Abstract
Plasmids play key roles in the spreading of many traits, ranging from antibiotic resistance to varied secondary metabolism, from virulence to mutualistic interactions, and from defense to antidefense. Our understanding of plasmid mobility has progressed extensively in the last few decades. Conjugative plasmids are still often the textbook image of plasmids, yet they are now known to represent a minority. Many plasmids are mobilized by other mobile genetic elements, some are mobilized as phages, and others use atypical mechanisms of transfer. This review focuses on recent advances in our understanding of plasmid mobility, from the molecular mechanisms allowing transfer and evolutionary changes of plasmids to the ecological determinants of their spread. In this emerging, extended view of plasmid mobility, interactions between mobile genetic elements, whether involving exploitation, competition, or elimination, affect plasmid transfer and stability. Likewise, interactions between multiple cells and their plasmids shape the latter patterns of transfer through transfer-mediated bacterial predation, interference, or eavesdropping in cell communication, and by deploying defense and antidefense activity. All these processes are relevant for microbiome intervention strategies, from plasmid containment in clinical settings to harnessing plasmids in ecological or industrial interventions.
Graphical Abstract
Graphical Abstract.
Introduction
Plasmids [1] are extracellular molecules of DNA that replicate autonomously in a regulated and controlled way, keeping an approximate stoichiometry with the chromosome. This distinguishes them from other mobile genetic elements (MGEs) that may have a plasmid-like structure during horizontal transfer between cells, such as excised genomic islands, filamentous or virulent bacteriophages (phages), but that are usually not replicating as plasmids in pace with the cell cycle. Plasmids profoundly shape bacterial evolution and adaptation. Their capacity for genetic exchange has transformed our understanding of microbial ecology and has broad implications in fields ranging from infectious disease management to synthetic biology. Since plasmids carry genes that confer adaptive traits—such as antibiotic resistance, virulence, and metabolic versatility—plasmids facilitate the rapid spread of advantageous traits across diverse microbial communities, thereby driving genetic diversity and ecological fitness in bacteria [2]. Bacteria and Archaea access a vast gene reservoir through horizontal gene transfer (HGT), and plasmids constitute a substantial portion of this pool [3]. As an example, 46% of the complete bacterial sequences in the RefSeq dataset contain plasmids (Fig. 1A-C), with a median of two per host. These plasmids are scattered across the Prokaryotic taxonomy with higher numbers among the most sampled bacteria. But the frequency of plasmids is uneven across phyla, with some showing a clear majority of genomes with plasmids (Pseudomonadota), and others a minority (Bacillota) (Fig. 1B). Plasmid transfer underpins the open pangenomes observed in many microbial species (reviewed in [4–6]). Of particular concern, plasmid-mediated spread of antibiotic resistance is driving the current emergence of multidrug resistant bacteria with serious implications in public health [7]. The advances in genomics over the last decades have expanded our understanding of the evolutionary dynamics of plasmids, showing how they interact with chromosomal elements and other MGEs, including transposons and integrons. Such interactions enable plasmids to recruit and disseminate resistance and virulence determinants, sometimes from environmental reservoirs, into clinical settings [3].
Figure 1.
Plasmid distribution in the RefSeq database. The NCBI RefSeq database (release 229, 3 March 2025) includes 47 368 complete bacterial genome sequences, of which 21 801, distributed across 28 phyla, contain a total of 58 156 plasmids (A). For the 10 bacterial phyla with the highest number of plasmid-containing members, the abundance of genomes with and without plasmids is shown (B). The number of plasmids per bacterial host ranges from 0 to 28 and decreases exponentially (C). Plasmid size exhibits a bimodal distribution (when log-transformed) (D).
Several classification systems have been created to organize plasmid diversity by focusing on particular functions, including the use of replication or partition incompatibility types [8, 9], relaxases [10, 11], or conjugative pilus [12] (reviewed in [13]). Recently, the identification of plasmid taxonomic units (PTUs) provided a framework for categorizing plasmids as cohesive evolutionary entities akin to bacterial species [14]. PTUs group plasmids based on all conserved genetic markers, including those defining their type of mobility, allowing researchers to better understand their evolutionary dynamics and functional implications. Closely related plasmids also tend to have similar size, a trait that is highly variable among plasmids (Fig. 1D). The adaptive traits associated with a PTU shape the ecological roles and stability of plasmids in microbial communities.
Conjugation is regarded as the most frequent mechanism of plasmid transfer. Imaging techniques [15] and dynamic studies of conjugation [16], have recently provided unprecedented insights into the mechanics and regulation of plasmid transfer. These findings highlight previously unrecognized components and steps within the conjugation process, shedding light on factors that may affect plasmid stability, transfer rate, and host range. The traditional model of conjugation as a simple process of exchange between two cells mediated by a conjugative pilus under the control of a single plasmid has thus evolved to reflect a highly regulated, adaptive process that varies significantly across bacterial species, environmental contexts, and PTUs [14].
Beyond traditional conjugation mechanisms, plasmid biology encompasses a remarkable diversity of mobility strategies. Among the most intriguing recent discoveries is the abundance of elements that blur the boundary between plasmids and bacteriophages. These “phage-plasmids” combine the reproductive and transmission strategies of both, leveraging phage-like pathways to bypass the spatial and environmental constraints of standard conjugation (assessed in [17]). By forming phage particles and packaging their genetic material, these elements can transfer genes across microbial populations in a manner akin to viral infection, significantly broadening plasmid ecological and evolutionary impact. The dual functionality of phage-plasmids positions them as key players in the horizontal dissemination of adaptive traits, such as antibiotic resistance and stress response capabilities, within microbial communities.
In addition to classical conjugation and the emerging role of phage-plasmids, plasmid mobility encompasses mechanisms that rely on the functions of other MGEs for transmission. These mechanisms include plasmids that encode genetic information allowing to use helper plasmid conjugative systems for mobilization (reviewed in [18]). These processes are facilitated by the presence of numerous plasmids in the same cell, up to 28 different ones among the available genomes (Fig. 1). Plasmids can also be mobilized by phage transduction, whereby they are packaged and transferred by viral particles [19, 20], conduction, where plasmids transfer by recombining with coresident transmissible element via transposable elements (reviewed in [21]), and vesicle-mediated transfer (reviewed in [22]). All these forms of indirect mobility extend the functional versatility of MGEs, enabling the horizontal transfer of genetic material without requiring fully autonomous transfer machinery. This “hitchhiking” behavior has raised questions about the adaptability and survival strategies of plasmids in situations where autonomous transfer is not possible. By leveraging pre-existing systems, all these transmissible elements lower their costs while maintaining adaptability and evolutionary significance, further underscoring the complexity and plasticity of plasmid-mediated gene transfer.
The specificity of plasmid–host interactions is another critical aspect for plasmid mobility, with implications for understanding the selective pressures that influence plasmid spread and persistence. Studies on plasmid receptor interactions highlighted the role of host cell surface components in determining plasmid compatibility (reviewed in [23]). Quorum sensing also regulates plasmid transfer, thereby coordinating this process with host population density and environmental signals (reviewed in [24]). This raises questions about whether plasmids exhibit a preference for certain hosts and how microbial community composition might shape plasmid dissemination.
Finally, the dynamic nature of plasmid evolution is highlighted by the modification of plasmids into various forms throughout their life cycle. Some plasmids evolve to become integrative elements [25], to transfer genes in the host genome [26], or lose genes to emerge as small plasmids with limited functionality [27]. Similarly, plasmids may emerge from integrative elements or increase in size by acquiring genes from chromosomes, although these events have been much less well studied. These transformations highlight the plasticity of plasmid architecture and raise intriguing questions about the evolutionary pressures driving these changes.
In summary, plasmids are central players in bacterial evolution, not only because of their genetic content but also due to the varied mechanisms by which they move between hosts. While classical modes of HGT—conjugation, transformation, and transduction—have long defined our understanding of plasmid dissemination, it is now clear that many plasmids exploit additional, often unconventional, strategies to ensure their mobility. This includes mechanisms of communication or sensing the environment and the microbial community for potential hosts and competitor plasmids. We refer to this broader repertoire as the extended mobility of plasmids—a concept that encompasses the full diversity of mechanisms allowing plasmids to disseminate across microbial communities.
The mechanics, physiology, and diversity of conjugation
Conjugation from the side of the donor cell is primarily driven by self-transmissible MGEs that encode the machinery necessary for DNA processing and transfer. The ‘shoot and pump model’ is still used to illustrate the basic steps in conjugation [28] (reviewed in [2, 29]). The plasmid journey toward a new host cell has several critical steps and bottlenecks, encompassing exploration and contact initiation, invasion, establishment, control, and assimilation (reviewed in [30]). The process begins with the formation of the relaxosome, a multiprotein complex associated with the origin of transfer (oriT) on the plasmid. A key component is the relaxase enzyme, which nicks one strand of the plasmid DNA at the oriT site and forms in most cases a covalent bond with the 5′ end of the nicked DNA. The relaxase-DNA complex is then transported to the recipient cell through a Type IV secretion system (T4SS), a multiprotein apparatus that spans the donor’s inner and outer membranes also called Mating Pair Formation (MPF) system. Coupling proteins within the system ensure that the relaxase and its attached DNA are recognized and translocated by the T4SS. The structures of several T4SS involved in conjugation were recently revealed, providing clues on their similarities, differences and assembly processes (reviewed in [31, 32]). The most frequent localization of the F-plasmid transfer machinery in donor Escherichia coli cells is on the side at quarter positions [33]. Its conjugative pili are highly dynamic structures capable of extending and retracting to form contacts with potential recipients probing a volume around the cell that can exceed 100 times the one of the cell itself [30]. A longstanding controversy surrounded the transfer of the DNA through the conjugative pilus, when there is one. Live-cell microscopy has finally allowed to settle it by demonstrating that the F pilus serves as a conduit for DNA transfer during conjugation between physically distant bacteria [34–36].
The acquisition of plasmids by the recipient cell is less well understood. The F-plasmid DNA tends to arrive at the polar regions of the recipient cell [33]. Once in the recipient cytoplasm, the single DNA strand is circularized and serves as a template for replication and establishment of the full double stranded DNA plasmid. Direct visualization of F plasmid conjugation revealed that conversion of the single-stranded DNA (ssDNA) into double-stranded DNA (dsDNA) is fast and occurs at specific subcellular localizations [33]. This conversion determines the timing of production of most plasmid-encoded proteins that are only expressed if the DNA is double stranded. Yet, the leading strand incoming region of some plasmids contains single stranded promoters, allowing immediate expression of certain genes [33, 37, 38] (see below for implications in plasmid defense). The full process of plasmid acquisition has costs that are still poorly understood and should not be mistaken for the costs of subsequent carriage. Some of these acquisition costs are associated with the activation of responses to the incoming ssDNA, e.g. the SOS response [39]. Other costs are more drastic, involving recipient cell death by lethal zygosis. This process was observed in matings involving donors with the integrated F plasmid (Hfr strains) [40, 41], and more recently with plasmids RP4 and R388 [42]. A combination of modeling and experiments revealed a tradeoff between the lag time a recipient cell requires to recover from conjugation and its subsequent growth rate [43], where bacteria with longer lag phases end up having higher growth rates. This results in unexpected ecological dynamics, where plasmids with intermediate fitness costs can outcompete both low- and high-cost variants. These intriguing results highlight that we need a deeper comprehension of plasmid acquisition costs [44] to better understand the spread of plasmids in microbial populations.
If the machinery is available, conjugation is a robust process that does not require gene expression to occur. This is why dead donors can conjugate, although relatively poorly [45]. The recipient cell provides the machinery for plasmid establishment and replication and does not strictly require specific nonessential genes for plasmid acquisition [46], unlike in natural transformation. Nevertheless, the efficiency of conjugation is profoundly influenced by environmental conditions such as nutrient availability, temperature, and stress factors including antibiotics (reviewed in [15, 47]). Conjugation rates are heterogeneous even within clonal populations, driven by variations in plasmid expression and host factors [48, 49].
Novel approaches are shedding light on the dynamics and environmental dependencies of plasmid conjugation. The use of time-lapse fluorescence microscopy revealed that conjugation events are not uniformly distributed within bacterial populations; rather, they occur in spatially confined ‘hotspots’ where donor and recipient cells form transient but highly efficient transfer clusters [36]. These findings challenge the traditional view of random plasmid transfer and suggest that microenvironmental factors and cell-to-cell interactions play crucial roles in shaping conjugation networks. Plasmid invasion by conjugation in biofilms depends on local cell densities where spatial proximity of bacteria facilitates transfer [50]. Social behaviors, like quorum sensing, regulate the expression of conjugation machinery in donors (reviewed in [51, 52]). Importantly, recent work indicates that at high cell densities conjugation becomes limited by engagement time rather than merely by the frequency of donor–recipient encounters [53]. Additionally, the presence of external stressors, such as sub-lethal antibiotic concentrations, can trigger plasmid-mediated stress responses, promoting conjugative activity [54, 55]. Studies with clinical plasmids revealed the key role of plasmid evolution within specific genetic backgrounds in shaping their rates of transfer [56]. Finally, as described more abundantly below, the transfer of conjugative plasmids is affected by cells’ MGEs, and notably by the presence of other plasmids [18, 21, 57].
Conjugation systems are very ancient and phylogenetic reconstructions of the T4SS suggest they originated in diderm bacteria [58]. Their diversification has created lineages that are broadly associated with bacterial super-phyla (left panel of Fig. 2). This means that while transfer of conjugative plasmids across phyla has been observed, the successful establishment of such plasmids is rare. Four MPF types are found in Pseudomonadota (previously Proteobacteria) and closely related phyla (F, I, T, G), one system is specific to Cyanobacteria (C) and another to Bacteroidota (B), two others being found among monoderms (Bacteria and Archaea: FA, FATA). These studies suggest that diderm T4SS were initially co-opted by bacterial monoderms from where they were subsequently transferred to Archaea. More recently, several conjugative T4SS were co-opted to secrete virulence factors into eukaryotes in bacterial pathogens [59] or toxins into competing bacteria [60] (reviewed in [61]). Some of these systems perform both functions (conjugation and secretion of effectors) [62]. It is important to note that the transfer channel encoded by plasmid F (MPFF), which has been the basis for many cellular studies, differs significantly from that of plasmid R388 (MPFT), the primary model for structural studies (Table 1). MPFF is genetically more complex than MPFT and their functions may differ markedly. For example, R388 conjugative pilus is not able to retract [63]. In some Firmicutes, conjugation does not require a pilus and uses aggregation proteins to keep cells in direct contact [64]. Of importance, a given relaxase can interact with different types of MPF systems [65, 66], resulting in diverse patterns of co-occurrence between types of mobilizable and conjugative plasmids (right panel of Fig. 2) and suggesting, at best, some loose co-evolution between the relaxase and the T4SS. In contrast, there is tight co-evolution between oriT and the cognate relaxases, since only relaxases with relatively high protein identity seem to recognize a given oriT [67]. Further research is needed to determine whether the dynamics observed in conjugative systems of types MPFF and MPFT apply to the many other systems that remain poorly known. These diverse MPF types have variable components of which few have identifiable homology across types [68]. Comparative studies across these systems will be essential in revealing their structural, mechanistic, and functional diversity.
Figure 2.
Association between relaxase MOB classes and MPF systems. Left panel: The schematic phylogenetic tree illustrates the evolutionary relationships between MPF types based on the VirB4 phylogeny, highlighting their diversification within diderms and subsequent transfer to monoderms. Examples of conjugative systems representing each MPF type are displayed at the right. The phylogenetic tree was adapted from [58]. Right panel: The chord diagram displays a ring featuring the nine relaxase MOB classes and eight MPF types. The width of each sector corresponds to the abundance of MOB or MPF in an NCBI dataset of 12 148 plasmids. Links indicate co-occurrence of MOB and MPF within the same plasmid. The data were taken from Supplementary Table S1 of [27].
