Abstract
Quinolones are potent antibacterial agents that specifically target bacterial DNA gyrase and topoisomerase IV. Widespread use of these agents has contributed to the rise of bacterial quinolone resistance. Previous studies have shown that quinolone resistance arises by mutations in chromosomal genes. Recently, a multiresistance plasmid was discovered that encodes transferable resistance to quinolones. We have cloned the plasmid-quinolone resistance gene, termed qnr, and found it in an integron-like environment upstream from qacEΔ1 and sulI. The gene product Qnr was a 218-aa protein belonging to the pentapeptide repeat family and shared sequence homology with the immunity protein McbG, which is thought to protect DNA gyrase from the action of microcin B17. Qnr had pentapeptide repeat domains of 11 and 28 tandem copies, separated by a single glycine with a consensus sequence of A/C D/N L/F X X. Because the primary target of quinolones is DNA gyrase in Gram-negative strains, we tested the ability of Qnr to reverse the inhibition of gyrase activity by quinolones. Purified Qnr-His6 protected Escherichia coli DNA gyrase from inhibition by ciprofloxacin. Gyrase protection was proportional to the concentration of Qnr-His6 and inversely proportional to the concentration of ciprofloxacin. The protective activity of Qnr-His6 was lost by boiling the protein and involved neither quinolone inactivation nor independent gyrase activity. Protection of topoisomerase IV, a secondary target of quinolone action in E. coli, was not evident. How Qnr protects DNA gyrase and the prevalence of this resistance mechanism in clinical isolates remains to be determined.
Since the introduction of nalidixic acid in the 1960s, quinolones have been increasingly used to treat a variety of infectious diseases. This widespread use has been followed by increasing bacterial resistance (1). Previous studies have shown that quinolone resistance arises by mutations in the chromosomal genes for type II topoisomerases, the targets of quinolone action (2), and by changes in expression of efflux pumps and porins that control the accumulation of these agents inside the bacterial cell (3).
In bacteria, the type II topoisomerases DNA gyrase and topoisomerase IV change the topology of DNA. These enzymes cleave both strands of DNA to allow one double-stranded DNA molecule to pass through another and then reseal the break. DNA gyrase wraps DNA around itself, and so introduces negative superhelical twists into DNA, whereas topoisomerase IV does not, and consequently relaxes DNA. Quinolones inhibit these enzymes by stabilizing the complex between DNA and DNA gyrase or topoisomerase IV and thus block progression of polymerase and DNA replication (4, 5).
Both enzymes are tetramers. DNA gyrase comprises two GyrA and two GyrB subunits. Mutations in the N-terminal domain of GyrA or the C-terminal portion of GyrB augment quinolone resistance, typically 4- to 8-fold (6). In Escherichia coli, mutations in topoisomerase IV do not confer resistance by themselves but enhance resistance attributable to GyrA, increasing it another 4- to 8-fold (2).
Transmissible resistance to quinolones, long thought not to exist (7), has recently been discovered on a plasmid from a resistant clinical isolate (8). The plasmid augments resistance to nalidixic acid, ciprofloxacin, and other fluoroquinolones 4- to 8-fold and supplements resistance due to gyrA, gyrB, and porin or efflux pump mutations (L. Martínez-Martínez, A. Pascual, I. García, J.H.T. and G.A.J., unpublished data). In plasmid-containing strains no change in quinolone accumulation, outer membrane porins, or drug inactivation could be detected, implying a resistance mechanism different from those currently known. Here we report the cloning of qnr, the novel plasmid-encoded quinolone resistance gene, the amplification and purification of Qnr, its gene product, and the demonstration that Qnr directly protects E. coli DNA gyrase from quinolone inhibition.
Materials and Methods
Bacterial Strains and Growth Conditions.
Strains were routinely grown at 37°C in Luria–Bertani broth except where noted. Culture plates contained tryptic soy agar (TSA) or Mueller–Hinton medium. Selective media contained ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), kanamycin (25 μg/ml), or nalidixic acid (4 μg/ml) as required. E. coli strains used were HB101 (ara-14 galK2 Δ(gpt-proA)62 hsdSB20 lacY1 leuB6 mcrBB mtl-1 recA rpsL20 supE44 thi-1 xyl-5), J53 Azir (met pro azide- resistant) (9), and XL1-Blue (endA1 gyrA96 hsdR17 lac recA1 relA1 supE44 thi [F′ proAB lacIqZ ΔM15 Tn10]).
