis (specifically analysis of covariance). Statistical significance refers to a two-tailed analysis where P < 0.05.
Results
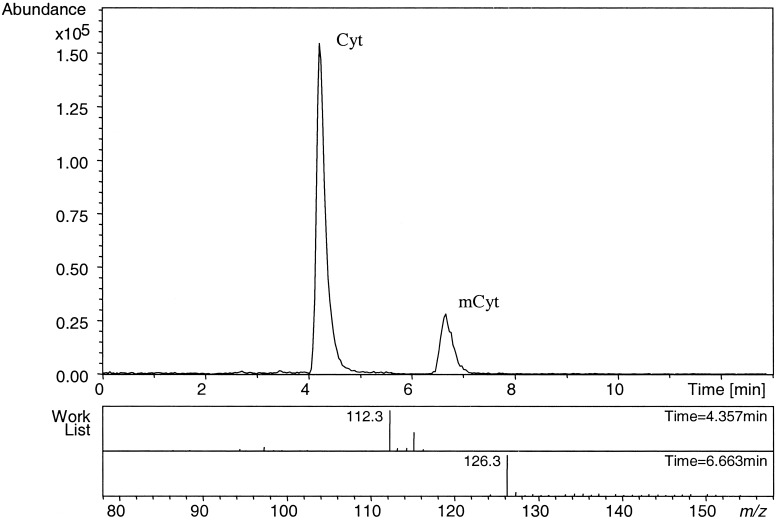
Fig. 1 shows a typical MS chromatogram of hydrolyzed DNA from peripheral blood mononuclear cells. The first peak at m/z = 112, eluting after 4.5 ± 0.5 min, corresponds to Cyt. The second peak, at m/z = 126 and eluting at 6.5 ± 0.5 min, corresponds to mCyt.
Figure 1.
Typical MS chromatogram of hydrolyzed peripheral blood mononuclear cell DNA. The first peak at m/z = 112, eluting after 4.5 ± 0.5 min, corresponds to Cyt. The second peak, at m/z = 126 and eluting at 6.5 ± 0.5 min, corresponds to mCyt.
A linear regression analysis, pooling both genotypes, demonstrated highly significant direct relationships between plasma and RBC folate and DNA methylation (P < 0.01). However, such relationships were driven entirely by the data from the T/T individuals and were not significant among the C/C individuals (see below). An indirect relationship was observed between DNA methylation and tHcy (P < 0.01). We found no significant correlation between DNA methylation and vitamin B6 or vitamin B12 status.
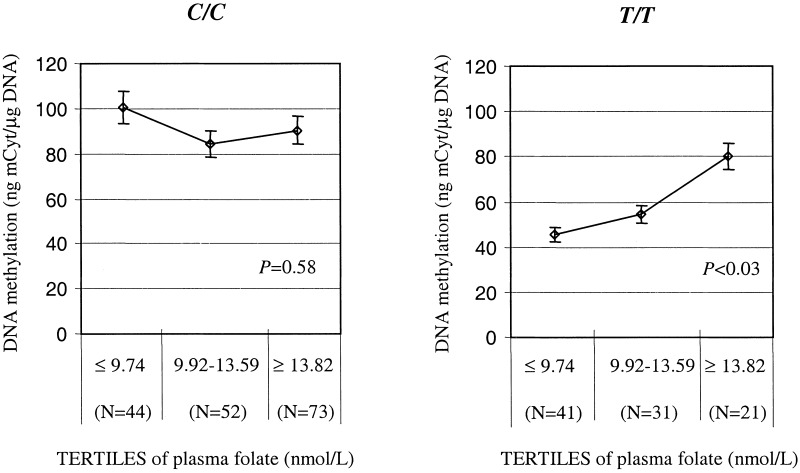
The characteristics of the population according to MTHFR genotype are described in Table 1. As shown, no differences were found in age, sex, vitamin B6, and vitamin B12 levels between the MTHFR C677T wild type (C/C) and the homozygous mutant (T/T) groups. Plasma folate, as well the RBC folate concentrations, were lower in the T/T as compared with the C/C group (P < 0.0001). Total plasma homocysteine was higher in the T/T than in the C/C group (P < 0.0001). These differences between the two MTHFR genotypes also extend to DNA methylation. The mean level of mCyt in the DNA from the T/T group was approximately half of that found in the C/C group (P < 0.0001; Table 1). This difference between genotypes is most evident when folate status was taken into account. Among those whose plasma folate status is above the median value (12 nmol/liter), genomic DNA methylation was similar in the two MTHFR genotypes (Table 2). However, when plasma folate status was below the median, the level of DNA methylation was considerably lower among the subjects homozygous for the C677T mutation than for the wild-type genotype. Moreover, the level of genomic DNA methylation was a 50% lower in this group compared with other T/T individuals (P < 0.0001), with levels of folate equal or above the median or with the C/C group in the lower range of plasma folate (Table 2). Further illustration