Table 1.
Widely used model mobile plasmids
| Plasmid | Size (bp) | PTU | MOB class | MPF type | Mobility type | Comments |
|---|---|---|---|---|---|---|
| ColE1 | 6646 | PTU-E1 | MOBP | - | Mobilizable | ColE1 plasmid family oriV is the basis for many modern cloning and expression vectors, including those for eukaryotic transfection; replication and mobilization extensively studied; model for antisense RNA-based regulation of plasmid replication |
| RSF1010 | 8688 | PTU-Q1 | MOBQ | - | Mobilizable | Broad host range plasmid; replicates in many Gram-negative and some Gram-positive bacteria; replication and mobilization extensively studied; model for strand-displacement replication mechanism |
| pMV158 | 5541 | - | MOBV | - | Mobilizable | Broad host range; replicates in Gram-negative and Gram-positive bacteria; replication and mobilization extensively studied; model for rolling-circle replication plasmids |
| pBBR1 | 2687 | - | MOBV | - | Mobilizable | Broad host range plasmid; replicates in Gram-positive and Gram-negative bacteria; engineered for constructing shuttle vectors |
| F | 99 159 | PTU-FE | MOBF | MPFF | Conjugative | Classic model for conjugation in E. coli; the mini-F replicon is used in bacterial artificial chromosomes and bacmids |
| RP4 | 60 099 | - | MOBP | MPFT | Conjugative | Broad host range conjugative plasmid; its oriT is used in shuttle vectors to transfer DNA between different bacterial phyla via conjugation; its replication, conjugation, and regulatory networks have been widely studied |
| R388 | 33 913 | PTU-W | MOBF | MPFT | Conjugative | Broad host range conjugative plasmid; its conjugation, regulatory networks, and segregation have been widely studied; model for structural biology studies on conjugative systems |
| R46 | 50 969 | PTU-N1 | MOBF | MPFT | Conjugative | Broad host range conjugative plasmid; its derivative, pKM101, has been widely studied for its conjugative transfer and error-prone DNA repair capabilities |
| R64 | 120 826 | PTU-I1 | MOBP | MPFI | Conjugative | Model for shufflon and type IV pilus role in conjugation |
| pTi-C58 | 214 233 | PTU-Rhi10 | MOBQ, MOBP | MPFT, MPFT | Conjugative | Natural tool for gene transfer to plants |
| pOXA-48 | 61 881 | PTU-L/M | MOBP | MPFI | Conjugative | Clinically relevant carbapenem-resistant plasmid; plasmid-associated fitness cost and transmission in hospital settings have been extensively studied |
| R27 | 180 461 | PTU-HI1A | MOBH | MPFF | Conjugative | Model for thermosensitive conjugative transfer |
| pIP501 | 30 599 | - | MOBV | MPFFATA | Conjugative | Model conjugative plasmid from Gram-positive bacteria; broad host range, replicates in many Gram-positive and Gram-negative bacteria; replication and conjugation have been extensively studied |
| pCF10 | 67 673 | PTU-Lab36 | MOBP | MPFFATA | Conjugative | Model for pheromone-inducible conjugation |
| pMW2 | 20 650 | - | - | - | pOriT | Example of pOriT plasmids |
| N15 | 46 375 | - | - | - | Prophage-plasmid (P-P) | Model for linear P-P |
| P1 | 94 800 | PTU-Y | - | - | P-P | Model phage-plasmid; widely used to transfer segments of the E. coli genome between strains via transduction |
When plasmids are also phages
Bacteriophages (phages) are bacterial viruses. They can be made of RNA, ssDNA or, more commonly, dsDNA [69]. Phages produce viral particles where they can package their own DNA. Upon infecting a sensitive bacterial cell many dsDNA phages (called temperate) make a lysis-lysogeny decision by either immediately producing progeny (lysis) or integrating the bacterial genome as prophages for several generations (lysogeny). There is still a frequent misperception that temperate phages necessarily integrate in the chromosome when they integrate the genome. Yet, phage-plasmids (or prophage-plasmids, P-Ps) are MGEs that transfer between cells as phages, while remaining in genomes as prophages under a replicative extra-chromosomal state, like other plasmids (Fig. 3A, reviewed in [70]). This replicative state in lysogeny should not be mistaken for the replication stage of the phage during lytic cycle, since the former takes place only about once per replicon per cell cycle. The best studied P-P is P1, which was the first phage isolated by Bertani from E. coli (in 1951) and became a popular tool for generalized transduction in molecular biology laboratories across the world [71] (Table 1). This P-P has become a model system to study molecular processes such as restriction–modification (R–M), site-specific recombination, phase-variation, plasmid replication, and plasmid partition (reviewed in [72, 73]). The unrelated phage N15 is a model system for linear plasmids (reviewed in [74]) (Table 1). Other known or putative P-Ps include the crassphage [75], the most abundant phage family in the human gut, Borrelia burgdorferi φBB1 [76] and derivative elements which encode adhesins that are important virulence factors in Lyme disease, and Vibrio cholerae’s VP882, which is a model system for quorum-sensing mediated regulation of the lysis-lysogeny decision [77]. Like many other phages, most P-Ps produce tailed viral particles, used to package their DNA into host cells.
Figure 3.
(A) Mechanism of mobility of phage-plasmids. (B) Distribution of plasmids per type in Bacteria and in E. coli (as described in [102]): conjugative (pCONJ), mobilizable with a relaxase (pMOB), mobilizable having an oriT (pOriT, only represented for E. coli), and phage-plasmid. “?” represents other, mostly uncharacterized, plasmids. Frequency of each type among all plasmids is proportional to the surface of the rectangle. (C) Comparison of plasmid mobility by conjugation (left) or within phage particles (right). P-Ps can produce very large progeny, but at the cost of cell death. Conjugative plasmids usually have broader host ranges than temperate phages and are more tolerant to extensive gene gain or loss. P-Ps may disperse faster, especially in aquatic environments, since their transfer does not require cell-to-cell contact.
P-Ps remained in the shadow for many decades, possibly because few labs studied both phages and plasmids. Hence, where a phage biologist saw a phage that seemed to behave like a plasmid, the plasmid biologist saw a plasmid with phage-like genes. The recent increase in interest by both types of MGEs and the availability of huge numbers of bacterial genomes enabled the discovery of many novel P-Ps, which account for around 5%–7% of all sequenced plasmids and a similar fraction of all sequenced phages [17] (Fig. 3B). P-Ps encode the genetic information required to start replication of their DNA during the lytic cycle, produce phage particles, package the DNA on the viral particles and lyse the host cells. They also encode the genes for plasmid replication and partition during the lysogenic cycle, and the regulatory elements to coordinate these functions and to transition between lysogeny and the lytic cycle. No P-P has yet been found to be also conjugative. This is not overly surprising considering that no prophage integrated in a bacterial chromosome has been found to be conjugative, although it was reported that prophages may be mobilizable by conjugation encoded in integrative and conjugative elements (ICEs) [78].
Most P-Ps found in current databases can be clustered in less than a dozen large families [17]. Each family is found within a bacterial genus or class, has a characteristic genome size, and equally characteristic ratios of plasmid-like and phage-like genes. This shows that many P-P families are ancient and not derived from recent fusions of plasmids and phages. Like other plasmids, their replication initiators make them unstable in the presence of closely related elements, allowing them to be grouped in incompatibility types. For example, P1 is the archetype of IncY plasmids that includes closely related phage-plasmids [79, 80]. Most of the known large families of P-Ps have at least one element that was shown experimentally to be both a temperate phage and a plasmid [17]. Yet, many small families of putative P-Ps still wait for experimental validation. It is also unclear if lysis-lysogeny decisions in P-Ps resemble those of integrative temperate phages. For example, phage lambda decision towards lysogeny is dependent on the multiplicity of infection, whereas that of P1 is independent [81]. On the other hand, both prophages start the lytic cycle upon induction with mitomycin C.
The dual nature of phage-plasmids, being phages and plasmids, has consequences for their evolution. The other plasmids rarely exchange genes with the other phages. In contrast, P-Ps often exchange genes with both other plasmids and other phages, possibly because they encode genes of both types of elements which facilitates recombination [82]. Accordingly, and contrary to other phages, a sizeable fraction of P-Ps has antibiotic resistance genes of clinical relevance, presumably acquired from conjugative plasmids [83]. These genes are transferred with the rest of the genome in viral particles and provide a resistance phenotype by lysogenic conversion. P-Ps, like other plasmids, encode a wide variety of other accessory functions that may enhance host fitness in specific circumstances [17, 83]. Because P-Ps are phages, and contrary to other plasmids, P-Ps actively induce cell death by lysis to disperse in the environment. A recent study revealed that recurrent loss-of-function mutations in the lytic cycle repressor of a Roseobacter P-P leads to continuous productive phage infection of the bacterial population, in which lytic and lysogenic P-Ps coexist [84]. Hence, the interplay between phage infection and plasmid genetics may result in unique outcomes for P-Ps.
Conjugation versus viral transfer
Around 25% of the plasmids in the database are conjugative and ∼5%–7% are recognizable P-Ps (Fig. 3B). Hence, P-Ps are rarer than conjugative plasmids, but not that rarer, especially considering that we may still be missing a lot of them (Fig. 3B). One must thus better understand the consequences associated with the two types of mobility, and how they shape the cost of plasmid transfer, its host range, genetic plasticity, and its ability to disperse in microbial communities (Fig. 3C).
The costs of transfer of conjugative plasmids and P-Ps are very different for the host [85]. While plasmid conjugation can be expensive because it requires the production and function of the MPF [16], and conjugative pili may increase the likelihood of phage infections [86], P-P transfer is deadly to the donor cell because, with the exception of filamentous phages, cell lysis is necessary to liberate phage progeny (Fig. 3C). This fitness effect may be less severe when lysis occurs in a small fraction of the bacterial population, allowing the survival of most lysogens. Lysis of a small subset of the population may even be favorable for the population of lysogens because the novel P-Ps may infect and kill the competitors that are sensitive to the phage (lysogens usually being protected from super-infection) [87–89].
The gains for the plasmid also differ depending on the mechanism of transfer. Each event of transfer by conjugation increases the number of plasmids in the population by one, whereas a lytic cycle produces dozens of phage particles carrying the P-P. There are thus key differences in transmission between the two types of plasmids: conjugative plasmids are less costly and reproduce at lower rates whereas P-Ps are deadly and reproduce at high rates (even if the original host dies in the process). On the other hand, conjugation delivers plasmids directly at host cells, whereas P-Ps particles still have to find a sensitive host before infecting another cell. From an ecological perspective, P-Ps generate vast numbers of progeny in a short period of time, of which only a few survive. This may provide an advantage for colonization, but a disadvantage when communities are mature. In contrast, conjugative plasmids generate a small progeny per cycle, but more of them survive because conjugation targets directly susceptible bacterial recipient cells. It is likely, even if it remains to be demonstrated, that ecological conditions determine which mechanism is most efficient for plasmid transfer.
The range of bacterial hosts permissive to plasmid transfer is much larger in conjugative plasmids than in P-Ps. Conjugative plasmids generally have broad host ranges, as shown by the fact that 81% of mobilizable and conjugative PTUs are capable of crossing genus barriers [14], even if decreasing taxonomic relatedness is associated with lower conjugation frequencies [90]. In contrast, most temperate phages have narrow host ranges, often infecting only a few strains within a species or closely related species [91]. The P1 P-P is somewhat unusual in that it was shown to infect multiple genera in Enterobacteria thanks to its encoding multiple tail fibers, the expression of which is under the control of the Cin site-specific recombinase and results in broader host range than the typical temperate phage [73]. Nevertheless, this is different from the broad host range of some plasmids (e.g. IncP-1 plasmids, [92]) where the same pilus is used to transfer plasmids into very different bacterial and even eukaryotic cells [93, 94].
The different transmission mechanisms affect plasmid genetic plasticity. In contrast to conjugative elements that may transfer vast amounts of DNA per event of transfer [95], the amount of transferred DNA by a P-P is limited by the volume of its capsid [17, 96, 97]. This constraint restricts the range of viable changes in the size of P-Ps to a few percent of their genomes, since beyond this threshold the P-P DNA can no longer fit the capsid [96, 97]. Therefore, conjugative plasmids more easily accommodate novel genes than P-Ps (Fig. 3C). For example, given the rate of conjugation of the F plasmid, an increase of 10 kb in the plasmid size, corresponding to an increase of 10% of its genome, only increases the transfer time by ca. 15 s [33, 98]. Hence, increase in plasmid size will much more likely block the transfer of P-Ps than those transferring by conjugation, which may allow the latter to vary their gene repertoires more freely.
The mechanism of mobility also shapes the patterns of plasmid dissemination (Fig. 3B). The differences between the two mechanisms in the requirement for close contact between cells, essential for conjugation, but not for phages, implicate the latter may disperse faster in the environment and quickly access distant communities. This difference in the dispersion mode may be particularly important for bacterial communities in aquatic environments, where phages are indeed exceedingly abundant [99, 100]. On the other hand, high dispersion also entails opportunity costs when the phage disperses out of a community with sensitive hosts to which it is adapted and entails risks when it ends up in habitats lacking in hosts (for a review of dispersal costs, see [101]). In conclusion, the cost, range, plasticity, and dispersion of plasmids depends on their mechanisms of mobility. This is likely to affect the accessory traits carried by plasmids because these are often associated with specific ecological conditions, although more work needs to be carried out to clarify this issue.
Hitchers
Plasmids encoding a full set of functions required for conjugation or phage reproduction are autonomous in their mobility, i.e. they only require host core functions, like transcription or replication, to transfer between cells. Yet, a majority (ca. 70%) of plasmids are not autonomously transmissible [2, 103], but can be mobilized by conjugation encoded in other MGEs (including plasmids), or by phages through transduction (Fig. 4). They are called hitchers because they require other elements to transfer (the helpers). Hitchers are not just hitchikers in that they actively attach themselves to conjugation machineries in the cell. They were extensively reviewed recently [18]. To attain a high frequency of mobility by conjugation or transduction, many hitcher plasmids evolved mechanisms to interact efficiently with the helper’s machinery (Fig. 4). For example, some encode specific relaxases and oriT whereas others encode just an oriT and all remaining machinery is provided by the helper. The consequence of these processes is that the probability of transfer of a hitcher plasmid depends on the content of the bacteria in other MGEs [18, 104]. Accordingly, plasmids in bacteria with few plasmids are conjugative, whereas bacteria with many plasmids have an over-representation of mobilizable ones [103].
Figure 4.
Mechanisms allowing the mobility of plasmids that do not encode a complete machinery for self-transfer. Mechanisms of transfer may involve mobilization by a conjugative element (top three rows) where a hitcher plasmid recruits an MPF and eventually also a relaxase from other plasmids. Phages may transfer plasmids by transduction either because they make generalized transduction and randomly package bacterial DNA in some of their capsids or because plasmids have evolved sequences to favor recognition by the phage packaging system. Plasmids may also transfer by other processes such as conduction (co-integration in a transferable plasmid), natural transformation, or within vesicles (although the latter mechanism is yet poorly understood).