Cloning and Nucleotide Sequence Analysis.
Plasmid pMG252 was isolated from an E. coli J53 derivative by using a plasmid maxi kit (Qiagen, Valencia, CA), digested with EcoRI, ligated to EcoRI-restricted phagemid pBC SK (cat) (Stratagene), and introduced into E. coli HB101 by electroporation with selection on TSA plates containing chloramphenicol and nalidixic acid to produce plasmid pMG253. Using an Applied Biosystems Prism 3700 DNA Analyzer, we initiated cycle sequencing of purified pMG253 with primers complementary to the vector near its multiple cloning site and continued by primer walking over both DNA strands. Further analysis and comparisons were performed with Vector NTI Suite software (InforMax, Bethesda). Subclones were prepared from HindIII (pMG254) or ApaI digests of pMG253 religated into pBC SK.
Expression and Purification of Qnr.
The qnr gene was amplified from pMG254 by PCR using primers 5′-GGCCATGGATATTATTGATAA and 5′-GGATCCGGGCAGCACTATTACTCC. After digestion with NcoI and BamHI the DNA segment was ligated into vector pQE-60 (bla) (Qiagen), placing qnr under the control of the phage T5 promoter/lac operator combination and adding coding sequence for a C-terminal His6 tag. Proper qnr placement in the recombinant plasmid, pMG255, was confirmed by sequencing. Plasmid pMG255 was introduced into E. coli XL-1 Blue containing pREP4 (lacI neo)(Qiagen) and the strain was grown in 100 ml of Circlegrow broth (Qbiogene, Carlsbad, CA) with ampicillin and kanamycin to mid logarithmic phase, induced with isopropyl 1-thio-β-d-galactopyranoside at a final concentration of 1 mM, and allowed 4 h further growth until harvesting by centrifugation and storage at −70°C until protein isolation. Isolation was carried out with nickel nitrilotriacetate (Ni-NTA) agarose (Qiagen) following the manufacturer's recommendations. The next steps were carried out at 4°C. The pellet was resuspended in 2 ml lysis buffer (50 mM NaH2PO4/300 mM NaCl/1 mM imidazole, pH 8.0). Aliquots were added to 3 FastProtein Blue (Qbiogene) tubes, vortexed for 5 min, and centrifuged for 1 min at 10,000 × g. The supernatant was collected and incubated in a 1.5-ml tube with Ni-NTA resin at a 4:1 ratio by volume on a rocking platform for 2 to 3 h to allow the His6-tagged protein to bind to the resin. After centrifugation for 6 min at 3,000 × g, the pellet was resuspended in 1 ml of wash buffer (50 mM NaH2PO4/300 mM NaCl/20 mM imidazole, pH 8.0) and incubated on a rocking platform for 20 min. Two subsequent washes were performed, the first at 4°C and the second at room temperature, each with 10-min incubation and frequent inverting. Four elutions followed using 150 μl of elution buffer (50 mM NaH2PO4/300 mM NaCl/250 mM imidazole, pH 8.0). For each elution, samples were incubated and inverted at room temperature for 10 min, then centrifuged at 3,000 × g for 5 min. Samples were collected as separate elution fractions and dialyzed immediately in Slide-A-Lyzer Mini dialysis units (Pierce) for 22 h at 4°C in 50 mM Tris, pH 7.5. After dialysis, glycerol was added to the samples at 10% final concentration, and they were stored at −20°C. Protein concentration was determined by using a Protein Assay kit (Bio-Rad).
Preparation of Relaxed pBR322 Substrate.
Relaxed pBR322 (from New England Biolabs or Life Technologies, Rockville, MD) was prepared by incubation with calf thymus topoisomerase I (Life Technologies), following the manufacturer's recommendations.
DNA Gyrase Assay.