of these differences is shown in Fig. 2. When the population sample was divided into tertiles of plasma folate according to C/C and T/T genotype, a graded effect of folate in determining levels of DNA methylation was detected only among the T/T individuals. The level of DNA methylation among those T/T individuals in the lowest tertile of folate compared with the highest was approximately a 35% lower and was statistically significant (P < 0.03; Fig. 2). Among those with the C/C genotype, however, DNA methylation was not different across the tertiles.
Table 1.
Characteristics of the population according to MTHFR genotype
| C/C (n = 187) | T/T (n = 105) | P value | |
|---|---|---|---|
| Age | 59.77 ± 9.96 | 59.92 ± 11.39 | N.S. |
| Sex, % male | 80.21 | 77.14 | N.S.* |
| Plasma folate, nmol/liter | 12.71 (9.81–11.96) | 10.42 (9.58–11.33) | <0.0001 |
| RBC folate, nmol/g Hb | 1267.49 ± 684.28 | 833.55 ± 435.84 | <0.0001 |
| Vitamin B12, pmol/liter | 297 (280–315) | 292 (274–316) | N.S. |
| Vitamin B6, nmol/liter | 30.38 (27.93–33.04) | 33.85 (30.17–37.97) | N.S. |
| tHcy, μmol/liter | 14.70 (13.90–15.54) | 21.26 (19.64–22.98) | <0.0001 |
| Genomic DNA methylation status, ng mCyt/μg DNA | 62.24 (54.32–71.31) | 32.23 (24.78–41.92) | <0.0001 |
Values are expressed as mean ± SD for age and RBC folate. Plasma folate, vitamin B12, vitamin B6, tHcy, and DNA methylation status are presented as geometric means (antilogarithms of the transformed means) and 95% confidence intervals are reported in parentheses with two-tailed P values. Statistical difference was evaluated by Student's t test except when differently indicated. N.S., not statistically significant.
Statistical difference was evaluated by χ2 test.
Table 2.
Genomic DNA methylation status (ng mCyt/μg DNA) according to MTHFR genotype and plasma folate levels (stratification by folate values above and below the median)
| Plasma folate ≥ 12 nmol/liter | Plasma folate ≤ 12 nmol/liter | P value | |
|---|---|---|---|
| C/C (n = 187) | 64.00 (52.72–77.63) | 63.05 (51.47–77.25) | N.S. |
| T/T (n = 105) | 67.56 (50.45–90.38) | 26.21 (18.81–36.49) | <0.0001 |
| P value | N.S. | <0.0001 |
DNA methylation status is presented as geometric mean (antilogarithm of the transformed means) and 95% confidence intervals are reported in parentheses with two-tailed P values. Statistical difference was evaluated by Student's t test. N.S., not statistically significant.
Figure 2.
Correlation between DNA methylation and levels of plasma folate divided into tertiles and according by MTHFR genotype: C/C (Left) and T/T (Right).