Hitchers can be mobilized by conjugative helpers in three ways (upper panels in Fig. 4). In plasmids encoding a relaxase and an oriT, the relaxase interacts with the oriT of the element as described above for conjugative plasmids [105, 106]. The resulting nucleoprotein filament is then transferred by a conjugative system encoded by a helper. This process depends on the interaction of the relaxase of the mobilizable plasmid with the conjugative system of the helper. This can occur with the coupling protein of the conjugative system, or directly with the T4SS if the hitcher plasmid encodes a coupling protein [107, 108]. These physical interactions seem easy to mimic, since some mobilizable plasmids can be transferred by widely different conjugative systems [65, 108]. A more peculiar case was reported in Bacillus subtilis [109], where the ICE ICEBs1 can mobilize plasmids lacking typical conjugative relaxases, by interacting with the plasmid replicative relaxase. Other mobilizable plasmids only have an oriT for their mobility (they may encode many other unrelated genes) and are called pOriT [103, 110]. They may recruit a relaxase from a mobilizable element and a conjugative system from a conjugative element, or they may recruit both functions from the same helper. In E. coli the latter account for a little >60% of all pOriT [103]. This still leaves almost 40% of the plasmids depending on two different and compatible helpers to transfer between cells. The probability that a pOriT transfers into a cell with such a combination of MGEs is unknown, but their sheer abundance suggests that such combinations are not rare. One possibility is that plasmids are often co-transferred between donors and recipients, therefore allowing the pOriT to find the same compatible helper plasmid in the recipient. While this does not seem to have been thoroughly studied for co-transfer of hitchers and helpers, it was previously shown that different conjugative plasmids residing in the same cell tend to co-transfer [111].
Plasmids may also be mobilized by phages [19, 20], in a process called transduction [112] (Fig. 4). Generalized transduction results from a lack of specificity in the phage packaging machinery that leads to the encapsidation of random pieces of bacterial DNA. The P1 phage-plasmid is a model system to study generalized transduction and can be used in the laboratory to transduce plasmids or parts of them [113]. It was thought that generalized transduction is an inevitable “mistake” by the phage resulting from imprecise recognition of DNA sequences by its packaging machinery. Yet, recent works suggest transduction may have little cost to the phage (only a few capsids per replication cycle) and provide it with some advantages. Co-transfer of the phage and the bacterial DNA could for example allow the former to encounter in the recipient cells other adaptive functions for the host replication [114]. In this context, a low rate of transduction is no longer seen as a DNA packaging “mistake” but could instead be selected for. The intriguing similarity between the distribution of sizes of plasmids and those of phage genomes, has led to suggestions that co-occurrence of plasmids and transducing phages in the same cell often results in the transfer of the former by generalized transduction in phage capsids [20]. Transduction is especially likely for small plasmids because these will fit more easily the size of phage capsids [115] even if this may sometimes require they are packaged as concatemers [116]. It is also more frequent for high copy number plasmids because of their high concentration in cells [19].
Plasmids may further increase their rate of transduction by encoding DNA motifs similar to those used by the phages to recognize their own DNA. Some MGEs have developed this strategy to high degrees of sophistication and have become hitchers, which in this case are called phage satellites [117]. Contrary to common misperception, many satellites are not degenerate phages. Instead, they are MGEs that have evolved the ability to hijack phage particles and use them to transfer. At least one phage satellite has been shown to be a plasmid [118], but many others may exist and remain unknown. If plasmids often have phage packaging sites, it is likely that some are satellites of P-Ps.
Interactions between hitchers and helpers can diminish the latter’s capacity to transfer and pave the way to genetic conflicts. Some mobilizable plasmids can be costly to their helper conjugative elements by either diminishing or even blocking their dissemination [119, 120]. Phage satellites, by hijacking phage particles, diminish phage progeny [117]. Other hitcher plasmids have little, if any, impact in the transfer of the helper conjugative elements [110]. While mobilizable plasmids are in general smaller than conjugative ones [2, 103], there is no clear mechanistic reason for this [besides the fact that the loci encoding the transfer machinery can be large (15–40 kb)]. For example, the pSymB plasmid of Sinorhizobium meliloti is mobilized in trans by the pSymA even though it is over 1.6 Mb in length [121].
Lazy mobility
At least three other mechanisms allow plasmids to transfer between cells without having to encode specific mobility functions (“lazy” mobility, three lower panels in Fig. 4). Some plasmids can transfer by co-integrating with mobile plasmids (conduction) (reviewed in [21]). The two elements are transferred by conjugation in a single DNA molecule, a process that is different from the case of hitchers described above, where hitchers and helpers are transferred separately. Fusion between plasmids can occur by a variety of mechanisms, including the action of transposable elements [122] or homologous recombination [123]. Sometimes, these mechanisms allow the transfer of large nonmobilizable plasmids, such as the mobilization by conduction with a PTU-E21 plasmid of the large (>220 kb) Klebsiella pneumoniae virulence plasmid [124]. In the recipient cell the co-integrates may or may not reverse to the initial state, resulting in either a large recombinant plasmid or the two original plasmids. Many processes of conduction described in the literature involve integration mediated by transposable elements [125, 126]. It is not clear if conduction is a side effect of the action of these elements or if there are mechanisms that evolved specifically to facilitate conduction. It is also not clear how often the plasmid fusion may revert to the two original plasmids after transfer.
Plasmids may be present in the extra-cellular environment, e.g. following lysis of the donor cell. Recipient cells in a state of competence for natural transformation may uptake these plasmids when they attach to a structure in the cell envelope (typically to a type IV filament, for reviews, see [127, 128]). During DNA uptake by transformation, the DNA is made single-stranded and cut into fragments of a few kilobases before entering the cytoplasm. Hence, transformation of plasmids is thought to be more efficient for small high copy number plasmids [129–131], for which the likelihood of acquisition and re-assembly of the whole plasmid sequence (from one or multiple pieces) in the recipient cell is highest. While transfer of plasmids by natural transformation in the environment was described long ago [132], its importance remains unclear.
Small plasmids were also described to be transferred in outer membrane vesicles [133, 134]. This process is not yet fully understood. For example, it remains to be elucidated how plasmids arrive at the vesicle from the cytoplasm in the donor cell and from the periplasm to the cytoplasm in the recipient cell. The frequency of transfer of plasmids in vesicles seems to increase with their copy number and decrease with their size, but also depends on other unknown characteristics of plasmids [135]. Vesicles may favor transfer between neighbouring cells, since their diffusion away from cells is rare in microcolonies [136]. In summary, transfer of plasmids by transformation and by outer membrane vesicles favors small plasmids which usually have high copy number [137–139].
Is there a plasmid paradox?
Plasmids persist in bacterial populations in spite of conjugation rates that are often found to be low and costs to the host that are often high—a phenomenon termed the plasmid paradox [140–142]. The accumulation of data has started to solve the riddle (reviewed in [143]). A lot of the apparent plasmid paradox relies on the relatively low transfer rates given plasmid carriage costs observed in the laboratory. But quantifying conjugation rates in nature and their ecological implications remains challenging given the dynamic nature of microbial communities, host heterogeneity, and the regulation of conjugation [144, 145]. The quantification of plasmid carriage costs across environments is also still in its infancy. Nonetheless, recent works suggest that frequent conjugative transfer, combined with the selective advantages provided by plasmid-encoded traits, can sustain plasmid prevalence over evolutionary timescales. In support of this view, plasmid-mediated antibiotic resistance was shown to remain resilient in microbial communities due to conjugation dynamics that counteract plasmid loss, even when antibiotic selection is reduced [55]. The environmental plasmid pQBR57 was shown to transfer until fixation in conditions where it provides no fitness gain [146]. This may occur even in hosts where the plasmid has low persistence by a source-sink dynamics where plasmids are constantly transmitted from hosts where their presence is stable [146, 147]. The costs of plasmid carriage are also extremely variable and this may favor their persistence (reviewed in [85]). For example, the resistance plasmid pOXA-48 (Table 1) can have widely different, positive or negative, carriage costs in the absence of antibiotics depending on the genetic background [148]. A recent analysis of 40 plasmids of clinical origin revealed that most plasmids operate at a rate of conjugation that is below the one resulting in major growth effects [149]. In nonmobile plasmids, stability may arise even in the absence of antibiotics from the coordination of plasmid gene transcription and replication dynamics [150]. These works underscore that plasmid persistence is governed by a delicate balance of conjugation dynamics, selective pressures, and host adaptation.
Transmission costs can also influence the probability of plasmid persistence in bacterial populations. MGEs are subject to virulence–transmission trade-offs, where high transmission is regarded as a marker of high virulence because of its costs [151]. Hence, plasmids with high rates of horizontal transmission will tend to have lower rates of vertical transmission. The extended mobility of plasmids shapes these trade-offs. For example, P-Ps transfer at high rates as phages and have thus a very large transmission cost, relative to conjugative plasmids, potentially resulting in very different population dynamics. Hitchers have lower transmission costs than phages or conjugative plasmids, because they use the transmission mechanisms of the helpers. The marginal costs of their transfer to the donor bacterium is thus very low [18]. Finally, the existence of multiple regulatory mechanisms favoring plasmid transfer in specific circumstances, such as high density of potential hosts, may contribute to lower transmission costs and alleviate the vertical-horizontal transmission trade-off, making the spread of plasmids across microbial populations very far from paradoxical.
Do plasmids pick their hosts, or vice-versa?
Long term plasmid survival depends on achieving a delicate balance between promoting their own transfer and minimizing the burden placed on their host [149, 152]. For example, experimental evolution of the antibiotic resistance plasmid R1 rapidly led to higher conjugation rates under appropriate selection pressure (antibiotics) and when confronted with high immigration rates of susceptible hosts, but this implicated higher costs to the carrying cells [153]. Sophisticated regulatory systems have evolved to favor plasmid mobility when it is more likely to be successful (reviewed in [154, 155]). As a result, many conjugative plasmids are naturally repressed for transfer. Recent mathematical models suggest that reduced transfer rates due to host control have limited effects on plasmid prevalence [156]. For F-like plasmids, repression of transfer operons is primarily mediated by the FinOP fertility inhibition system [157], which ensures that only a small fraction of bacterial cells harboring F-like plasmids express the genes of its conjugative apparatus at any given time. However, newly formed transconjugants are derepressed, allowing a temporary burst of plasmid transfer from these cells [158]. In competition experiments, repression of the R1 plasmid resulted in a significantly lower fitness cost to the host than the derepressed mutant [159], highlighting the advantage of repressing horizontal transfer in many circumstances. Even naturally derepressed plasmids, such as RP4 and R388 (Table 1), maintain tight control over their transfer genes, with plasmid regulators functioning in negative feedback loops [160–162]. When plasmid DNA is transferred into a recipient cell, the absence of these negative regulators triggers transient gene expression of the mobility genes. This burst facilitates rapid plasmid propagation [163], though it temporarily impairs the host’s growth rate until regulatory equilibrium is restored [161]. Other regulatory mechanisms act at the level of the MPF pilus assembly. For example, R388 pilus assembly is greatly stimulated by the presence of recipient cells, through mechanisms that remain to be determined [63].
Some conjugative elements use sophisticated cell-cell communication systems to express the functions necessary for transfer when recipient cells lacking the element are abundant (reviewed in [164, 165]). The Rhizobium leguminosarum symbiotic plasmid pRL1JI responds to the high concentration of an N-acyl-homoserine lactone by inducing conjugation [166]. Enterococcus faecalis produces a specific pheromone whose concentration is thus a proxy for cell concentration. Plasmid pCF10 inhibits both pheromone activity and its export by E. faecalis [165] (Table 1). In these cases, conjugation will preferably take place when potential recipient cells lacking the plasmid are abundant relative to those carrying the plasmid [167, 168]. The ICE ICEBs1 also employs intercellular peptide signaling to regulate excision and mating under conditions that optimize its transmission efficiency [169]. It encodes an activator of excision and transfer, and a peptide that inhibits this activity. Coordinated regulation ensures ICEBs1 excision and transfer when host cells are surrounded by potential recipients that lack the element. Recent works have shown similar strategies in many conjugative plasmids [170] and temperate phages, including phage-plasmids [171], which can regulate the lysis-lysogeny decision using quorum-sensing [172, 173]. Upon infecting a new host cell, the Vibrio parahaemolyticus temperate phage-plasmid φVP882 assesses host cell density to make the decision whether to lyse or lysogenize. It shows a strong preference for establishing lysogeny at low cell density, when hosts are potentially rare, while favoring host cell lysis at high cell density [77, 174], when future hosts may be more abundant. Other mechanisms also favor plasmid transfer between closely related bacteria. This is the case of natural transformation that can be regulated by quorum-sensing mechanisms [175] or by the recognition of species-specific motifs in the incoming DNA [176, 177].
A recent meta-analysis highlighted the key role of the recipient cell in determining the rates of transfer [47]. Accordingly, mechanistic studies have shown how the composition and physiological state of the recipient cell envelope influences the success and specificity of plasmid transfer. Early studies identified the outer membrane protein OmpA and core lipopolysaccharide (LPS) as critical factors in recipient cells for acquiring F plasmids via conjugation, particularly under liquid culture conditions [46, 178]. These effects are less important for other plasmids like R100-1 [179] or R388 [46]. Capsules were recently shown to negatively affect plasmid conjugation rates [180–182]. The effect is symmetric, similar in the donor and in the recipient cells, and increases in direct relation to the capsule volume [183], likely because larger capsules physically separate bacteria, hindering plasmid transfer. MPFF plasmids, which encode longer retractable pili, seem unaffected by the capsule suggesting that barriers to conjugation may be surpassed by some types of MPF [183]. Conjugation of ICEBs1 into B. subtilis is influenced by the phospholipid composition of the donor and recipient cell membranes [184, 185], while wall teichoic acids were found to be essential in both donor and recipient cells for efficient transfer [186]. The impact of the cell envelope is even more crucial for the transfer of DNA in phage particles. In tailed phages, such as most P-Ps, tail fibers or other receptor binding proteins interact specifically with receptors at the cell envelope [187]. If this interaction does not take place, e.g. because the receptor is lacking, mutated or hidden, the phage cannot infect the cell [91]. Hence, P-Ps target specific types of cells, which limits their host range even when, as mentioned above for P1, they have multiple variants of tail fibers. For example, the prophages induced from K. pneumoniae tend to infect bacteria with the same capsule serotype [188]. Hence, key traits of the cell envelope, such as LPS and capsule serotypes, shape the plasmid transfer network and their effects depend on the specific mechanisms of transfer.
Some outer membrane proteins from recipient cells play a crucial role in mating pair stabilization, enhancing DNA transfer [178]. TraN, an outer membrane protein encoded by F-like plasmids, is a key player in the process of mating pair stabilization by converting initial pairings with OmpA and LPS into stable complexes capable of resisting shear forces (the so-called kissing complex) [189, 190]. The interaction of TraN with OmpA takes place at its N-terminal domain [191], where structural variations across different F-like plasmids are primarily located [192]. These variations define four distinct TraN structural variants, each specifically interacting with a different outer membrane receptor in recipient cells (OmpA, OmpW, OmpK36, and OmpF) (upper panel of Fig. 5). Minor variations in the recipient outer membrane porins can profoundly affect conjugation efficiency and specificity [192, 193]. By stabilizing mating pairs, these interactions facilitate DNA transfer. Moreover, TraN variants tend to be associated with certain species, indicating that mating pair stabilization plays a role in shaping the plasmid transfer networks [192] (recently reviewed in [23]).
Figure 5.
Interacting partners that promote targeted conjugation. The left part of the panels represents conditions where conjugation is promoted, while the right part includes scenarios where conjugation is impeded. The upper panel shows the interaction between TraN and a specific Omp versus nonspecific partner. The middle panel depicts the interaction of a specific PilV adhesin with a compatible LPS versus an incompatible LPS. The lower panel illustrates the plasmid transfer to an empty recipient versus a recipient already containing a copy of the plasmid.
Some plasmids have evolved other strategies to facilitate their transfer to specific recipient cells and to quickly adjust their specificity. Donor cells bearing plasmid R64 (Table 1) show recipient specificity during liquid mating, with transfer frequencies varying significantly depending on the combinations of recipient bacterial strains and the C-terminal segments of the PilV adhesin of the donor cell’s Type IV pilus [194, 195] (middle panel of Fig. 5). These segments recognize distinct core regions of the LPS on the surface of recipient cells during liquid mating [196, 197]. PilV variants arise through site-specific recombination mediated by the Rci recombinase, a member of the tyrosine site-specific recombinase family (reviewed in [198]). This system, called shufflon, has been identified in PTU-I1 [199], PTU-I2 [200], and PTU-B/O/K/Z [201, 202] plasmids, all of which encode a type IV pilus required for liquid mating in addition to the conjugative pilus [203]. Multiple shufflon arrangements are detectable even in plasmid DNA isolated from a culture derived from a single colony [204], suggesting that donor cells rapidly switch PilV variants to adapt to suitable recipient cells.