DNA supercoiling activity was assayed (10) in buffer containing 17 mM Tris⋅HCl (pH 7.5), 19 mM KCl, 6 mM MgCl2, 8 μg/ml tRNA (Sigma), 5 mM DTT, 357 μg/ml BSA, 5% glycerol, 1.8 mM spermidine, and 1.4 mM ATP. Purified E. coli DNA gyrase (Enzyco, Denver) was diluted in buffer consisting of 50 mM Tris⋅HCl (pH 7.5), 5 mM DTT, 1 mM EDTA, 150 mM NaCl, and 38% glycerol. The final concentration of gyrase in the reaction mix was 0.93 nM unless specified otherwise. For each reaction, 0.25 μg of relaxed pBR322 DNA was used. Ciprofloxacin was added where appropriate. The reaction was carried out at 25°C for 1 h and stopped by adding EDTA to final concentration 51 mM. The resulting topoisomers were resolved by agarose gel electrophoresis. All 35 μl of each reaction was loaded onto a 1% agarose gel in TBE buffer (89 mM Tris/89 mM boric acid/2 mM EDTA, pH 8.3) and run at 3.5 V/cm for 14–16 h. Gels were stained with 0.6 μg/ml ethidium bromide for 30 min, destained in water, and visualized under UV light. Inhibition or protection was calculated from band intensity using 1D Image Analysis Software, Version 3.0 (Kodak Digital Science, Rochester, NY). Inhibition was defined by the difference in intensity between the most supercoiled band in lanes containing gyrase alone and lanes containing gyrase and ciprofloxacin. Protection was defined by the difference in intensity of the most supercoiled band in lanes containing Qnr, gyrase, and ciprofloxacin, and lanes containing only gyrase and ciprofloxacin. Percent protection was expressed by the ratio of protection to inhibition.
Topoisomerase IV Assay.
DNA relaxation activity was assayed (11) in buffer containing 40 mM Tris⋅HCl (pH 7.5), 20 mM KCl, 6 mM MgCl2, 10 mM DTT, 50 μg/ml BSA, 1 mM spermidine, and 1 mM ATP. Purified E. coli topoisomerase IV (Enzyco) was diluted in buffer with 50 mM Tris⋅HCl (pH 7.5), 5 mM DTT, 1 mM EDTA, 150 mM NaCl, and 40% glycerol. The final concentration of topoisomerase IV was 20.3 nM. For each reaction 0.5 μg of supercoiled pBR322 DNA was added, and ciprofloxacin was added at 8 μg/ml where indicated. The reaction was carried out at 37°C for 30 min and stopped with EDTA at a final concentration of 65 mM. All 20 μl of the mixture was loaded onto a TBE 1% agarose gel, and the resulting topoisomers were separated and visualized by electrophoresis as for the gyrase assay.
Bioassay.
The gyrase assay was performed as described above, except with no enzyme added to the reaction. Qnr, BSA, or water was added to the DNA/quinolone mixture. From each assay 70 μl was added to a sterile paper disk that was placed on a Mueller–Hinton agar plate newly streaked with a lawn of quinolone-susceptible strain J53 Azir. After incubation overnight at 37°C the zone of inhibition around the disk was measured.
Results
Cloning the qnr Gene.
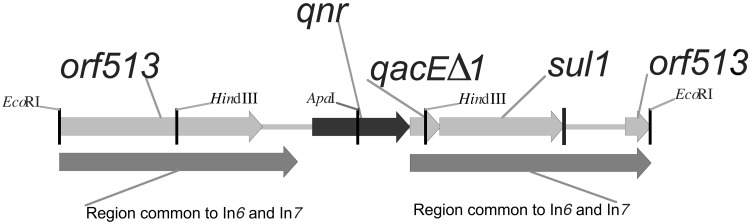
An EcoRI digest of plasmid pMG252 carrying qnr was ligated into EcoRI digested vector pBC SK, and recombinant plasmids were transformed into E. coli HB101, selecting for resistance to 4 μg/ml nalidixic acid and 25 μg/ml chloramphenicol. Each of four transformants contained an ≈4-kb insert of DNA. The insert in one such plasmid, termed pMG253, was sequenced, and a map of the resulting annotated structure is shown in Fig. 1. More than 3 kb of the cloned fragment consisted of DNA found previously in integrons In6 and In7 (12), including sulI preceded by qacEΔ1 and immediately upstream from these an ORF of 657 nucleotides (submitted to GenBank as accession no. AY070235), with the whole bounded by two copies of an element originally termed orf341 but recently renamed orf513 when the reading frame was extended (GenBank L06418). In In6 the sulI gene was not expressed (13), and similarly pMG253 did not confer sulfonamide resistance. To localize qnr further, a HindIII subclone (Fig. 1) of pMG253 was prepared and shown to confer quinolone resistance. On the other hand, a subclone using ApaI, which has one site within the 657-nucleotide ORF and another in the multiple cloning region of the pBC SK vector, lost quinolone resistance, supporting the 657-nucleotide ORF as the site of qnr.