The results of the analyses when RBC folate was used as a measure of folate status precisely paralleled the observations made with plasma folate (Table 3). When both the C/C and the T/T individuals are divided into groups according to the median RBC folate value (1.13 nmol/folate/g Hb), only the T/T group with folate below the median had diminished DNA methylation (P < 0.0001). No such differences were found in the C/C group (Table 3). At RBC folate levels above the median, DNA methylation was not influenced by the MTHFR genotype.
Table 3.
RBCs folate forms distribution and DNA methylation status according to MTHFR C677T genotype and stratification in low and high RBCs folate values
| Low
RBC folate
|
P value | High RBC
folate
|
P value | |||
|---|---|---|---|---|---|---|
| C/C | T/T | C/C | T/T | |||
| Total folate, nmol/g Hb | 0.81 ± 0.20 | 0.68 ± 0.27 | 0.003 | 1.69 ± 0.70 | 1.48 ± 0.27 | N.S. |
| RBC methyl-THF, % of total | 98.8 ± 5.7 | 67.3 ± 29.0 | <0.0001 | 99.4 ± 1.1 | 69.6 ± 30.9 | 0.002 |
| Genomic DNA methylation status, ng mCyt/μg DNA | 64.07 (49.89–81.45) | 21.93 (14.73–32.45) | <0.0001 | 57.97 (45.60–73.55) | 57.39 (29.96–109.94) | N.S. |
Low and high RBCs folate status is intended for levels below and above the total RBCs median value (1.13 nmol/g hemoglobin), respectively. Values are expressed as mean ± SD for RBCs folate levels. DNA methylation is presented as geometric mean (antilogarithm of the transformed mean) and 95% confidence intervals are reported in parentheses with two-tailed P values. Statistical difference was evaluated by Student's t test. N.S., not statistically significant.
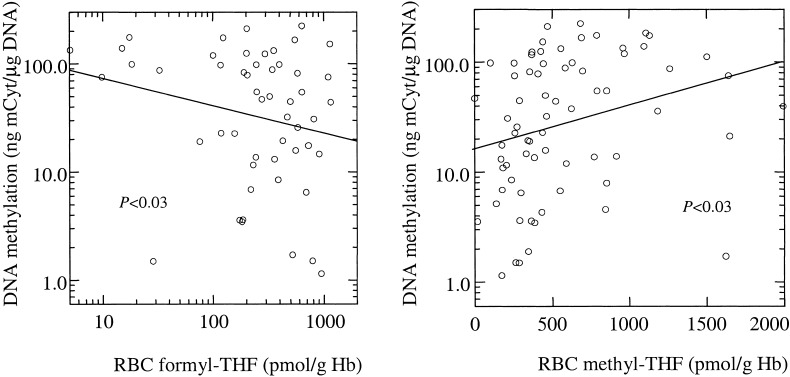
The assessment of RBC folate vitamer distribution demonstrated that the folate contained in RBCs from individuals carrying the C/C genotype is comprised entirely of 5-methylTHFs, whereas the RBC folate from individuals with the homozygous mutant T/T genotype consists of 30% formylated THF polyglutamates, confirming what we have described (27). Furthermore, the profile of folate coenzymes within the cell of T/T individuals bears a close relationship with peripheral blood mononuclear cell DNA methylation as shown in Fig. 3: a regression analysis within the T/T group revealed an inverse relationship between the proportion of formyl-THF and DNA methylation (P < 0.03), as well as a positive association between DNA methylation and methyl-THF proportions (P < 0.03).
Figure 3.
Correlation between DNA methylation (expressed in log-scale) and levels of different coenzymatic forms of RBC folate in MTHFR T/T subjects. An inverse relationship was detected between DNA methylation and formylated tetrahydrofolate polyglutamates (formyl-THF; Left). A positive correlation was detected between DNA methylation and methyltetrahydrofolate polyglutamates (methyl-THF; Right).
Discussion
Controlling the epigenetic patterns of DNA methylation is fundamental in the regulation of gene expression (1, 3, 5, 13, 39). Methylation at 5′ position of cytosine is a major epigenetic mechanism for controlling DNA gene expression and silencing (40–42) and is considered crucial in the maintenance of genomic structural integrity (43). Although essential for normal development, DNA methylation may become misdirected and lead to carcinogenesis (44–46) or other abnormal conditions (2, 43). It is therefore of considerable interest to identify the endogenous and exogenous factors that determine the patterns of methylation.