Targeted conjugation by selective recognition of compatible recipients serves to identify incompatible recipients, such as those already carrying a copy of the plasmid. Exclusion occurs in two forms: entry exclusion, which blocks DNA translocation into the recipient cell; and surface exclusion, which prevents cell to cell contact, thereby destabilizing the mating pair [205] (see lower panel of Fig. 5). A different strategy, immunity against superinfection, is common among temperate bacteriophages [206]. For example, the P1 P-P impedes the entry of similar elements in the cell [207]. Similar strategies were reported for ICEBs1 [208]. Genes responsible for both types of exclusion are encoded within the transfer region of the conjugative element (reviewed in [209]). Entry exclusion is mediated by an inner membrane protein in the recipient cell [210–214]. In some cases (e.g. R27 and pKPC_UVA01), this protein must also be present in the donor cells [211, 215]. The donor cell partners are all VirB6 homologs: TraG for elements encoding a type F mating pair formation (MPFF) [216–219], TraY for MPFI-encoding elements [220], and ConG for ICEBs1, which harbors MPFFA [221]. Exclusion is typically highly specific to its cognate plasmid, though exceptions exist [215].
Surface exclusion is restricted to a subset of MPFF plasmids [205, 211–223]. It is mediated by the outer membrane protein TraT, which functions in the recipient cell. Subtle amino acid variations influence the specificity of the protein in surface exclusion [224]. In F-like plasmids, surface exclusion specificity has been attributed to interactions with different proteins: the tip of the pilus on the donor cell, which may block its engagement with a receptor site on the recipient cell surface [225]; and OmpA in the recipient cell, which could potentially mask its interaction with the donor partner [226]. In PTU-C plasmids, TraN, located in the donor cell, has been identified as the surface exclusion target [223], but it does not serve this role in F-like plasmids [190]. Recently, cryo-electron microscopy structures of the surface exclusion protein TraT from pKpQIL and the F plasmid were determined [227], but their exact mechanism of action has yet to be elucidated. Pheromone-responsive conjugative plasmids exhibit a different type of surface exclusion. In these plasmids, a surface-exposed protease protrudes far out from the cell wall, reducing aggregation and transfer through the activity of the protease domain [228].
All these studies show that, although pervasive, conjugation does not occur indiscriminately. Instead, there are characteristics in the conjugative pilus, in phage particle, in the recipient or in the donor cell that shape the success and the efficiency of transfer. So, do plasmids pick their hosts? Or are they just constrained in their transfer to novel hosts? It is sometimes difficult to distinguish between the two hypotheses: whether plasmids select for specific conditions and targets of transfer or whether they face obstacles to transfer in the other conditions. For some mechanisms the answer seems to fall within the hypothesis that plasmids exert a choice, e.g. the use of quorum-sensing mechanisms to decide when to transfer (by phage-plasmids or conjugative plasmids). In contrast, the phage-plasmid dependency on cell receptors is more usually regarded as a constraint because phage attachment is strictly necessary for infection. In many cases the answer may be mixed, requiring further studies from the angle of evolutionary biology. A promising case of plasmids exerting a partner choice to conjugate concerns the Tra–Omp interactions described above. In this case, the specificity of the interactions changes a lot across plasmids [192], suggesting that high specificity evolved to allow the plasmid to actively pick specific hosts for conjugation. The matter of mating choice has implications for understanding the evolution of plasmid host range in natural populations and for predicting the evolutionary paths of synthetic plasmids (and assess their risks). Such plasmid–host associations operate through a key-lock mechanism, where plasmids evolve specific traits (the “keys”) to recognize and transfer into compatible recipient cells (the “locks”). In turn, recipient cells present structural and physiological features that determine which plasmids they can accept or exclude. This interaction can be viewed from two perspectives: either plasmids actively “pick” suitable hosts through targeted mechanisms, or recipient cells effectively “select” which plasmids they acquire based on compatibility constraints. In this framework, both plasmids and hosts exert agency in shaping HGT outcomes.
Plasmid antagonistic interactions
Plasmids can be costly to bacteria [85], and may compete with other MGEs. As a result, many bacteria evolved mechanisms to counter plasmid transfer. Some target plasmids specifically, but the majority also blocks other MGEs and virulent phages (extensively reviewed in [229, 230]). Here, we will focus specifically on the known impact of these systems on plasmid transfer. R–M systems were the earliest recognized barrier to DNA entry [231] and are the most widespread bacterial defense mechanisms [232]. Types I, II, and III R–M systems encode both a methylase and an endonuclease function, with the latter targeting unmethylated DNA [233]. In contrast, type IV systems encode an endonuclease that targets modified DNA [234]. In all cases, R–M functions as an innate immune response, as it does not distinguish among MGEs nor does it learn from past encounters. R–M reduces plasmid transfer up to 105-fold [235], but only when donor and recipient bacteria have different epigenetic markers, e.g. different patterns of DNA methylation. Hence, bacteria with compatible R–M systems exchange more genetic material than those with incompatible ones [236]. The level of restriction-based defense correlates with the number of recognition sites present on the plasmid [235]. Small plasmids tend to avoid restriction sites, thereby evading restriction [237]. In contrast, some large plasmids, for which full avoidance of restriction-sites is difficult to attain, encode orphan DNA methylases [237], which facilitate plasmid establishment in new hosts by inhibiting restriction enzyme activity [235, 238, 239] [first panel (left) of Fig. 6]. Interestingly, recent studies suggest that R–M targets are depleted in the leading regions of conjugative plasmids [240].
Figure 6.
Antiplasmid defense systems and plasmid antidefense systems. The first panel illustrates the action of plasmid-encoded methylases (left) and antirestriction proteins (right) in protecting the transferred DNA from the restriction activity of R–M systems in the recipient cell. The second panel depicts the antiplasmid immunity activity of CRISPR-Cas systems (left) and the antidefense activity of Acr proteins (right). The third panel shows the defense systems pAgo/DmdDE and DmdABC (Lamassu) (left) and the fertility inhibition activity exerted by a plasmid against the transfer of a co-resident plasmid (right). The fourth panel represents the counterattack deployed by T6SS against bacteria harboring conjugative plasmids (left) and the repression of chromosome-encoded T6SS by a transcription factor encoded in the conjugative plasmid (right).
R–M systems are encoded in approximately 3.1% of the plasmids [237]. These R–M systems can promote post-segregational killing of the host cell when the plasmid encoding them is displaced by an incompatible plasmid lacking the same R–M system [241]. Similarly, plasmids carrying toxin–antitoxin systems reduce the transmission of incompatible plasmids lacking such post-segregational killing systems [242]. Cell death eliminates both the competitor plasmid and the host, thereby favoring the persistence of plasmids encoding the post-segregational killing system [243].
CRISPR-Cas are adaptive immune systems that store small sequences (spacers) from previously encountered MGEs (protospacers) in an array (the CRISPR) [244]. This array is then used to recognize further infections. The majority of CRISPR spacers with known protospacers originate from bacteriophage and prophage sequences, while nonviral spacers predominantly derive from gene families involved in conjugative transfer and plasmid replication [245] [second panel (left) of Fig. 6]. Plasmid protospacers are unevenly distributed over plasmid regions, exhibiting higher frequency in the genes encoding mobility, a pattern that varies with the MOB type [246]. A class 1, type III-A CRISPR-Cas system from Staphylococcus epidermidis was the first shown to provide sequence-specific immunity against both phages and incoming plasmids via conjugation and transformation [247]. Plasmid clearance occurs through transcript depletion and plasmid DNA degradation [248–250]. Similarly, a class 2, type II-A CRISPR-Cas system in Streptococcus thermophilus was shown to induce plasmid loss [251], demonstrating for the first time spacer acquisition in response to plasmid invasion and in vivo CRISPR RNA (crRNA)-guided cleavage of dsDNA. The impact of CRISPR-Cas on HGT is undetectable at large time scales, possibly because it is a specific system that will only target a small number of MGEs at a given time [252, 253]. Yet, certain CRISPR-Cas systems are associated with reduced gene gain rate and low MGE abundance [253, 254]. There are also contradictory results concerning their effect on the spread of antibiotic resistance [255–258].
The first functional evidence of a type IV CRISPR-Cas system revealed that Pseudomonas aeruginosa strain PA83 uses crRNA-guided interference to clear plasmid DNA and block its uptake in vivo [259]. This CRISPR-Cas type is nearly exclusively associated with plasmids and its spacers show a pronounced preference for plasmid-derived protospacers. This suggests that type IV CRISPR-Cas systems specialised in mediating conflicts between plasmids [260–263]. Type IV systems lack the adaptation module responsible for spacer acquisition and the effector nuclease [262]. Nevertheless, they can leverage the host CRISPR-Cas adaptation machinery to modify the plasmid CRISPR array [264]. Unlike other CRISPR-Cas systems, its interference mechanism does not rely on cleavage of the targeted DNA but functions similarly to dead Cas9 [265, 266]. In this system, the Csf4/DinG helicase binds the dsDNA-bound complex and unwinds target DNA [259, 267], which blocks the DNA from interacting with gene expression machineries, negatively affecting plasmid transfer and stability. This effect is especially pronounced when core plasmid functions are silenced, thereby limiting plasmid transfer [264]. Other CRISPR-Cas systems are enriched in plasmids (reviewed in [268]). Among them, type V-M has been shown to inhibit plasmid replication by repressing transcription of essential plasmid genes, without cleaving the target DNA [269].
Several defense systems discovered in the last few years target plasmids with different degrees of specificity. For example, the prokaryotic Argonaute (pAgo) system degrades multi-copy DNA [270], which includes many small plasmids [178, 179] [third panel (left) of Fig. 6]. More specific systems have been found in pandemic V. cholerae strains, which are often, but not always [271], devoid of plasmids [272, 273]. In V. cholerae O1 El Tor, small, multicopy plasmids were found to be unstable, a phenomenon attributed to two DNA-defense modules: Lamassu/DmdABC and DmdDE [274]. The production of DmdABC causes significant but incomplete plasmid loss, whereas DmdDE leads to rapid and complete plasmid elimination through DNA degradation. The DmdABC system also induces plasmid-dependent cell toxicity in the presence of large conjugative plasmids, resulting in their loss [274]. Stem-loop hairpin structures in plasmids were identified as triggers of DmdABC activity, leading to bacterial death [275] [third panel (left) of Fig. 6]. Other proteins belonging to the SMC family, the one of DmdC, have been identified as antiplasmid defense systems, e.g. by inhibiting plasmid segregation [276]. Another system similar to the SMC complex, the Wadjet system, protects B. subtilis from plasmid transfer [277], by identifying and cleaving closed-circular DNA <100 kb [278]. In Corynebacterium glutamicum, Wadjet affects the maintenance of low-copy-number plasmids through its nuclease activity [279]. DNA loop extrusion promotes selective restriction of plasmids by Wadjet [280, 281], (recently reviewed [282]). The DmdDE system protects against plasmids through DNA-guided DNA targeting by the pAgo protein DmdE and subsequent degradation by the helicase-nuclease [282–285]. This type of plasmid instability may have consequences for AMR dissemination, as it indirectly favors the mobilization of plasmid-encoded AMR genes to the chromosome, thereby further stabilizing the resistance genes [216, 286]. These works revived significant interest in plasmid-specific defense systems and the next few years will likely reveal novel ones, especially with the advent of artificial intelligence-assisted defense prediction tools [287, 288].
A peculiar type of antiplasmid transfer mechanism is provided by type 6 secretion systems (T6SS), contractile injection machines that deliver toxins into target cells [289]. The P. aeruginosa H1-locus T6SS launches a lethal counterattack targeting conjugating donor cells carrying the broad-host-range IncP-1 conjugative plasmid RP4 or the PTU-N1 plasmid pKM101 [290] (Table 1). This tit-for-tat response is triggered by the MPF system encoded by conjugative plasmids in the donor cell [fourth panel (left) of Fig. 6]. In this specific case, the social interactions between bacteria directly shape the success of plasmid transfer. In another context, the anticonjugation activity of T6SS is counterbalanced: multidrug resistance plasmids of Acinetobacter baumannii encode transcriptional repressors of the chromosomal T6SS [291]. As a result, these potential conjugation donors are less effective predators, thereby facilitating plasmid transfer [fourth panel (right) of Fig. 6].
Other plasmid conflicts are mediated by fertility inhibition systems distinct from FinOP. They interfere with the transfer of a co-resident plasmid and may thus serve as competitive tools for colonizing new hosts (reviewed in [292]). These diverse fertility inhibition factors operate at different levels, but their mechanisms of action remain controversial [293–295] [third panel (right) of Fig. 6].
Research in the last few years revealed strong linkage between defence systems and MGEs: many of the defence systems found in bacterial genomes are actually encoded within MGEs and are thus implicated in MGE–MGE interactions (reviewed in [296]). As a result, while individual defence systems may diminish plasmid transfer, their presence at high frequency may be caused by the presence of plasmids. This may explain why associations between horizontal gene transfer and the presence of defense systems in bacterial genomes are taxa-dependent and in many cases non-significant [253].
Plasmids strike back
Plasmids have evolved numerous responses to defense systems encoded by bacteria or other MGEs [297]. Many of them are recorded in databases such as dbAPIS [298] and AntiDefenseFinder [299], which serve as tools to detect and annotate homologous antidefense systems. Genes known as “alleviation of restriction of DNA” or ard genes (reviewed in [300]), block the restriction sites [301] or inhibit the enzymes that restrict DNA, either by DNA mimicry [302] or by depleting the nuclease through ClpX [302, 303] [first panel (right) of Fig. 6]. One of the earliest known antirestriction systems is DarAB encoded by phage-plasmid P1 [304]. Initially identified in phages and prophages [305], anti-CRISPR (Acr) proteins have been detected in diverse plasmids [306, 307]. Acrs exhibit a variety of inhibitory mechanisms, including blocking crRNA loading, preventing target DNA recognition, and hindering DNA cleavage (reviewed in [308]) [second panel (right) of Fig. 6]. A different host defense evasion (hde) mechanism was identified in PTU-C conjugative plasmids and SXT/R391 elements [309]. This mechanism promotes evasion of the type I CRISPR-Cas system in V. cholerae by repairing dsDNA breaks through recombination between short sequence repeats, similar to what occurs in the lambda Red recombination system.
Plasmid counter-defenses must act quickly upon transfer to the new bacterium. Yet, transcription of plasmid genes by conjugation usually requires conversion of the incoming ssDNA to dsDNA, which takes time. The incoming ssDNA becomes an adequate substrate for transcription by forming stem-loop structures allowing the establishment of a dsDNA-like promoter [37, 38, 310]. These single-stranded promoters enable early protein synthesis of genes located in the so-called leading region [311]. Their subsequent conversion into dsDNA turns off this unconventional form of gene expression, a process that occurs in as little as four minutes for the F plasmid [33]. The leading regions of conjugative plasmids are highly enriched with antidefense genes [312]. They include SOS inhibitors, antirestriction proteins, anti-CRISPR proteins, DNA methyltransferases, toxin–antitoxin systems, and ssDNA binding proteins. The positioning of antidefense genes in the leading region is essential for efficiently neutralizing recipient defense systems [312]. Notably, several proteins encoded in the leading region act themselves as substrates of the conjugative T4SS and are thus translocated from the donor to the recipient cell, mitigating the SOS response in the recipient [313]. Given that a significant portion of the most prevalent gene families in leading regions are still uncharacterized, it is likely that novel antidefense related functions will be discovered there in the coming years.