Figure 1.
Map of the EcoRI fragment of pMG252 encoding quinolone resistance. The qnr gene is immediately upstream from qacEΔ1, which encodes resistance to quaternary ammonium compounds, and sulI, an unexpressed gene for sulfonamide resistance. Two copies of orf513, found in integrons In6 and In7, surround these genes. Restriction sites mentioned in the text are also shown.
Characteristics of the Qnr Protein.
The product of the putative qnr gene was a protein (Qnr) belonging to the pentapeptide repeat family (14). Qnr contained 39 leucine and phenylalanine-rich degenerate pentapeptide repeats that formed two distinct domains linked by a glycine (Fig. 2). The consensus sequence of the repeat was A/C D/N L/F X X, where X could be any amino acid. The third position was highly conserved, with 80% of the residues being leucine or phenylalanine. In the first position, 54% of the amino acids were alanine or cysteine, whereas in the second, 49% were aspartate or asparagine. Proteins in this family characteristically interact with other proteins (15).
Figure 2.
The amino acid sequence of Qnr written to emphasize the pentapeptide repeating unit with a consensus sequence of A/C D/N L/F X X. The protein can be divided into domains of 11 and 28 units connected by a single glycine. The conserved amino acid residues are highlighted.
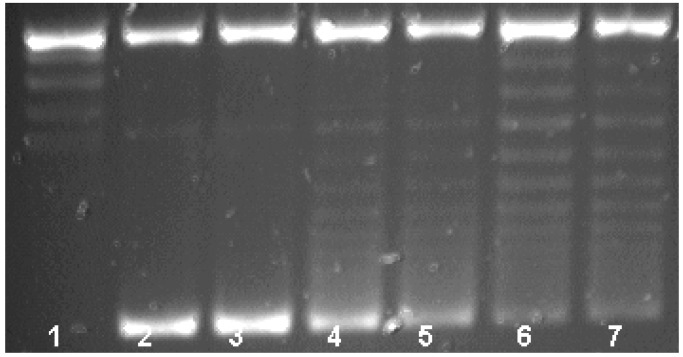
Qnr Protection of DNA Gyrase.
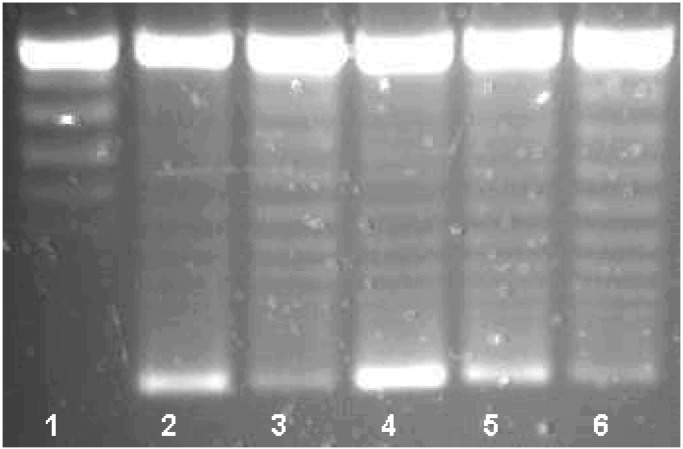
The qnr gene was inserted into the pQE-60 expression vector, adding a C-terminal His6 tag that allowed purification of the complex with nickel-nitrilotriacetic acid agarose. After elution with imidazole and extensive dialysis, the Qnr-His6 protein appeared homogeneous by SDS/PAGE and staining with Coomassie Blue. The supercoiling activity of DNA gyrase was measured by the ability of the enzyme to form supercoiled isomers from relaxed pBR322 DNA (Fig. 3). By agarose gel electrophoresis, a series of topoisomers was visualized, with the most supercoiled having the greatest mobility (Fig. 3, lane 2). Inhibition by 1.5 μM (0.5 μg/ml) ciprofloxacin was evidenced by reduced intensity of the most supercoiled isomer (Fig. 3, lane 3). When 2.01 μM Qnr-His6 was added, ciprofloxacin inhibition was reversed and the most supercoiled isomer band was actually more intense than with gyrase alone (Fig. 3, lane 4). With eluate from uninduced cells supplying 1.2 μM Qnr-His6, 62% protection was observed (Fig. 3, lane 5). No effect on ciprofloxacin inhibition was seen with an eluate from cells containing pQE-60 without the insert (Fig. 3, lane 6), with BSA at the same concentration as Qnr-His-6, or with Qnr-His6 itself after boiling (data not shown).