The present study shows that the level of 5-methylcytosine in genomic DNA from peripheral blood mononuclear cells is related to folate status. This relationship is further modified by an interaction of this nutrient with a common mutation in the one-carbon metabolism pathway that regulates the availability of methyl groups for methylation reactions. Homozygous mutants for the C677T polymorphism in the MTHFR gene exhibit a significantly lower level of mCyt in DNA but only under conditions of low folate status. At higher levels of folate, the amount of mCyt in DNA from T/T homozygous mutants does not differ from that among the C677T wild-type individuals. The interaction between folate status and MTHFR genotype has been also described in relation to plasma tHcy levels (47). Studies by us (21, 22) and others (48, 49) have shown that plasma tHcy levels are significantly higher in homozygotes for the C677T MTHFR mutation than in those with the wild-type genotype, but only among those mutants whose level of plasma folate is low (21, 22, 48, 49). At higher folate status, plasma tHcy levels are unaffected by the MTHFR genotype.
The presence of such a gene–nutrient interaction between the mutant MTHFR enzyme and folate status (50) is consistent with the recent study of Guenther et al. (51) in which the authors evaluated the biochemical structure of the mutant MTHFR and explained its propensity to lose its essential flavin cofactor. Guenther et al. used a thermolabile variant MTHFR model expressed in Escherichia coli to show that the C677T mutation results in the exposure of binding sites for the flavin adenine dinucleotide (FAD) cofactor, which otherwise would be embedded in a barrel-like structure. Such exposure results in a weakened enzyme/FAD complex, and hence the loss of activity. The presence of 5-methylTHF substrates is associated with conformational changes that strengthen the complex, thereby protecting the MTHFR against the loss of its flavin cofactor.
The finding of a low level of mCyt in peripheral blood mononuclear cell DNA with high tHcy observed in T/T subjects with a low folate status strongly implies the limited capacity of the mutant MTHFR for synthesizing sufficient 5-methylTHF to meet the demands for the de novo methionine methyl group supply. This is consistent with our previous report (27) that RBCs from homozygous mutant individuals for the C677T mutation have formylated tetrahydrofolate polyglutamates in addition to 5-methylTHF polyglutamates, which are the sole form of folate in RBCs from MTHFR C/C wild-type individuals (27).
In the present study, we observed an inverse relationship between tHcy and DNA methylation. Homocysteine is a product of the intracellular hydrolysis of S-adenosylhomocysteine (SAH). It then condenses with 5-methylTHF to form methionine or is transsulfurated in a B6-dependent reaction to form cysthathionine. It is therefore not surprising that tHcy inversely correlated with DNA because (i) elevation in tHcy is a reflection of folate depletion and (ii) elevation in total plasma homocysteine levels is an indication of elevated intracellular increased concentrations of SAH, which acts as an end product inhibitor of the S-adenosyl-L-methionine (SAM)-dependent methylation reactions. This finding is consistent with a recent report by Yi et al. (52), who identified plasma SAH as a strong predictor of the genomic methylation of nucleated blood cells.
We show in the current study that the modulation between the MTHFR variant and folate holds true for both plasma and RBC folate levels. The concentration of folate in plasma is generally accepted as an indicator of short-term changes of the nutrient, whereas RBC folate is considered a good index of body folate stores (53, 54). The present finding, however, suggests that plasma levels of folate in populations may be as reliable as RBC folate as an indicator of long-term folate status.
Whether other key vitamins involved in the one-carbon metabolic pathway influence DNA methylation has also been examined. In the present study, however, no association was found with either vitamin B12 or vitamin B6 levels.