Fast plasmid evolution and implications for their mobility
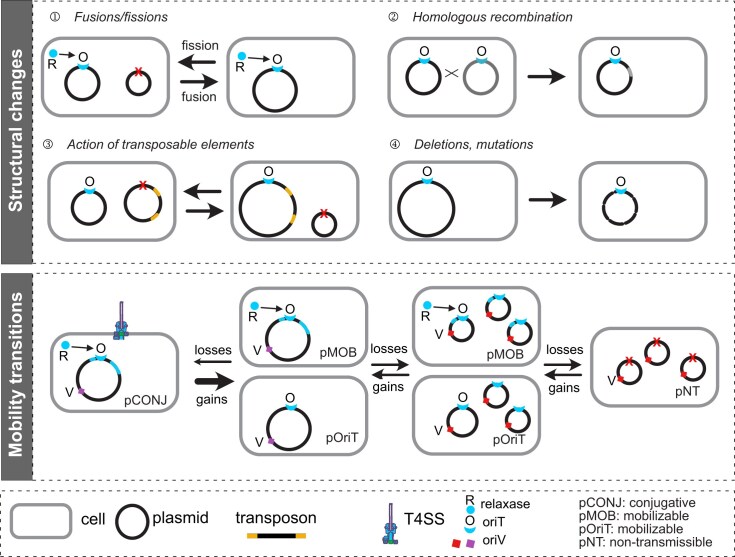
Plasmids are particularly prone to rapid change of their gene repertoires [27]. Conjugative plasmids are more variable than ICEs [314] and P-Ps more variable than integrative prophages [17]. The extra-chromosomal state of plasmids may favor higher genetic plasticity in multiple ways (Fig. 7). First, structural modifications in plasmids are not likely to affect core chromosomal genes whose integrity and expression are under strong purifying selection. Second, plasmid copy number fluctuates more widely than that of chromosomes, with small plasmids usually having both higher copy number and higher variability in copy number than large ones [138]. High copy number provides opportunities for changes in gene dosage, recombination rates, and effective accumulation of mutations [3, 315]. All these effects spur functional innovation. Finally, and possibly because of these traits, plasmids contain a higher number of large DNA repeats, transposable elements, and integrons than other MGEs [314]. All of them promote genetic modifications [316], while facilitating gene exchange with other plasmids and the host chromosome [317–319]. The p96 plasmid of K. pneumoniae is an interesting example where genome changes result in resistance gene dosage increasing up to 58-fold by processes of tandem amplification, 89-fold by increase in plasmid copy number, or up to 172-fold by recombining with high copy number plasmids [320]. While most of these outcomes are genetically unstable in the absence of selective pressure, they may vastly accelerate antibiotic resistance by providing higher gene expression and mutation supply. Accordingly, all could be observed among plasmids of E. coli bloodstream isolates [320].
Figure 7.
Key mechanisms of genetic change in plasmids (top panel) and transitions in terms of mobility for plasmids mobilizable by conjugation (lower panel). Plasmids genomes may change by several mechanisms (top), including fusions and fissions of plasmids, recombination, translocation of transposable elements and mutations and deletions of genetic material. Successions of gene gains and losses may result in transitions between types of mobility (lower panel). Those resulting in more limited mobility are more frequent because they only require gene deletions, but available evidence suggests they are counter-selected [27].
Rapid plasmid evolution implies that most available information on plasmid dynamics is at short time scales, as can be obtained from comparisons within incompatibility types, PTUs or epidemiological clones. Some plasmids have the reputation of rapidly changing their gene repertoires even at this narrow evolutionary scales [125] whereas others are described as more stable [321, 322]. Precise quantification of these rates of change is difficult beyond the epidemiological scale because of the lack of tools to date plasmid divergence, which is needed to normalize the number of changes per unit of time. One way to quantify plasmid dynamics is to trace the evolution of conserved genes while assuming that they represent the plasmid history. The analysis of plasmid evolution through the study of sequence divergence between homologous relaxases revealed that a few percent divergence in this protein (e.g. 5%) is associated with mobilizable plasmids that have fewer than 25% homologous genes [27]. In the longer term, as one compares plasmids with widely divergent relaxases, one cannot really trace the macro-evolution of plasmids because there is a loss of their individuality, i.e. there are so many differences between plasmids that no gene is a good proxy for the evolution of the plasmid lineage (each functional module has a different natural history). This could be regarded as the plasmid version of the Theseus’s paradox, where the nature of an object both remains similar and completely changes by gradual replacement of all its parts [323].
Structural changes in plasmids can dramatically impact their mechanisms of mobility (Fig. 7). For example, the ancestral plasmid of MOBF12 was probably conjugative [27]. Since then, there were many independent events of loss of essential conjugation-related genes leading to plasmids that cannot conjugate autonomously. A systematic analysis of such transitions revealed that it is much more common for autonomously conjugative plasmids to become just mobilizable by loss of all or part of the MPF genes, than the converse [27]. This is probably because gene deletions are more likely events than the acquisition of a functional MPF. The existence of many conjugative plasmids in bacterial genomes and the observation that most transitions from conjugative to mobilizable are recent, suggests these events are often deleterious and end up being purged by natural selection. This results in a source-sink evolutionary dynamics between conjugative and mobilizable plasmids, where the former constantly generate novel versions of the latter which frequently get purged by natural selection. But in some cases, such changes have produced novel plasmid families. For example, pO157 is a virulence F-like plasmid that lost the genes for conjugation and the oriT, but remains frequent in a lineage of E. coli because it carries virulence genes [324].
Mobility is associated with plasmid size and therefore plasmid cargo potential, the number of accessory functions it may encode, is affected by changes in the mobility type. It has often been observed that mobilizable and putatively nontransmissible plasmids are on average smaller than conjugative plasmids. Remarkable exceptions are found among plasmids on the way to become secondary chromosomes or chromids (reviewed in [325, 326]). Interestingly, plasmids that recently lost the ability to conjugate autonomously and became mobilizable by conjugation tend to have sizes intermediate between the conjugative and the mobilizable ones, suggesting that, upon loss of autonomous mobility, there is usually a selection pressure to reduce the size of the plasmid [27]. The reasons for this trend are not completely clear. It is possible that loss of autonomous mobility favors vertical transmission, which may in turn select for smaller, more streamlined plasmids with higher copy number. Some of the plasmid genes, especially those of adaptive value, may be transferred from the plasmid to the host chromosome. Yet, the quantitative impact of such a process in bacterial evolution remains controversial [26, 327]. Transposon-mediated events may have a key role in the process of plasmid size reduction by promoting deletions and exchange between replicons by promoting translocations [328]. For example, lab evolution of a PTU-Q1 plasmid revealed frequent gene deletions resulting in smaller plasmids that require the presence of the longer (complete) version of the plasmid to be mobilized and even replicate. Intriguingly, the emergence of these plasmids decreases carriage cost [329]. Whether such plasmids persist in lineages for long periods is unknown. Again, this may be a source-sink process where a viable mobilizable plasmid constantly produces less autonomous plasmids that tend to die out. Genome databases contain plasmids lacking any protein coding gene or just the ones coding for replication initiators. If these are dying plasmids that were once mobile remains to be investigated.
The acquisition and exchanges of genes can result in even more radical transitions of plasmid mobility. Some P1-like phage-plasmids have lost mobility-associated genes and are now either just phages (integrating in the chromosome) or just plasmids (incapable of developing a lytic cycle). Cases of the former were first reported over 50 years ago [330]. P-Ps that lost the ability to transfer as phages include the resistance plasmid RCS47 (derived from a P1-like P-P [331]), several plasmids of Salmonella enterica [332, 333], and several of the cp32 elements carrying virulence factors in B. burgdorferii [334]. Most such transitions are found only sporadically and may be on the way to being purged by natural selection [82]. But one lineage of plasmids derived from P1-like P-Ps has become frequent in enterobacterial genomes after its ancestor acquired several antibiotic resistance genes and lost key phage genes. It is possible that acquisition of the latter rendered the element too large to be packaged in the viral particle and resulted in the loss of phage functions. Interestingly, concomitant genetic exchanges led to the acquisition of oriTs by conjugation in most of these plasmids [82]. This lineage of plasmids has thus, in a short time span, changed from transferring as a phage to being mobilizable by conjugation. This illustrates the remarkable plasticity of plasmids’ gene repertoires and its impact on plasmid transfer.
Plasmids may also integrate the chromosome and become integrative elements [335]. Even if these events have often been described in the literature, their frequency is unknown. A particularly famous example concerns the integration of F in the chromosome with the creation of Hfr strains (see above). Conversion of a conjugative plasmid into an ICE probably requires further fine-tuning of the integration and excision functions (and their regulation). Another study identified several past conversions of conjugative plasmids into ICEs when the former conjugate into hosts where they cannot autonomously replicate [314]. Integration in the chromosome allows the element to replicate with it, removing the requirement of the element to replicate autonomously as a plasmid. Similar events may have occurred for other types of plasmids. Conversions in the opposite sense (from ICE to a conjugative plasmid) are also possible. A phylogenetic analysis of the MPFT conjugative system has shown that ICEs and conjugative systems have intermingled phylogenies suggesting many interconversions along natural history [12].
The interplay between plasmid plasticity and mobility may be particularly important for the spread of antibiotic resistance (reviewed in [336]). This trait is often transferred horizontally in plasmids which lacked such genes 70 years ago [337]. Since then, a planetary process of transfer of antibiotic resistance genes to human-associated bacteria has taken place from distant bacteria, both in terms of taxonomy and biome. This process seems to have favored the plasmids changing their gene repertoires faster, since these contain the majority of antibiotic resistance genes [102]. Antibiotic resistance plasmids also tend to recombine more frequently [102], be transferrable by conjugation [102], and have broad host ranges [102, 338, 339], which presumably diminishes the number of transfer events required to transmit a trait from a soil bacterium to an enterobacterium in the hospital [336]. Within plasmids, resistance genes tend to regroup in islands that change quickly in composition [338], creating hotspots of variability on already very plastic MGEs. The existence of fast evolving plasmids may thus have a unique role in accelerating the spread of novel traits in microbial communities.
When a plasmid changes in terms of mobility, its accessory genes also experience a corresponding shift in mobility (the two being linked). Cargo gene mobility may change when a plasmid gene is translocated to another plasmid or to chromosomal regions with other mechanisms of mobility (or lacking them altogether). Such genes can now explore different transmission paths in bacterial communities, because plasmids and other MGEs diverge in their rates of transmission, host ranges, and costs to the host cell. Low cost and high plasticity may contribute to explain why mobilizable plasmids have the highest frequency of antibiotic resistance genes [102], especially if they can be mobilized by multiple conjugative plasmids to partly compensate their lack of autonomy regarding HGT.
Plasmid mobility and biotechnology
Despite notable progress in culturomics, our capacity to genetically engineer bacteria remains limited to a small fraction of the known and culturable bacterial species, with plasmids commonly employed as tools for this purpose (Table 1). At the same time, emerging research in microbiome science has sparked interest in manipulating microbial communities through plasmid-mediated gene transfer. Notably, synthetic conjugative elements have been developed to enable gene delivery to undomesticated bacteria [340], including the engineering of HGT events at the community level [341]. These innovations address two persistent bottlenecks in microbiome biotechnology: the restricted host range of culturing techniques and the lack of efficient genetic tools. Recent discoveries in plasmid mobility offer promising solutions to both, paving the way for technologies that increase the efficiency, specificity, and functional versatility of genetic transfer, whether to modify, reprogram, or eliminate target bacteria.
Cutting-edge biotechnology projects often necessitate the engineering of nondomesticated strains [342]. The initial step in achieving genetic modification is the delivery of DNA. Chemical and electrical transformation methods typically require extensive optimization and are impractical for DNA transfer outside the laboratory. As mentioned above, natural transformation is limited to strains with functional DNA uptake machinery and small plasmids [343]. Hence, most works have used viral particles, such as those containing engineered phagemids [344], or conjugation to deliver plasmids to the cells. Yet, the narrow host range of most phages may complicate the delivery of plasmids by transduction to distantly related bacteria. In contrast, the broad range of conjugative plasmids, which can vastly exceed its replication range [93, 94], has been leveraged to develop shuttle vectors incorporating origins of transfer from various conjugative plasmids. This enables the transfer of genetic material from E. coli to distant bacterial phyla and thus their genetic manipulation [345–347].
Beyond delivery, the next challenge lies in specificity, ensuring DNA reaches only the intended targets. Therefore, a second step in enabling genetic modification is the precise delivery of genes to target bacteria within complex microbial communities, a process that can be influenced by different variables (reviewed in [348]). When plasmids are delivered via viral particles, their narrow host range naturally favors specificity [349]. In contrast, the low selectivity of transfer by conjugation, with DNA reaching diverse taxa, becomes a limitation in this context. A synthetic bacterial adhesion system was recently developed, utilizing mating assemblers composed of specific nanobodies engineered to bind their cognate natural antigens. The proper nanobodies are expressed on the surfaces of donor cells so that they bind the selected antigen in targeted recipients [350]. This strategy has been shown to increase conjugation rates between specific pairs of donor and recipient cells by several orders of magnitude in liquid environments, where conjugation is typically less efficient for most conjugative plasmids. Hence, this approach could be particularly useful for engineering microbiomes in liquid or semiliquid environments. It has already been used to enhance the targeted delivery of conjugative plasmids equipped with CRISPR-Cas9 systems [351].
Successful plasmid transfer requires overcoming bacterial immune defenses. For example, degradation of the incoming DNA by the activity of R–M systems is a major cause of genetic intractability of bacterial species. As defense and antidefense systems become better known, defense systems can be identified in the genomes of the target bacteria, so that the cognate antidefense systems can be deployed to impose plasmid transfer on the target bacteria. For example, the phage antirestriction protein Ocr, which overcomes the restriction barrier [352], is already being commercialized (TypeOneTM Restriction Enzyme Inhibitor) to increase bacterial transformation frequency in the laboratory. Similarly, in vitro methylation of the plasmid DNA, using cell-free transcription-translation systems that express the methyltransferases of the target host, has enhanced plasmid transformation in monoderms (typically Gram+) and diderm (typically Gram−) bacteria [353].
With these barriers addressed, new opportunities arise to manipulate or eliminate specific bacterial strains from populations. Several ongoing projects use conjugation to deliver inactivating or silencing tools, offering a targeted way to either eliminate problematic plasmids or suppress key bacterial functions without completely eradicating beneficial microbes. For example, the delivery of CRISPR-Cas has been explored to selectively target bacteria carrying genes of interest, such as virulence factors or antibiotic resistance genes (reviewed in [354, 355]). Ten years ago, engineered phagemids and mobilizable plasmids were first used as vehicles to deliver reprogrammed CRISPR-Cas9 systems to re-sensitize bacterial host populations by curing AMR-carrying plasmids [356, 357]. Since then, CRISPR-Cas systems have been widely used to selectively eliminate plasmid encoding antibiotic resistance genes, with particular emphasis on carbapenem resistance [358, 359] (recently reviewed in [360, 361]). Among the various delivery methods, conjugation has emerged as a very promising strategy for transferring CRISPR-Cas systems to antibiotic-resistant bacteria. However, its success is often constrained by the need for high in situ DNA transfer rates, which may be improved through optimization strategies such as experimental evolution [362].
Recent approaches have shifted focus from targeting the resistance genes to directly attacking their genomic platforms. For instance, the class I integron integrase has been targeted using a CRISPR-Cas9 system engineered into a broad host-range conjugative plasmid, leading to the removal of resistance plasmids with subsequent re-sensitization to multiple antibiotics whose resistance genes co-occur in integrons [363]. The K. pneumoniae endogenous type I-E CRISPR-Cas3 system targets conserved sequences in multidrug-resistance IncFII plasmids [364]. This system was engineered into a mobilizable vector and delivered to clinical K. pneumoniae isolates, efficiently curing the IncFII plasmids both in vitro and in vivo in a Galleria mellonella infection model [364]. This approach could also be used preventively, as a genetic firewall against acquiring unwanted traits (e.g. by colonizing bacteria with programmed CRISPR-Cas that prevent the uptake of predefined genetic traits). Indeed, K. pneumoniae harboring the CRISPR-Cas3-engineered platform successfully blocked the invasion of IncFII plasmids [364]. These advances mark a paradigm shift: rather than eradicating entire populations, microbiome engineering is moving toward precision interventions at the strain, plasmid, or gene level.