Figure 3.
Qnr reversal of DNA gyrase inhibition. Reaction mixtures contained no enzyme (lane 1) or 0.93 nM DNA gyrase (lanes 2–6) and ciprofloxacin at concentration 1.5 μM (0.5 μg/ml) (lanes 3–6). The assays were performed in the presence of 2.01 μM Qnr from cells induced at a final concentration of 1 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG) (lane 4) or 1.2 μM Qnr from uninduced cells (lane 5), and from IPTG-induced cells transformed with the expression vector pQE60 alone (lane 6).
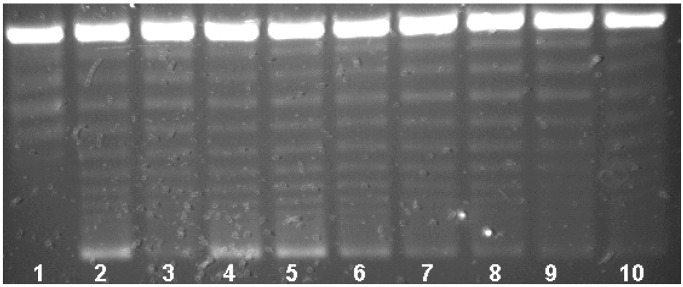
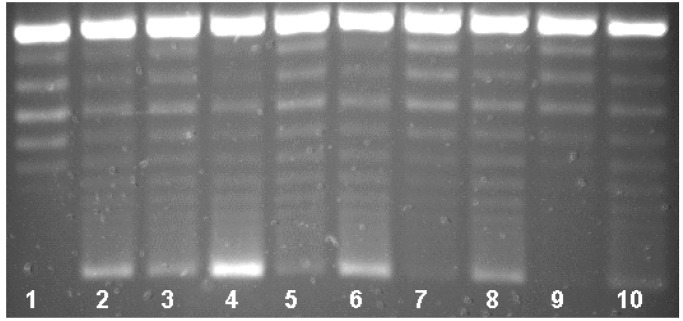
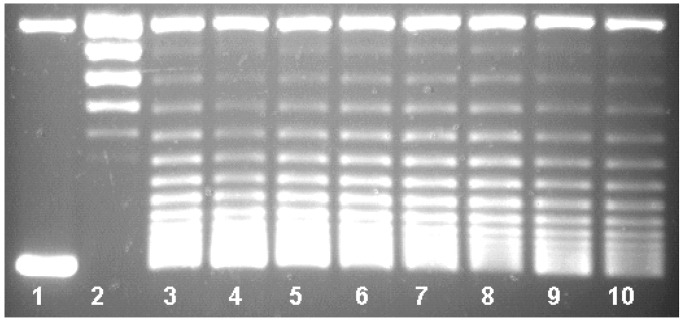
Qnr-His6 protection of DNA gyrase was proportional to the concentration of Qnr-His6 in the reaction mixture. As shown in Fig. 4, Qnr-His6 at 2.170 μM conferred 96% protection from inhibition by 1.5 μM ciprofloxacin (lane 4). At 725 nM, Qnr-His6 protection was 75% (Fig. 4, lane 5), at 242 nM protection was 31% (lane 6), and at 81 nM or below (lanes 7–10), no protection was observed. Compiling data from six assays, the amount of Qnr required to reverse 50% of gyrase inhibition by 1.5 μM (0.5 μg/ml) ciprofloxacin was 320 nM Qnr. Protection was also inversely proportional to the concentration of ciprofloxacin. In Fig. 5 the same concentration of Qnr-His6 (2.01 μM) protected completely (actually appearing to stimulate activity) against inhibition by 0.75 μM ciprofloxacin (lane 4) or 1.5 μM ciprofloxacin (lane 6), but was progressively less effective against inhibition by 3.0 μM (lane 8) or 6.0 μM (lane 10) ciprofloxacin.
Figure 4.
Stoichiometry of Qnr protection. Reactions contained no enzyme (lane 1) or 0.93 nM DNA gyrase (lanes 2–10) and 1.5 μM ciprofloxacin (lanes 3–10). The assays were performed in the presence of Qnr at concentrations of 2,170 nM (lane 4), 725 nM (lane 5), 242 nM (lane 6), 81 nM (lane 7), 27 nM (lane 8), 9 nM (lane 9), and 3 nM (lane 10).