A feature of the present study that strengthens confidence in the observations pertains to the development of a LC/MS method to directly measure genomic DNA methylation. Most existing methods for assessing genomic DNA methylation suffer from: (i) being semiquantitative rather than quantitative, (ii) assessing DNA methylation indirectly, and (iii) possessing large intra- as well as interassay variation. The LC/MS method affords much greater reproducibility and precision because it is based on the direct, quantitative measurement of methylated cytosine bases in DNA digests using the specificity of a MS technique (31).
This study in MTHFR C677T variants suggests that folate may have an important role in modulating an epigenetic feature of DNA that controls gene expression (2). However, it remains to be determined whether the DNA hypomethylation seen in peripheral blood mononuclear cell DNA also occurs in other tissues, and particularly in those that have rapid rates of proliferation and are thereby more susceptible to the adverse effect of low folate status—e.g., colon, bone marrow, and others (55–59).
Both hypo- or hyper-gene-specific methylation has been described in relation to impaired folate status. As we previously showed in animal studies, folate deficiency induces hypomethylation within critical regions of the p53 gene (60)—although, under dietary folate/methyl depletion, DNA methyltransferase activity in rats has been described to be up-regulated (61, 62), leading to hypermethylation at specific loci (63). In future studies it will be crucial to identify DNA methylation anomalies in specific loci in humans.
Acknowledgments
This material is based on work supported by the U.S. Department of Agriculture, under agreement No. 58-1950-9-001. This work was also supported in part by grants from Cancer Research Foundation of America (to S.-W.C.); Regione Veneto, Ricerca Finalizzata Venezia-Italia (to D.G.); the Cariverona Foundation (to O.O. and R.C.); and the Ministero dell'Universita' e della Ricerca Scientifica e Tecnologica of Italy (to O.O. and R.C.).
Abbreviations
- LC
liquid chromatography
- MTHFR
methylenetetrahydrofolate reductase
- 5-methylTHF
5-methyltetrahydrofolate
- tHcy
plasma total homocysteine
- RBC
red blood cell
- mCyt
5-methylcytosine
- Cyt
cytosine
References
- 1.Jones P A, Gonzalgo M L. Proc Natl Acad Sci USA. 1997;94:2103–2105. doi: 10.1073/pnas.94.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robertson K D, Wolffe A P. Nat Rev Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 3.Jones P A, Takai D. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 4.Razin A, Riggs A D. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 5.Bird A P, Wolffe A P. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg A P. Nat Genet. 2001;27:9–10. doi: 10.1038/83825. [DOI] [PubMed] [Google Scholar]
- 7.Riggs A D. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 8.Holliday R, Pugh J E. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 9.Bird A P. Nature (London) 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 10.Bird A P. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 11.Antequera F, Bird A P. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feinberg A P, Vogelstein B. Nature (London) 1983;301:89–91. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 13.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 14.Bestor TH Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 15.Vilkaitis G, Merkiene E, Serva S, Weinhold E, Klimasauskas S. J Biol Chem. 2001;276:20924–20934. doi: 10.1074/jbc.M101429200. [DOI] [PubMed] [Google Scholar]