Beyond CRISPR, other strategies have been developed to selectively eliminate target strains. Selective removal of targeted bacterial strains, by plasmid inactivation or host elimination, is expected to cause minimal ecological disturbances to the microbiome. This approach could circumvent many of the issues associated with antimicrobial therapies. Genetic modules based on toxin-intein systems delivered by conjugation have been shown to selectively kill V. cholerae and its antibiotic-resistant derivatives in mixed populations [365]. Similar approaches have been developed using phages that selectively target specific conjugative pili [86, 366, 367] (reviewed in [368]). It is yet unclear which strategy will be more successful: phages, with their higher infection and dispersion rates, or conjugation, which is less costly and more resilient to microevolution in the target host. Finally, some strategies bypass the plasmid elimination step by reprogramming type IV-A3 CRISPR-Cas systems to silence target genes of interest through a transcriptional repression mechanism [264].
Conjugation offers opportunities beyond gene delivery and has been harnessed for deploying genetic tools even in the absence of DNA transfer. As relaxases, and other proteins, are transferred during conjugation [369, 370], they have been fused with the endonuclease Cas12a and base editors to introduce specific mutations, bypassing the need to deliver and express a cas gene in the target cells [371]. Knowledge on the conjugation mechanisms has also promoted the search for conjugation inhibitors able to slow down the spread of plasmids carrying AMR genes [372–375] (reviewed in [376]). While it may be futile to expect complete stop of the spread of adaptive traits in natural populations, the use of such compounds when selection for spread is maximal, i.e. when under antibiotic therapy, could be a game changer in the current pandemic of antibiotic resistance.
Taken together, these strategies illustrate the growing potential of plasmid engineering. The ability to transfer, block, eliminate, or regulate genetic material exchange between bacteria has many potential applications to tackle environmental and medical challenges. Long viewed as molecular biologist’s best friends (see Table 1) and clinician’s worst enemies, plasmids now stand at the center of innovative engineering strategies. Engineers and evolutionary ecologists are joining the effort of microbiologists to make plasmids formidable tools for microbiome intervention.
Ongoing questions on plasmid mobility
While there is now a better understanding of plasmid mobility than 15 years ago, when we first reviewed the field [2], many questions remain to be answered.
How much do we know about the diversity of plasmid conjugation across Prokaryotes? While the discovery of conjugation pre-dates the naming of plasmids [1], there is yet much to understand in terms of plasmid mobility by this mechanism. In Archaea, the conjugation system derives from those of bacterial monoderms, as reported a long time ago [377, 378]. There are now structural details of a conjugative pilus in Sulfolobales, the so-called crenarchaeal exchange of DNA system (Ced). Yet, this system is dedicated to the import of and not the export of DNA [379]. Only one plasmid of an Euryarchaeon was demonstrated to be conjugative. This confirms original predictions that these elements are homologous to those of bacteria and can provide long-range transfer, including to a Crenarchaeon [380]. It also shows how much remains to be understood about plasmid conjugation in Archaea. For example, we still ignore the relaxases used by these systems, or, if they do not exist, which is the mechanism that specifically brings ssDNA to the conjugative pilus. Among bacteria, even highly studied phyla remain poorly known in terms of conjugation and present intriguing novel systems (reviewed in [381]). Mycobacteria have peculiar outer membranes, and this may explain why a conjugative plasmid was found to depend on a minimalistic T4SS and on a T7SS to transfer DNA [382]. In Streptomyces spp. conjugation can also take place by a very simple system transferring dsDNA within neighbouring cells in mycelia [383, 384]. In Cyanobacteriota, a conjugative system was first mentioned over 40 years ago [385]. Computational studies showed many homologous systems across the plasmids of the phylum, defining a specific type of MPF (MPFC), that is amongst the most divergent from the model conjugative systems [58, 68]. Yet, there is a lack of functional studies on these systems, which remain hypothetical (but see [386]). Similarly, we need information on the conjugation systems of most of the recently uncovered bacterial phyla [387], which are gradually becoming more understood [388].
How different are phage-plasmids in shaping plasmid mobility? Many novel P-P families have been uncovered [17], but most remain to be tested experimentally. Their characterization will be important to test hypotheses on the advantages and costs of phage-driven versus conjugation driven plasmid mobility. P-Ps reproduce quickly, disperse in the environment, and kill donor cells. We suspect, although we do not have definitive evidence, that this influences the types of accessory traits they encode and the environments in which they are most prevalent. Given their potential costs to the cellular genome, and to the latter MGEs, they are probably more targeted by defense systems than other plasmids.
What other mechanisms are involved in plasmid mobility? Hitchers are now being uncovered at a fast pace. These elements require mechanisms of mobility by conjugation or transduction encoded by other MGEs. They have evolved sophisticated mechanisms to hijack such processes, and to counter-act the responses by helpers. Given the fast pace at which they are being unraveled, many may yet remain to be uncovered. Some mechanisms of transfer providing lazy mobility are well known, e.g., natural transformation, but their impact remains to be quantified. For example, natural transformation and vesicles allow the transfer of small plasmids in the lab, but their relevance in natural environments is unknown. Similarly, the impact of conduction and transduction in the spread of plasmids in natural populations is unclear. For conduction it is unknown if plasmids are often re-established (re-split) after transfer. There may also be radically novel transfer mechanisms to be uncovered. For example, the plasmid pR1SE from an haloarchaea encodes proteins that are found in regularly shaped membrane vesicles enclosing the plasmid DNA which upon cell release can infecting other cells, resembling a viral transmission [389].
May the existence of alternative mechanisms of mobility contribute to explain the transfer of putatively nontransmissible plasmids? We refer here to plasmids lacking recognizable genetic elements of helpers and hitchers. Even in the best studied model organisms, E. coli and Staphylococcus aureus, >10% of the plasmids seem to lack any genetic element known to drive mobilization by conjugation or transduction [103]. In most species, there are huge numbers of plasmids lacking recognizable genetic elements allowing to associate them with conjugation or with phage-driven mobility. Maybe they harbor novel families of oriT, maybe they are phage-plasmid satellites, maybe they encode novel mechanisms of transfer, or maybe they just lazily transfer at low rates using less-specific mechanisms (generalized transduction, transformation, vesicles).
Is plasmid transmission indiscriminate? Plasmid survival hinges on a fine-tuned balance between promoting its horizontal transfer and minimizing the fitness cost to host cells. The expression of mobility genes can be promoted by cues that environmental conditions are favorable (induction of conjugation by quorum sensing) or that the host cell is in danger (induction of P-Ps under DNA damage). Understanding the conditions favoring horizontal transfer of plasmids will allow a better characterization of plasmid epidemiology and of the cost of their mobility. One other important and related aspect of the extended regulation of plasmid mobility is the possibility of plasmids picking their hosts (or being picked by them). The physiological and structural traits of recipient cells play a crucial role in determining transfer efficiency and specificity, raising questions about whether they represent a functional constraint or a set of options shaped by natural selection. Together, these findings challenge the idea of indiscriminate plasmid spread, suggesting that conjugation often discriminates between targets and is tightly regulated in response to environmental variables. The extent to which a plasmid “chooses” a suitable recipient, and the recipient “accepts” the plasmid, remains an open question.
How do plasmid–plasmid interactions influence their evolutionary stability and ecological spread? Functional dependencies, intense antagonism, and competition between plasmids makes their mobility dependent on the presence of other plasmids in the cell. Some plasmids actively reduce the conjugative spread of co-resident plasmids—for example, through fertility inhibition systems or type IV CRISPR-Cas mechanisms. In turn, plasmids have evolved countermeasures such as anti-CRISPR proteins, antirestriction systems, and early expressed genes in the leading region that help neutralize rival plasmids or bypass recipient defenses shortly after conjugation. These known antagonistic co-evolutionary dynamics may represent just the tip of the iceberg of inter-plasmid conflict and their impact remains to be fully understood.
Can the definition of plasmid be extended to other elements? Some virulent phages have been observed to remain in cells for long periods of time in a state that seems intermediate between lysogeny as a phage-plasmid and pseudo-lysogenization (where the phage does not replicate) [390]. Filamentous phages produce chronic infections where a plasmid form is continuously replicated, encapsidated, and actively transported outside the functional cell (without cell lysis) [391]. Some ICEs are replicative, i.e. upon induction they excise and replicate as plasmids [392]. We have excluded these elements from our plasmid definition, and from this review, because their replication is not usually in pace with the cell cycle. Yet, they all illustrate the capacity of MGEs to cross the boundaries of our definitions.
How much of what we have learned about plasmid mobility in the laboratory is directly applicable to understand plasmid mobility in natural populations? The analysis of environmental bacteria using metagenomics methods will be key to answer this question. The use of proximity ligation or fusion polymerase chain reaction and epigenetic markers to query the presence and location of plasmids in environmental samples will allow a better understanding of the distribution and mobility of plasmids in nature (reviewed in [393]). These technologies were shown to be capable of identifying plasmid transfers in nature during serial sampling [394]. Their development could help understand plasmid mobility and co-mobility. For example, a recent study found numerous transfers within wastewater communities, pinpointing the mobilizable PTU-Q1 plasmids as those having the broadest host range in the samples [395]. Even analysis of pre-existing metagenomics data is providing interesting findings. pBI143 is a mobilizable plasmid that transfers between Bacteroidales and is present in the human gut with abundances surpassing those of the crassphage [396]. Analysis of conjugation in natura is also bringing surprises. A recent study showed that among 13 conjugative plasmids, few attained high conjugation rates in vivo and half revealed no conjugation at all [397]. Another study revealed the dissemination of a carbapenemase-encoding conjugative plasmid in a hospital setting where plasmid transfer occurred in the gut of almost all colonized patients in certain hospital locations [398]. The importance of understanding plasmid mobility in this context cannot be underestimated: a study of patients carrying more than one bacterial host with a variant of the pOXA-48 resistance plasmid revealed that in almost all cases this was due to recent conjugation since the elements were identical across hosts [399]. The ability to trace the spread of plasmids across bacteria in real-life contexts will ultimately allow to quantify the effective rates of plasmid transfer in nature. This will open the possibility of understanding plasmid mobility variation in function of ecological variables, such as presence of antibiotics, pollutants, stress response or microbial communication. Identifying the plasmid transfer networks in nature will also allow to understand how much of the spread of plasmids is determined by transfer itself and how much by their phenotypic impact in the bacterial host that allows their subsequent vertical transmission.
Acknowledgements
The authors dedicate this review to the memory of two remarkable scientists, Laura Frost and Manolo Espinosa, whose lifelong research significantly advanced our understanding of plasmid mobility. E.P.C.R. thanks the past and present members of the Microbial Evolutionary Genomics laboratory for the hard work and great ideas that led to this review, with particular emphasis on contributions by Eugen Pfeifer, Manuel Ares-Arroyo, Charles Coluzzi, Julien Guglielmini, and Yiqing Wang. Special thanks to Manuel Ares-Arroyo for comments on an earlier version of the manuscript.
Author contributions: Maria Pilar Garcillan Barcia (Conceptualization [equal], Writing—original draft [equal], Writing—review & editing [equal]), Fernando de la Cruz (Conceptualization [equal], Writing—original draft [equal], Writing—review & editing [equal]), Eduardo P C Rocha (Conceptualization [equal], Visualization [equal], Writing—original draft [equal], Writing—review & editing [equal]).
Contributor Information
Maria Pilar Garcillán-Barcia, Instituto de Biomedicina y Biotecnología de Cantabria (Consejo Superior de Investigaciones Científicas – Universidad de Cantabria), 39011 Santander, Cantabria, Spain.
Fernando de la Cruz, Instituto de Biomedicina y Biotecnología de Cantabria (Consejo Superior de Investigaciones Científicas – Universidad de Cantabria), 39011 Santander, Cantabria, Spain.
Eduardo P C Rocha, Institut Pasteur, Université Paris Cité, CNRS UMR3525, Microbial Evolutionary Genomics, 75015 Paris, France.
Conflict of interest
None declared.
Funding
Work in EPCR lab is supported by the Institut Pasteur, the CNRS, the INCEPTION project [PIA/ANR-16-CONV-0005], and the Laboratoire d’Excellence IBEID Integrative Biology of Emerging Infectious Diseases [ANR-10-LABX-62-IBEID]. Work in MPB-B lab is funded by the Spanish Ministry of Science and Innovation [grant MCIN/AEI/10.13039/501100011033 PID2020-117923GB-I00].
Data availability
No new data were generated or analysed in support of this research.