Figure 5.
Dependence of Qnr protection on ciprofloxacin concentration. Reactions contained no enzyme (lane 1) or 0.93 nM DNA gyrase (lanes 2–10) and Qnr at 2.01 μM (lanes 4, 6, 8, 10). The assays were performed in the presence of ciprofloxacin at 0.75 μM (lanes 3 and 4), 1.5 μM (lanes 5 and 6), 3.0 μM (lanes 7 and 8), and 6.0 μM (lanes 9 and 10).
Qnr-His6 was tested further for activity with DNA gyrase in the absence of the inhibitor, for intrinsic supercoiling activity, and for ability to degrade ciprofloxacin. Fig. 6 shows that Qnr-His6 did not stimulate gyrase activity in the absence of ciprofloxacin. Furthermore, Qnr-His6 by itself lacked the ability to supercoil pBR322 (data not shown). Incubation of 1.7 μM Qnr-His6 with 1.5 μM ciprofloxacin also had no effect on quinolone activity by bioassay, ruling out drug inactivation as the mechanism for protection.
Figure 6.
Lack of supercoiling activity. Reactions contained no enzyme (lane 1) or DNA gyrase at concentrations of 0.93 nM (lanes 2 and 3), 0.62 nM (lanes 4 and 5), or 0.46 nM (lanes 6 and 7). The assays were performed in the presence of Qnr at 3.12 μM (lanes 3, 5, and 7).
Qnr-His6 Effect on Topoisomerase IV.
In contrast to the protective effect on DNA gyrase inhibition, no effect of Qnr-His6 on topoisomerase inhibition by ciprofloxacin was seen. Fig. 7 demonstrates relaxation of supercoiled pBR322 in the presence of purified topoisomerase IV, its inhibition by ciprofloxacin (note the higher concentration required), and lack of reversal by Qnr-His6.
Figure 7.
Failure of Qnr to protect topoisomerase IV. Reactions contained no enzyme (lane 1) or 20.3 nM topoisomerase IV (lanes 2–10) and 24.1 μM ciprofloxacin (lanes 3–10). Qnr was present at 6.5 μM (lane 4), 3.4 μM (lane 5), 1.7 μM (lane 6), 0.58 μM (lane 7), 0.19 μM (lane 8), or 0.06 μM (lane 9), and BSA was added at 3.2 μM (lane 10).
Discussion
Until recently, chromosomal mutations that modified the targets of quinolone action or diminished quinolone accumulation were the only known mechanisms for bacterial resistance to quinolones. Transfer of nalidixic acid resistance from Shigella dysenteriae to E. coli via a plasmid was claimed in 1987 (16), but further studies showed that putative donor strains contained gyrA mutations (17) and that a plasmid was not directly involved (18). In 1998 Martínez-Martínez et al. (8) reported that a multiresistant isolate of Klebsiella pneumoniae from a urine culture collected in Birmingham, Alabama, contained a broad host range plasmid that in E. coli transconjugants increased resistance to nalidixic acid from 4 to 32 μg/ml, and to ciprofloxacin from 0.008 to 0.25 μg/ml. The plasmid, termed pMG252, facilitated the selection of mutants with even higher levels of resistance (8) and augmented resistance due to defined DNA gyrase, porin, or efflux pump mutations from 4- to 8-fold (L. Martínez-Martínez, A. Pascual, I. García, J.H.T. and G.A.J., unpublished data). Because pMG252 did not alter host porin expression and did not reduce quinolone accumulation, the possibility of a novel resistance mechanism was suggested.
To pursue this mechanism further, the qnr gene from pMG252 was cloned, sequenced, and then amplified by PCR and inserted into an expression vector that added a C-terminal polyhistidine tag that facilitated purification with nickel nitrilotriacetate agarose. Purified Qnr-His6 was shown to protect E. coli DNA gyrase from inhibition by ciprofloxacin. Gyrase protection was proportional to the concentration of Qnr-His6, inversely proportional to the concentration of ciprofloxacin, lost by boiling, and shown not to involve quinolone inactivation or independent gyrase activity. Topoisomerase IV, a secondary target of quinolone action in E. coli, was not, however, protected from ciprofloxacin inhibition. Whether Qnr-His6 interferes with the binding of quinolone to DNA gyrase or destabilizes the formation of cleavable complexes between quinolone, gyrase, and double-strand cleaved DNA remains to be determined.