- 16.Clark S J, Harrison J, Frommer M. Nat Genet. 1995;10:20–27. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- 17.Okano M, Bell D W, Haber D A, Li E. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 18.Selhub J. Annu Rev Nutr. 1999;19:217–246. doi: 10.1146/annurev.nutr.19.1.217. [DOI] [PubMed] [Google Scholar]
- 19.Goyette P, Sumner J S, Milos R, Duncan A M, Rosenblatt D S, Matthews R G, Rozen R. Nat Genet. 1994;2:195–200. doi: 10.1038/ng0694-195. [DOI] [PubMed] [Google Scholar]
- 20.Frosst P, Blom H J, Milos R, Goyette P, Sheppard C A, Matthews R G, Boers G J H, denHeijer M, Kluijtmans L A J, van den Heuvel L P, Rozen R. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 21.Jacques P F, Bostom A G, Williams R R, Ellison R C, Eckfeldt J H, Rosenberg I H, Selhub J, Rozen R. Circulation. 1996;93:7–9. doi: 10.1161/01.cir.93.1.7. [DOI] [PubMed] [Google Scholar]
- 22.Girelli D, Friso S, Trabetti E, Olivieri O, Russo C, Pessotto R, Faccini G, Pignatti P F, Mazzucco A, Corrocher R. Blood. 1998;91:4158–4163. [PubMed] [Google Scholar]
- 23.Welch G N, Loscalzo J. N Engl J Med. 1998;338:1042–1050. doi: 10.1056/NEJM199804093381507. [DOI] [PubMed] [Google Scholar]
- 24.Ueland P M, Refsum H, Beresford S A, Vollset S E. Am J Clin Nutr. 2000;72:324–332. doi: 10.1093/ajcn/72.2.324. [DOI] [PubMed] [Google Scholar]
- 25.van der Put N M, Steegers-Theunissen R P, Frosst P, Trijbels F J, Eskes T K, van den Heuvel L P, Mariman E C, den Heyer M, Rozen R, Blom H J. Lancet. 1995;346:1070–1071. doi: 10.1016/s0140-6736(95)91743-8. [DOI] [PubMed] [Google Scholar]
- 26.Botto L D, Yang Q. Am J Epidemiol. 2000;9:862–877. doi: 10.1093/oxfordjournals.aje.a010290. [DOI] [PubMed] [Google Scholar]
- 27.Bagley P, Selhub J. Proc Natl Acad Sci USA. 1998;95:13217–13220. doi: 10.1073/pnas.95.22.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern-Lathrop L, Mason J B, Selhub J, Choi S-W. Cancer Epidemiol Biomarkers Prev. 2000;9:849–853. [PubMed] [Google Scholar]
- 29.Kim Y I, Christman J K, Fleet J C, Cravo M L, Salomon R N, Smith D, Ordovas J, Selhub J, Mason J B. Am J Clin Nutr. 1995;61:1083–1090. doi: 10.1093/ajcn/61.4.1083. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y I, Giuliano A, Hatch K D, Schneider A, Nour M A, Dallal G E, Selhub J, Mason J B. Cancer. 1994;74:893–899. doi: 10.1002/1097-0142(19940801)74:3<893::aid-cncr2820740316>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 31.Oakeley E J. Pharmacol Ther. 1999;84:389–400. doi: 10.1016/s0163-7258(99)00043-1. [DOI] [PubMed] [Google Scholar]
- 32.Girelli D, Russo C, Ferraresi P, Olivieri O, Pinotti M, Friso S, Manzato F, Mazzucco A, Bernardi F, Corrocher R. N Engl J Med. 2000;343:774–780. doi: 10.1056/NEJM200009143431104. [DOI] [PubMed] [Google Scholar]
- 33.Araki A, Sako Y. J Chromatogr. 1987;422:43–52. doi: 10.1016/0378-4347(87)80438-3. [DOI] [PubMed] [Google Scholar]
- 34.Kimura M, Kanehira K, Yokoi K. J Chromatogr. 1996;722:296–301. doi: 10.1016/0021-9673(95)00354-1. [DOI] [PubMed] [Google Scholar]
- 35.Bagley P J, Selhub J. Clin Chem. 2000;46:404–411. [PubMed] [Google Scholar]
- 36.Crain P F. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 37.Zambonin C G, Palmisano F. Rapid Comm Mass Spectrom. 1999;13:2160–2165. doi: 10.1002/(SICI)1097-0231(19991115)13:21<2160::AID-RCM768>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 38.Banks JF, Jr, Shen S, Whitehouse C M, Fenn J B. Anal Chem. 1994;66:406–414. doi: 10.1021/ac00075a015. [DOI] [PubMed] [Google Scholar]