References
- 1. Lederberg J Plasmid (1952–1997). Plasmid. 1998; 39:1–9. 10.1006/plas.1997.1320. [DOI] [PubMed] [Google Scholar]
- 2. Smillie C, Pilar Garcillan-Barcia M, Victoria Francia M et al. Mobility of plasmids. Microbiol Mol Biol Rev. 2010; 74:434–52. 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodríguez-Beltrán J, DelaFuente J, León-Sampedro R et al. Beyond horizontal gene transfer: the role of plasmids in bacterial evolution. Nat Rev Micro. 2021; 19:347–59. 10.1038/s41579-020-00497-1. [DOI] [PubMed] [Google Scholar]
- 4. Tettelin H, Riley D, Cattuto C et al. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008; 11:472–7. 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 5. Arnold BJ, Huang I, Hanage WP Horizontal gene transfer and adaptive evolution in bacteria. Nat Rev Micro. 2021; 30:206–18. [DOI] [PubMed] [Google Scholar]
- 6. Brockhurst MA, Harrison E, Hall JP et al. The ecology and evolution of pangenomes. Curr Biol. 2019; 29:R1094–103. 10.1016/j.cub.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 7. Partridge SR, Kwong SM, Firth N et al. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018; 31:e00088-17. 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Novick RP Plasmid incompatibility. Microbiol Rev. 1987; 51:381–95. 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carattoli A, Bertini A, Villa L et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005; 63:219–28. 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 10. Francia MV, Varsaki A, Garcillan-Barcia MP et al. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 2004; 28:79–100. 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 11. Robertson J, Nash JHE MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genomics. 2018; 4:e000206. 10.1099/mgen.0.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guglielmini J, Quintais L, Garcillan-Barcia P et al. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011; 7:e1002222. 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garcillán-Barcia MP, Redondo-Salvo S, de la Cruz F Plasmid classifications. Plasmid. 2023; 126:102684. 10.1016/j.plasmid.2023.102684. [DOI] [PubMed] [Google Scholar]
- 14. Redondo-Salvo S, Fernández-López R, Ruiz R et al. Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat Commun. 2020; 11:3602. 10.1038/s41467-020-17278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Virolle C, Goldlust K, Djermoun S et al. Plasmid transfer by conjugation in gram-negative bacteria: from the cellular to the community level. Genes. 2020; 11:1239. 10.3390/genes11111239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prensky H, Gomez-Simmonds A, Uhlemann A et al. Conjugation dynamics depend on both the plasmid acquisition cost and the fitness cost. Mol Syst Biol. 2021; 17:e9913. 10.15252/msb.20209913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pfeifer E, Moura de Sousa JA, Touchon M et al. Bacteria have numerous distinctive groups of phage-plasmids with conserved phage and variable plasmid gene repertoires. Nucleic Acids Res. 2021; 49:2655–73. 10.1093/nar/gkab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ares-Arroyo M, Coluzzi C, Moura de Sousa JA et al. Hijackers, hitchhikers, or co-drivers? The mysteries of mobilizable genetic elements. PLoS Biol. 2024; 22:e3002796. 10.1371/journal.pbio.3002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rodríguez-Rubio L, Serna C, Ares-Arroyo M et al. Extensive antimicrobial resistance mobilization via multicopy plasmid encapsidation mediated by temperate phages. J Antimicrob Chemother. 2020; 75:3173–80. 10.1093/jac/dkaa311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Humphrey S, San Millán Á, Toll-Riera M et al. Staphylococcal phages and pathogenicity islands drive plasmid evolution. Nat Commun. 2021; 12:5845. 10.1038/s41467-021-26101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dionisio F, Zilhão R, Gama JA Interactions between plasmids and other mobile genetic elements affect their transmission and persistence. Plasmid. 2019; 102:29–36. 10.1016/j.plasmid.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 22. Domingues S, Nielsen KM Membrane vesicles and horizontal gene transfer in prokaryotes. Curr Opin Microbiol. 2017; 38:16–21. 10.1016/j.mib.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 23. Frankel G, David S, Low WW et al. Plasmids pick a bacterial partner before committing to conjugation. Nucleic Acids Res. 2023; 51:8925–33. 10.1093/nar/gkad678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee P, Eldakar O, Gogarten J. et al. Bacterial cooperation through horizontal gene transfer: trends in Ecology & Evolution. Trends Ecol Evol. 2022; 37:223–32. 10.1016/j.tree.2021.11.006. [DOI] [PubMed] [Google Scholar]
- 25. Johnson CM, Grossman AD Integrative and conjugative elements (ICEs): what they do and how they work. Annu Rev Genet. 2015; 49:577–601. 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kadibalban AS, Landan G, Dagan T The extent and characteristics of DNA transfer between plasmids and chromosomes. Curr Biol. 2024; 34:3189–3200. 10.1016/j.cub.2024.06.030. [DOI] [PubMed] [Google Scholar]
- 27. Coluzzi C, Garcillán-Barcia MP, de la Cruz F et al. Evolution of plasmid mobility: origin and fate of conjugative and nonconjugative plasmids. Mol Biol Evol. 2022; 39:msac115. 10.1093/molbev/msac115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Llosa M, Gomis-Ruth FX, Coll M et al. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002; 45:1–8. 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 29. Fernandez-Lopez R, Redondo S, Garcillan-Barcia MP et al. Towards a taxonomy of conjugative plasmids. Curr Opin Microbiol. 2017; 38:106–13. 10.1016/j.mib.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 30. Fraikin N, Couturier A, Lesterlin C The winding journey of conjugative plasmids toward a novel host cell. Curr Opin Microbiol. 2024; 78:102449. 10.1016/j.mib.2024.102449. [DOI] [PubMed] [Google Scholar]
- 31. Waksman G Molecular basis of conjugation-mediated DNA transfer by gram-negative bacteria. Curr Opin Struct Biol. 2025; 90:102978. 10.1016/j.sbi.2024.102978. [DOI] [PubMed] [Google Scholar]
- 32. Costa TRD, Patkowski JB, Macé K et al. Structural and functional diversity of type IV secretion systems. Nat Rev Micro. 2024; 22:170–85. 10.1038/s41579-023-00974-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Couturier A, Virolle C, Goldlust K et al. Real-time visualisation of the intracellular dynamics of conjugative plasmid transfer. Nat Commun. 2023; 14:294. 10.1038/s41467-023-35978-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Beltrán L, Torsilieri H, Patkowski JB et al. The mating pilus of E. coli pED208 acts as a conduit for ssDNA during horizontal gene transfer. mBio. 2023; 15:e02857–23. 10.1128/mbio.02857-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goldlust K, Ducret A, Halte M et al. The F pilus serves as a conduit for the DNA during conjugation between physically distant bacteria. Proc Natl Acad Sci USA. 2023; 120:e2310842120. 10.1073/pnas.2310842120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Babic A, Lindner AB, Vulic M et al. Direct visualization of horizontal gene transfer. Science. 2008; 319:1533–6. 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- 37. Bates S, Roscoe RA, Althorpe NJ et al. Expression of leading region genes on IncI1 plasmid ColIb-P9: genetic evidence for single-stranded DNA transcription. Microbiology. 1999; 145:2655–62. 10.1099/00221287-145-10-2655. [DOI] [PubMed] [Google Scholar]
- 38. Nasim MT, Eperon IC, Wilkins BM et al. The activity of a single-stranded promoter of plasmid ColIb-P9 depends on its secondary structure. Mol Microbiol. 2004; 53:405–17. 10.1111/j.1365-2958.2004.04114.x. [DOI] [PubMed] [Google Scholar]
- 39. Baharoglu Z, Bikard D, Mazel D Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 2010; 6:e1001165. 10.1371/journal.pgen.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Skurray RA, Reeves P Characterization of lethal zygosis associated with conjugation in Escherichia coli K-12. J Bacteriol. 1973; 113:58–70. 10.1128/jb.113.1.58-70.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ou JT Inhibition of formation of Escherichia coli mating pairs by f1 and MS2 bacteriophages as determined with a Coulter counter. J Bacteriol. 1973; 114:1108–15. 10.1128/jb.114.3.1108-1115.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gordils-Valentin L, Ouyang H, Qian L et al. Conjugative type IV secretion systems enable bacterial antagonism that operates independently of plasmid transfer. Commun Biol. 2024; 7:499. 10.1038/s42003-024-06192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ahmad M, Prensky H, Balestrieri J et al. Tradeoff between lag time and growth rate drives the plasmid acquisition cost. Nat Commun. 2023; 14:2343. 10.1038/s41467-023-38022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Curtsinger HD, Martínez-Absalón S, Liu Y et al. The metabolic burden associated with plasmid acquisition: an assessment of the unrecognized benefits to host cells. Bioessays. 2025; 47:2400164. 10.1002/bies.202400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heinemann JA, Ankenbauer RG Retrotransfer of IncP piasmid R751 from Escherichia coli maxicells: evidence for the genetic sufficiency of self-transferable plasmids for bacterial conjugation. Mol Microbiol. 1993; 10:57–62. 10.1111/j.1365-2958.1993.tb00903.x. [DOI] [PubMed] [Google Scholar]
- 46. Perez-Mendoza D, de la Cruz F Escherichia coli genes affecting recipient ability in plasmid conjugation: are there any?. BMC Genomics. 2009; 10:71. 10.1186/1471-2164-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sheppard RJ, Beddis AE, Barraclough TG The role of hosts, plasmids and environment in determining plasmid transfer rates: a meta-analysis. Plasmid. 2020; 108:102489. 10.1016/j.plasmid.2020.102489. [DOI] [PubMed] [Google Scholar]
- 48. Cyriaque V, Ibarra-Chávez R, Kuchina A et al. Single-cell RNA sequencing reveals plasmid constrains bacterial population heterogeneity and identifies a non-conjugating subpopulation. Nat Commun. 2024; 15:5853. 10.1038/s41467-024-49793-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hall JP, Brockhurst MA, Dytham C et al. The evolution of plasmid stability: are infectious transmission and compensatory evolution competing evolutionary trajectories?. Plasmid. 2017; 91:90–5. 10.1016/j.plasmid.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 50. Djermoun S, Rode DKH, Jiménez-Siebert E et al. Biofilm architecture determines the dissemination of conjugative plasmids. Proc Natl Acad Sci USA. 2025; 122:e2417452122. 10.1073/pnas.2417452122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Madsen JS, Burmølle M, Hansen LH et al. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol. 2012; 65:183–95. 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 52. Stalder T, Top E Plasmid transfer in biofilms: a perspective on limitations and opportunities. NPJ Biofilms Microbiomes. 2016; 2:16022. 10.1038/npjbiofilms.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rodriguez-Grande J, Ortiz Y, Garcia-Lopez D et al. Encounter rates and engagement times limit the transmission of conjugative plasmids. PLoS Genet. 2025; 21:e1011560. 10.1371/journal.pgen.1011560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Andersson DI, Hughes D Microbiological effects of sublethal levels of antibiotics. Nat Rev Micro. 2014; 12:465–78. 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 55. Lopatkin AJ, Huang S, Smith RP et al. Antibiotics as a selective driver for conjugation dynamics. Nat Microbiol. 2016; 1:16044. 10.1038/nmicrobiol.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Benz F, Hall AR Host-specific plasmid evolution explains the variable spread of clinical antibiotic-resistance plasmids. Proc Natl Acad Sci USA. 2023; 120:e2212147120. 10.1073/pnas.2212147120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. San Millan A, Heilbron K, MacLean RC Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J. 2014; 8:601–12. 10.1038/ismej.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guglielmini J, de la Cruz F, Rocha EPC Evolution of conjugation and type IV secretion systems. Mol Biol Evol. 2013; 30:315–31. 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gomez-Valero L, Rusniok C, Carson D et al. More than 18,000 effectors in the Legionella genus genome provide multiple, independent combinations for replication in human cells. Proc Natl Acad Sci USA. 2019; 116:2265–73. 10.1073/pnas.1808016116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Souza DP, Oka GU, Alvarez-Martinez CE et al. Bacterial killing via a type IV secretion system. Nat Commun. 2015; 6:6453. 10.1038/ncomms7453. [DOI] [PubMed] [Google Scholar]
- 61. Alvarez-Martinez CE, Christie PJ Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009; 73:775–808. 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vogel J, Andrews H, Wong S et al. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998; 279:873–6. 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 63. Vadakkepat AK, Xue S, Redzej A et al. Cryo-EM structure of the R388 plasmid conjugative pilus reveals a helical polymer characterized by an unusual pilin/phospholipid binary complex. Structure. 2024; 32:1335–47. 10.1016/j.str.2024.06.009. [DOI] [PubMed] [Google Scholar]
- 64. Bhatty M, Cruz MR, Frank KL et al. Enterococcusfaecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence. Mol Microbiol. 2015; 95:660–77. 10.1111/mmi.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Garcillan-Barcia MP, Cuartas-Lanza R, Cuevas A et al. Cis-acting relaxases guarantee independent mobilization of MOBQ4 plasmids. Front Microbiol. 2019; 10:2557. 10.3389/fmicb.2019.02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Meyer R Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 2009; 62:57–70. 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ares-Arroyo M, Nucci A, Rocha EP Expanding the diversity of origin of transfer-containing sequences in mobilizable plasmids. Nat Microbiol. 2024; 9:3240–53. 10.1038/s41564-024-01844-1. [DOI] [PubMed] [Google Scholar]
- 68. Guglielmini J, Neron B, Abby SS et al. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 2014; 42:5715–27. 10.1093/nar/gku194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Abedon ST, Calendar RL The Bacteriophages. 2005; 2nd edNew York: Oxford University Press. [Google Scholar]
- 70. Sayid R, van den Hurk AWM, Rothschild-Rodriguez D et al. Characteristics of phage-plasmids and their impact on microbial communities. Essays Biochem. 2024; 68:583–92. 10.1042/EBC20240014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bertani G Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951; 62:293–300. 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yarmolinsky MB Bacteriophage P1 in retrospect and in prospect. J Bacteriol. 2004; 186:7025–8. 10.1128/JB.186.21.7025-7028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Łobocka M, Rose D, Plunkett G. et al. Genome of bacteriophage P1. J Bacteriol. 2004; 186:7032–7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ravin NV N15: the linear phage–plasmid. Plasmid. 2011; 65:102–9. 10.1016/j.plasmid.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 75. Schmidtke DT, Hickey AS, Liachko I. et al. Analysis and culturing of the prototypic crAssphage reveals a phage-plasmid lifestyle. bioRxiv21 March 2024, preprint: not peer reviewed 10.1101/2024.03.20.585998. [DOI]
- 76. Faith DR, Kinnersley M, Brooks DM et al. Characterization and genomic analysis of the Lyme disease spirochete bacteriophage ϕbb-1. PLoS Pathog. 2024; 20:e1012122. 10.1371/journal.ppat.1012122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Silpe JE, Bassler BL A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Cell. 2019; 176:268–80. 10.1016/j.cell.2018.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Huang J, Dai X, Wu Z et al. Conjugative transfer of streptococcal prophages harboring antibiotic resistance and virulence genes. ISME J. 2023; 17:1467–81. 10.1038/s41396-023-01463-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Abeles AL, Snyder KM, Chattoraj DK P1 plasmid replication: replicon structure. J Mol Biol. 1984; 173:307–24. 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- 80. Sternberg N, Powers M, Yarmolinsky M et al. Group Y incompatibility and copy control of P1 prophage. Plasmid. 1981; 5:138–49. 10.1016/0147-619X(81)90015-9. [DOI] [PubMed] [Google Scholar]
- 81. Zhang K, Pankratz K, Duong H et al. Interactions between viral regulatory proteins ensure an MOI-independent probability of lysogeny during infection by bacteriophage P1. mBio. 2021; 12:e01013-21. 10.1128/mbio.01013-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pfeifer E, Rocha EP Phage-plasmids promote recombination and emergence of phages and plasmids. Nat Commun. 2024; 15:1545. 10.1038/s41467-024-45757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pfeifer E, Bonnin RA, Rocha EPC Phage-plasmids spread antibiotic resistance genes through infection and lysogenic conversion. mBIO. 2022; 13:e01851-22. 10.1128/mbio.01851-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Shan X, Szabo RE, Cordero OX Mutation-induced infections of phage-plasmids. Nat Commun. 2023; 14:2049. 10.1038/s41467-023-37512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. San Millan A, Craig MacLean R Fitness costs of plasmids: a limit to plasmid transmission. Microbiol Spectr. 2017; 5:65–79. 10.1128/microbiolspec.MTBP-0016-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Quinones-Olvera N, Owen SV, McCully LM et al. Diverse and abundant phages exploit conjugative plasmids. Nat Commun. 2024; 15:3197. 10.1038/s41467-024-47416-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bossi L, Fuentes JA, Mora G et al. Prophage contribution to bacterial population dynamics. J Bacteriol. 2003; 185:6467–71. 10.1128/JB.185.21.6467-6471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Brown SP, Le Chat L, De Paepe M et al. Ecology of microbial invasions: amplification allows virus carriers to invade more rapidly when rare. Curr Biol. 2006; 16:2048–52. 10.1016/j.cub.2006.08.089. [DOI] [PubMed] [Google Scholar]