Currently, the closest relative to the qnr gene is a gene of unknown function (TIGR locus CC1891) from Caulobacter crescentus that has 44.7% nucleotide identity. Both genes code for proteins belonging to the pentapeptide repeat family, which at present includes more than 90 members. This functionally diverse family is defined by tandem five-residue repeats with the motif A (D/N) L X X, where X can be any amino acid (14). Such proteins have been found in many bacterial genera, but seem most common in cyanobacteria, and may be cytoplasmic or membrane bound. One such protein is horseshoe shaped, with α-helices at its outer circumference and β-strands forming a parallel β-sheet at its inner circumference (15), a structure well suited for a role in protein–protein interaction.
Two members of the pentapeptide family are especially relevant. One is McbG, a component of the system that protects bacteria synthesizing microcin B17 (MccB17) from self-inhibition. MccB17 is a 3.1-kDa posttranslationally modified peptide that blocks DNA replication (19) and that can, like ciprofloxacin, inhibit DNA gyrase supercoiling (20) and stabilize a gyrase-dependent DNA cleavage complex in the presence of ATP (21). Immunity involves three genes: mcbE and mcbF that are concerned with pumping MccB17 out of the cell, and mcbG that by itself can protect against exogenous MccB17 as though it blocked DNA gyrase inhibition (22). Furthermore, a plasmid carrying mcbEFG produced a 2- to 8-fold increase in resistance to certain fluoroquinolones (23), an observation attributed to the pump genes but seen also with a plasmid carrying only mcbG (D. Hooper and J. L. Yu, personal communication).
A second and recently described member of the pentapeptide family is MfpA, a protein cloned from the genome of Mycobacterium smegmatis in a search for an efflux pump contributing to quinolone resistance (24). A plasmid encoding MfpA increased resistance to ciprofloxacin 4-fold, whereas inactivation of the mfpA gene made plasmid-free M. smegmatis 4-fold more ciprofloxacin sensitive. The mechanism for this protection was not established, but MfpA had no effect on 14C-labeled ciprofloxacin accumulation and hence was not an efflux pump.
Fig. 8 shows the alignment between Qnr, McbG, and MfpA. Qnr and McbG have 19.6% amino acids in common, while Qnr and MfpA share 18.9% residues. Similar matches can be obtained with proteins from Bacillus subtilis (TIGR locus NT01BS1274), Enterococcus faecalis (TIGR locus EF0905), or Mycobacterium tuberculosis (TIGR locus MT3469). Because of the repeating pentapeptide backbone, interpretation of the relationship between members of this family is difficult. Nonetheless, Qnr could have evolved from an immunity protein designed to protect DNA gyrase from a naturally occurring inhibitor, or from a chromosomal gene of unknown function encoding a protein in the pentapeptide repeat family from Mycobacteria, Cyanobacteria, or other bacterial groups.
Figure 8.
clustal w alignment of Qnr, McbG (Swissprot P05530), and MfpA (24).
The qnr gene is located on plasmid pMG252 surrounded by a nucleotide sequence originally characterized in integrons In6 from plasmid pSa and In7 from pDGO100 (12). Each of these integrons has a common 3′-conserved region containing qacEΔ1 (conferring low level resistance to certain dyes and quaternary ammonium compounds and earlier known as orf4) (25) and sulI (coding for sulfonamide resistance but not expressed in these integrons due to a missing promoter) (13). The 3′-conserved region in pSa is preceded by a cat gene and in pDGO100 by a dhfr gene. Both elements also contain what was originally termed orf341 and now orf513, postulated to encode a recombinase involved in site-specific acquisition of resistance genes (13). Typically chromosomal β-lactamase genes that have recently been found on plasmids have also been mapped in similar structures and located between orf513 (341) and a 3′-conserved segment containing qacEΔ1 and sulI (26). Although a 59 bp element found in typical integron gene cassettes (27) is missing from all these inserted genes, and from qnr, a mechanism involving orf513 in site-specific gene acquisition seems inescapable.
By itself qnr provides such low level resistance that an E. coli strain carrying pMG252 would be classed as ciprofloxacin susceptible. Its clinical importance lies in the augmenting effect between qnr and other resistance mutations and the subsequent greater ease of selecting higher level quinolone resistance from a strain with qnr than one without its gene (8). How common the qnr mechanism is in clinical isolates is currently being investigated.