- 39.Lorincz M C, Groudine M. Proc Natl Acad Sci USA. 2001;98:10034–10036. doi: 10.1073/pnas.201392598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine A, Cantoni G L, Razin A. Proc Natl Acad Sci USA. 1991;88:6515–6518. doi: 10.1073/pnas.88.15.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 42.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 43.Wolffe A P, Matzke M A. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 44.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J P. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 45.Cameron E E, Bachman K E, Myohanen S, Herman J G, Baylin S B. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 46.Santini V, Kantarjian H M, Issa J P. Ann Intern Med. 2001;134:573–586. doi: 10.7326/0003-4819-134-7-200104030-00011. [DOI] [PubMed] [Google Scholar]
- 47.Ueland P M, Hustad S, Schneede J, Refsum H, Vollset S E. Trends Pharmacol Sci. 2001;22:195–201. doi: 10.1016/s0165-6147(00)01675-8. [DOI] [PubMed] [Google Scholar]
- 48.Verhoef P, Kok F J, Kluijtmans L A, Blom H J, Refsum H, Ueland P M, Kruyssen D A. Atherosclerosis. 1997;132:105–113. doi: 10.1016/s0021-9150(97)00084-1. [DOI] [PubMed] [Google Scholar]
- 49.Ma J, Stampfer M J, Hennekens C H, Frosst P, Selhub J, Horsford J, Malinow M R, Willett W C, Rozen R. Circulation. 1996;94:2410–2416. doi: 10.1161/01.cir.94.10.2410. [DOI] [PubMed] [Google Scholar]
- 50.Rosenberg I H, Rosenberg L E. Nutr Rev. 1998;56:s47–s53. doi: 10.1111/j.1753-4887.1998.tb01687.x. [DOI] [PubMed] [Google Scholar]
- 51.Guenther B D, Sheppard C A, Tran P, Rozen R, Matthews R G, Ludwig M L. Nat Struct Biol. 1999;6:359–365. doi: 10.1038/7594. [DOI] [PubMed] [Google Scholar]
- 52.Yi P, Melnyk S, Pogribna M, Pogribny I P, Hine R J, James S J. J Biol Chem. 2000;275:29318–29323. doi: 10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]
- 53.Herbert V. Am J Hematol. 1987;26:199–207. doi: 10.1002/ajh.2830260211. [DOI] [PubMed] [Google Scholar]
- 54.Bailey L B. J Nutr. 1990;11:1508–1511. doi: 10.1093/jn/120.suppl_11.1508. [DOI] [PubMed] [Google Scholar]
- 55.Skibola C F, Smith M T, Kane E, Roman E, Rollinson S, Cartwright R A, Morgan G. Proc Natl Acad Sci USA. 1999;96:12810–12815. doi: 10.1073/pnas.96.22.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toyota M, Ohe-Toyota M, Ahuja N, Issa J P. Proc Natl Acad Sci USA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi S W, Mason J B. J Nutr. 2000;130:129–132. doi: 10.1093/jn/130.2.129. [DOI] [PubMed] [Google Scholar]
- 58.Wiemels J L, Smith R N, Taylor G M, Eden O B, Alexander F E, Greaves M F. Proc Natl Acad Sci USA. 2001;98:4004–4009. doi: 10.1073/pnas.061408298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malone C S, Miner M D, Doerr J R, Jackson J P, Jacobsen S E, Wall R, Teitell M. Proc Natl Acad Sci USA. 2001;98:10404–10409. doi: 10.1073/pnas.181206898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim Y I, Pogribny I P, Salomon R N, Choi S W, Smith D E, James S J, Mason J B. Am J Pathol. 1996;149:1129–1137. [PMC free article] [PubMed] [Google Scholar]
- 61.Christman J K, Sheikhnejad G, Dizik M, Abileah S, Wainfan E. Carcinogenesis. 1993;14:551–557. doi: 10.1093/carcin/14.4.551. [DOI] [PubMed] [Google Scholar]
- 62.Pogribny I P, Poirier L A, James S J. Carcinogenesis. 1995;11:2863–2867. doi: 10.1093/carcin/16.11.2863. [DOI] [PubMed] [Google Scholar]
- 63.Pogribny I P, Miller B J, James S J. Cancer Lett. 1997;115:31–38. doi: 10.1016/s0304-3835(97)04708-3. [DOI] [PubMed] [Google Scholar]