Acknowledgments
This work was supported by Public Health Service Grant AI43312 from the National Institute of Allergy and Infectious Diseases (to G.A.J.).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY070235).
References
- 1.Hooper D C. Emerging Infect Dis. 2001;7:337–341. doi: 10.3201/eid0702.010239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drlica K, Zhao X. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poole K. Antimicrob Agents Chemother. 2000;44:2233–2241. doi: 10.1128/aac.44.9.2233-2241.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiasa H, Shea M E. J Biol Chem. 2000;275:34780–34786. doi: 10.1074/jbc.M001608200. [DOI] [PubMed] [Google Scholar]
- 5.Wentzell L M, Maxwell A. J Mol Biol. 2000;304:779–791. doi: 10.1006/jmbi.2000.4266. [DOI] [PubMed] [Google Scholar]
- 6.Sanders C C. Clin Infect Dis. 2001;32, Suppl. 1:S1–S8. doi: 10.1086/319369. [DOI] [PubMed] [Google Scholar]
- 7.Courvalin P. Antimicrob Agents Chemother. 1990;34:681–684. doi: 10.1128/aac.34.5.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez-Martínez L, Pascual A, Jacoby G A. Lancet. 1998;351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 9.Jacoby G A, Han P. J Clin Microbiol. 1996;34:908–911. doi: 10.1128/jcm.34.4.908-911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooper D C, Wolfson J S, McHugh G L, Winter M B, Swartz M N. Antimicrob Agents Chemother. 1982;22:662–671. doi: 10.1128/aac.22.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng H, Marians K J. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 12.Stokes H W, Tomaras C, Parsons Y, Hall R M. Plasmid. 1993;30:39–50. doi: 10.1006/plas.1993.1032. [DOI] [PubMed] [Google Scholar]
- 13.Valentine C R, Heinrich M J, Chissoe S L, Roe B R. Plasmid. 1994;32:222–227. doi: 10.1006/plas.1994.1059. [DOI] [PubMed] [Google Scholar]
- 14.Bateman A, Murzin A G, Teichmann S A. Protein Sci. 1998;7:1477–1480. doi: 10.1002/pro.5560070625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobe B, Deisenhofer J. Nature (London) 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 16.Munshi M H, Sack D A, Haider K, Ahmed Z U, Rahaman M M, Morshed M G. Lancet. 1987;ii:419–421. doi: 10.1016/s0140-6736(87)90957-3. [DOI] [PubMed] [Google Scholar]
- 17.Rahman M, Mauff G, Levy J, Couturier M, Pulverer G, Glansdorff N, Butzler J P. Antimicrob Agents Chemother. 1994;38:2488–2491. doi: 10.1128/aac.38.10.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashraf M M, Ahmed Z U, Sack D A. Can J Microbiol. 1991;37:59–63. doi: 10.1139/m91-009. [DOI] [PubMed] [Google Scholar]
- 19.Herrero M, Moreno F. J Gen Microbiol. 1986;132:393–402. doi: 10.1099/00221287-132-2-393. [DOI] [PubMed] [Google Scholar]
- 20.Zamble D B, Miller D A, Heddle J G, Maxwell A, Walsh C T, Hollfelder F. Proc Natl Acad Sci USA. 2001;98:7712–7717. doi: 10.1073/pnas.141225698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heddle J G, Blance S J, Zamble D B, Hollfelder F, Miller D A, Wentzell L M, Walsh C T, Maxwell A. J Mol Biol. 2001;307:1223–1234. doi: 10.1006/jmbi.2001.4562. [DOI] [PubMed] [Google Scholar]
- 22.Garrido M C, Herrero M, Kolter R, Moreno F. EMBO J. 1988;7:1853–1862. doi: 10.1002/j.1460-2075.1988.tb03018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomovskaya O, Kawai F, Matin A. Antimicrob Agents Chemother. 1996;40:1050–1052. doi: 10.1128/aac.40.4.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montero C, Mateu G, Rodriguez R, Takiff H. Antimicrob Agents Chemother. 2001;45:3387–3392. doi: 10.1128/AAC.45.12.3387-3392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulsen I T, Littlejohn T G, Rådström P, Sundström L, Sköld O, Swedberg G, Skurray R A. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verdet C, Arlet G, Barnaud G, Lagrange P H, Philippon A. Antimicrob Agents Chemother. 2000;44:222–225. doi: 10.1128/aac.44.1.222-225.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall R M, Collis C M. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]