- 89. Gama JA, Reis AM, Domingues I et al. Temperate bacterial viruses as double-edged swords in bacterial warfare. PLoS One. 2013; 8:e59043. 10.1371/journal.pone.0059043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Alderliesten JB, Duxbury SJN, Zwart MP et al. Effect of donor-recipient relatedness on the plasmid conjugation frequency: a meta-analysis. BMC Microbiol. 2020; 20:135. 10.1186/s12866-020-01825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. de Jonge PA, Nobrega FL, Brouns SJJ et al. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 2019; 27:51–63. 10.1016/j.tim.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 92. Norberg P, Bergstrom M, Jethava V et al. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat Commun. 2011; 2:268. 10.1038/ncomms1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Heinemann JA, Sprague GF Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989; 340:205–9. 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- 94. Buchanan-Wollaston V, Passiatore JE, Cannon F The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature. 1987; 328:172–5. 10.1038/328172a0. [DOI] [Google Scholar]
- 95. Bartke K, Garoff L, Huseby DL et al. Genetic architecture and fitness of bacterial interspecies hybrids. Mol Biol Evol. 2021; 38:1472–81. 10.1093/molbev/msaa307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nurmemmedov E, Castelnovo M, Medina E et al. Challenging packaging limits and infectivity of phage λ. J Mol Biol. 2012; 415:263–73. 10.1016/j.jmb.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 97. Feiss M, Fisher RA, Crayton MA et al. Packaging of the bacteriophage lambda chromosome: effect of chromosome length. Virology. 1977; 77:281–93. 10.1016/0042-6822(77)90425-1. [DOI] [PubMed] [Google Scholar]
- 98. Jacob F, Wollman EL Genetic and physical determinations of chromosomal segments in Escherichia coli. Symp Soc Exp Biol. 1958; 12:75–92. [PubMed] [Google Scholar]
- 99. Mizuno CM, Ghai R, Saghaï A et al. Genomes of abundant and widespread viruses from the Deep Ocean. mBio. 2016; 7:10.1128/mbio.00805-16. 10.1128/mbio.00805-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Luo E, Aylward FO, Mende DR et al. Bacteriophage distributions and temporal variability in the ocean’s interior. mBio. 2017; 8:10.1128/mbio.01903-17. 10.1128/mbio.01903-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bonte D, Van Dyck H, Bullock JM et al. Costs of dispersal. Biol Rev. 2012; 87:290–312. 10.1111/j.1469-185X.2011.00201.x. [DOI] [PubMed] [Google Scholar]
- 102. Coluzzi C, Rocha EPC The spread of antibiotic resistance is driven by plasmids among the fastest evolving and of broadest host range. Mol Biol Evol. 2025; 42:msaf060. 10.1093/molbev/msaf060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ares-Arroyo M, Coluzzi C, Rocha EP Origins of transfer establish networks of functional dependencies for plasmid transfer by conjugation. Nucleic Acids Res. 2023; 51:3001–16. 10.1093/nar/gkac1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Horne T, Orr VT, Hall JP How do interactions between mobile genetic elements affect horizontal gene transfer?. Curr Opin Microbiol. 2023; 73:102282. 10.1016/j.mib.2023.102282. [DOI] [PubMed] [Google Scholar]
- 105. Garcillán-Barcia MP, Pluta R, Lorenzo-Díaz F et al. The facts and Family secrets of plasmids that replicate via the rolling-circle mechanism. Microbiol Mol Biol Rev. 2021; 86:e00222-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Guzmán-Herrador DL, Llosa M The secret life of conjugative relaxases. Plasmid. 2019; 104:102415. 10.1016/j.plasmid.2019.102415. [DOI] [PubMed] [Google Scholar]
- 107. Hamilton CM, Lee H, Li P-L et al. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J Bacteriol. 2000; 182:1541–8. 10.1128/JB.182.6.1541-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Cabezón E, Sastre JI, de la Cruz F Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet. 1997; 254:400–6. 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 109. Lee CA, Thomas J, Grossman AD The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J Bacteriol. 2012; 194:3165–72. 10.1128/JB.00301-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. O’Brien FG, Yui Eto K, Murphy RJT et al. Origin-of-transfer sequences facilitate mobilisation of non-conjugative antimicrobial-resistance plasmids in Staphylococcus aureus. Nucleic Acids Res. 2015; 43:7971–83. 10.1093/nar/gkv755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Gama JA, Zilhão R, Dionisio F Co-resident plasmids travel together. Plasmid. 2017; 93:24–9. 10.1016/j.plasmid.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 112. Touchon M, de Sousa JAM, Rocha EP Embracing the enemy: the diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr Opin Microbiol. 2017; 38:66–73. 10.1016/j.mib.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 113. Thomason LC, Costantino N, Court DL Genome manipulation by P1 transduction. Curr Protoc Mol Biol. 2007; 79:1.17.1–8. 10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- 114. Fillol-Salom A, Alsaadi A, de Sousa JAM et al. Bacteriophages benefit from generalized transduction. PLoS Pathog. 2019; 15:e1007888. 10.1371/journal.ppat.1007888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hertwig S, Popp A, Freytag B et al. Generalized transduction of small Yersinia enterocolitica plasmids. Appl Environ Microb. 1999; 65:3862–6. 10.1128/AEM.65.9.3862-3866.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ubelaker MH, Rosenblum ED Transduction of plasmid determinants in Staphylococcus aureus and Escherichia coli. J Bacteriol. 1978; 133:699–707. 10.1128/jb.133.2.699-707.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. de Sousa JAM, Fillol-Salom A, Penadés JR et al. Identification and characterization of thousands of bacteriophage satellites across bacteria. Nucleic Acids Res. 2023; 51:2759–77. 10.1093/nar/gkad123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Schmid N, Brandt D, Walasek C et al. An autonomous plasmid as an inovirus phage satellite. Appl Environ Microb. 2024; 90:e00246-24. 10.1128/aem.00246-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Durand R, Huguet KT, Rivard N et al. Crucial role of Salmonella genomic island 1 master activator in the parasitism of IncC plasmids. Nucleic Acids Res. 2021; 49:7807–24. 10.1093/nar/gkab204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ortiz Charneco G, McDonnell B, Kelleher P et al. Plasmid-mediated horizontal gene mobilisation: insights from two lactococcal conjugative plasmids. Microb Biotechnol. 2024; 17:e14421. 10.1111/1751-7915.14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Blanca-Ordóñez H, Oliva-García JJ, Pérez-Mendoza D et al. pSymA-dependent mobilization of the Sinorhizobium meliloti pSymB megaplasmid. J Bacteriol. 2010; 192:6309–12. 10.1128/JB.00549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Bennett PM, Heritage J, Comanducci A et al. Evolution of R plasmids by replicon fusion. J Antimicrob Chemother. 1986; 18:103–11. 10.1093/jac/18.Supplement_C.103. [DOI] [PubMed] [Google Scholar]
- 123. Xie M, Chen K, Ye L. et al. Conjugation of virulence plasmid in clinical Klebsiella pneumoniae strains through formation of a fusion plasmid. Adv Biosys. 2020; 4:1900239. 10.1002/adbi.201900239. [DOI] [PubMed] [Google Scholar]
- 124. Xu Y, Zhang J, Wang M et al. Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 2021; 13:119. 10.1186/s13073-021-00936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. He S, Hickman AB, Varani AM et al. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio. 2015; 6:10.1128.00762-15. 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Harmer CJ, Moran RA, Hall RM Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio. 2014; 5:10.1128/mbio.01801-14. 10.1128/mbio.01801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Dubnau D, Blokesch M Mechanisms of DNA uptake by naturally competent bacteria. Annu Rev Genet. 2019; 53:217–37. 10.1146/annurev-genet-112618-043641. [DOI] [PubMed] [Google Scholar]
- 128. Johnston C, Martin B, Fichant G et al. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Micro. 2014; 12:181–96. 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 129. Jeffrey WH, Paul JH, Stewart GJ Natural transformation of a marineVibrio species by plasmid DNA. Microb Ecol. 1990; 19:259–68. 10.1007/BF02017170. [DOI] [PubMed] [Google Scholar]
- 130. Saunders CW, Guild WR Monomer plasmid DNA transforms Streptococcus pneumoniae. Mol Gen Genet. 1981; 181:57–62. 10.1007/BF00339005. [DOI] [PubMed] [Google Scholar]
- 131. Ohse M, Takahashi K, Kadowaki Y et al. Effects of plasmid DNA sizes and several other factors on transformation of Bacillus subtilis ISW1214 with plasmid DNA by electroporation. Biosci Biotechnol Biochem. 1995; 59:1433–7. 10.1271/bbb.59.1433. [DOI] [PubMed] [Google Scholar]
- 132. Frischer ME, Stewart GJ, Paul JH Plasmid transfer to indigenous marine bacterial populations by natural transformation. FEMS Microbiol Ecol. 1994; 15:127–35. 10.1111/j.1574-6941.1994.tb00237.x. [DOI] [Google Scholar]
- 133. Shen Z, Qin J, Xiang G et al. Outer membrane vesicles mediating horizontal transfer of the epidemic blaOXA-232 carbapenemase gene among Enterobacterales. Emerg Microbes Infect. 2024; 13:2290840. 10.1080/22221751.2023.2290840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Johnston EL, Zavan L, Bitto NJ et al. Planktonic and biofilm-derived Pseudomonas aeruginosa outer membrane vesicles facilitate horizontal gene transfer of plasmid DNA. Microbiol Spectr. 2023; 11:e05179–22. 10.1128/spectrum.05179-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Tran F, Boedicker JQ Genetic cargo and bacterial species set the rate of vesicle-mediated horizontal gene transfer. Sci Rep. 2017; 7:8813. 10.1038/s41598-017-07447-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Bos J, Cisneros LH, Mazel D Real-time tracking of bacterial membrane vesicles reveals enhanced membrane traffic upon antibiotic exposure. Sci Adv. 2021; 7:eabd1033. 10.1126/sciadv.abd1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Maddamsetti R, Wilson ML, Son H-I et al. Scaling laws of bacterial and archaeanplasmids. Nat Commun. 2025; 16:6023. 10.1038/s41467-025-61205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ramiro-Martínez P, Quinto Id, Lanza VF et al. Universal rules govern plasmid copy number. Nat Commun. 2025; 16:6022. 10.1038/s41467-025-61202-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Shaw LP, Chau KK, Kavanagh J et al. Niche and local geography shape the pangenome of wastewater- and livestock-associated Enterobacteriaceae. Sci Adv. 2021; 7:eabe3868. 10.1126/sciadv.abe3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Bergstrom CT, Lipsitch M, Levin BR Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics. 2000; 155:1505–19. 10.1093/genetics/155.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Simonsen L The existence conditions for bacterial plasmids: theory and reality. Microb Ecol. 1991; 22:187–205. 10.1007/BF02540223. [DOI] [PubMed] [Google Scholar]
- 142. Harrison E, Brockhurst MA Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012; 20:262–7. 10.1016/j.tim.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 143. Brockhurst MA, Harrison E Ecological and evolutionary solutions to the plasmid paradox. Trends Microbiol. 2022; 30:534–43. 10.1016/j.tim.2021.11.001. [DOI] [PubMed] [Google Scholar]
- 144. Huisman JS, Benz F, Duxbury SJ et al. Estimating plasmid conjugation rates: a new computational tool and a critical comparison of methods. Plasmid. 2022; 121:102627. 10.1016/j.plasmid.2022.102627. [DOI] [PubMed] [Google Scholar]
- 145. Kosterlitz O, Huisman JS Guidelines for the estimation and reporting of plasmid conjugation rates. Plasmid. 2023; 126:102685. 10.1016/j.plasmid.2023.102685. [DOI] [PubMed] [Google Scholar]
- 146. Hall JP, Wood AJ, Harrison E et al. Source-sink plasmid transfer dynamics maintain gene mobility in soil bacterial communities. Proc Natl Acad Sci USA. 2016; 113:8260–5. 10.1073/pnas.1600974113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Dionisio F, Matic I, Radman M et al. Plasmids spread very fast in heterogeneous bacterial communities. Genetics. 2002; 162:1525–32. 10.1093/genetics/162.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Valle A-d, L-S A., R-B R. et al. Variability of plasmid fitness effects contributes to plasmid persistence in bacterial communities. Nat Commun. 2021; 12:2653. 10.1038/s41467-021-22849-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Bethke JH, Ma HR, Tsoi R et al. Vertical and horizontal gene transfer tradeoffs direct plasmid fitness. Mol Syst Biol. 2023; 19:e11300. 10.15252/msb.202211300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Wein T, Hülter NF, Mizrahi I et al. Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat Commun. 2019; 10:2595. 10.1038/s41467-019-10600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Huisman JS, Bernhard A, Igler C Should I stay or should I go: transmission trade-offs in phages and plasmids. Trends Microbiol. 2025; 5:484–95. 10.1016/j.tim.2025.01.007. [DOI] [PubMed] [Google Scholar]
- 152. Turner P, Cooper SV, Lenski RE Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution. 1998; 52:484–95. 10.2307/2411070. [DOI] [PubMed] [Google Scholar]
- 153. Dimitriu T, Matthews AC, Buckling A Increased copy number couples the evolution of plasmid horizontal transmission and plasmid-encoded antibiotic resistance. Proc Natl Acad Sci USA. 2021; 118:e2107818118. 10.1073/pnas.2107818118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Frost LS, Koraimann G Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol. 2010; 5:1057–71. 10.2217/fmb.10.70. [DOI] [PubMed] [Google Scholar]
- 155. Wong JJW, Lu J, Glover JNM Relaxosome function and conjugation regulation in F-like plasmids – a structural biology perspective. Mol Microbiol. 2012; 85:602–17. 10.1111/j.1365-2958.2012.08131.x. [DOI] [PubMed] [Google Scholar]
- 156. Sheppard RJ, Barraclough TG, Jansen VAA The evolution of plasmid transfer rate in bacteria and its effect on plasmid persistence. Am Nat. 2021; 198:473–88. 10.1086/716063. [DOI] [PubMed] [Google Scholar]
- 157. Koraimann G, Teferle K, Markolin G et al. The FinOP repressor system of plasmid R1: analysis of the antisense RNA control of traJ expression and conjugative DNA transfer. Mol Microbiol. 1996; 21:811–21. 10.1046/j.1365-2958.1996.361401.x. [DOI] [PubMed] [Google Scholar]
- 158. Jacob F, Wollman EL Processes of conjugation and recombination in Escherichia coli. I. Induction by conjugation or zygotic induction. Ann Inst Pasteur. 1956; 91:486–510. [PubMed] [Google Scholar]
- 159. Haft RJ, Mittler JE, Traxler B Competition favours reduced cost of plasmids to host bacteria. ISME J. 2009; 3:761–9. 10.1038/ismej.2009.22. [DOI] [PubMed] [Google Scholar]
- 160. Bingle LEH, Thomas CM Regulatory circuits for plasmid survival. Curr Opin Microbiol. 2001; 4:194–200. 10.1016/S1369-5274(00)00188-0. [DOI] [PubMed] [Google Scholar]
- 161. Fernandez-Lopez R, Del Campo I, Revilla C et al. Negative feedback and transcriptional overshooting in a regulatory network for horizontal gene transfer. PLoS Genet. 2014; 10:e1004171. 10.1371/journal.pgen.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Miyakoshi M, Ohtsubo Y, Nagata Y et al. Transcriptome analysis of zygotic induction during conjugative transfer of plasmid RP4. Front Microbiol. 2020; 11:fmicb.2020.01125. 10.3389/fmicb.2020.01125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lundquist PD, Levin BR Transitory derepression and the maintenance of conjugative plasmids. Genetics. 1986; 113:483–97. 10.1093/genetics/113.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Dunny GM Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet. 2013; 47:457–82. 10.1146/annurev-genet-111212-133449. [DOI] [PubMed] [Google Scholar]
- 165. Dunny GM, Berntsson RP-A Enterococcal sex pheromones: evolutionary pathways to complex, two-signal systems. J Bacteriol. 2016; 198:1556–62. 10.1128/JB.00128-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Danino VE, Wilkinson A, Edwards A et al. Recipient-induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum-sensing relay. Mol Microbiol. 2003; 50:511–25. 10.1046/j.1365-2958.2003.03699.x. [DOI] [PubMed] [Google Scholar]
- 167. Clewell DB Sex pheromones and the plasmid-encoded mating response in Enterococcus faecalis. Bacterial Conjugation. 1993; Boston, MA: Springer US; 349–67. 10.1007/978-1-4757-9357-4. [DOI] [Google Scholar]
- 168. Dunny GM Genetic functions and cell–cell interactions in the pheromone-inducible plasmid transfer system of Enterococcus faecalis. Mol Microbiol. 1990; 4:689–96. 10.1111/j.1365-2958.1990.tb00639.x. [DOI] [PubMed] [Google Scholar]
- 169. Auchtung JM, Lee CA, Monson RE et al. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci USA. 2005; 102:12554–9. 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Bernard C, Li Y, Lopez P et al. Large-scale identification of known and novel RRNPP quorum-sensing systems by rrnpp_detector captures novel features of bacterial, plasmidic, and viral coevolution. Mol Biol Evol. 2023; 40:msad062. 10.1093/molbev/msad062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Felipe-Ruiz A, Marina A, Rocha EPC Structural and genomic evolution of RRNPPA systems and their pheromone signaling. mBio. 2022; 13:e02514-22. 10.1128/mbio.02514-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Duddy OP, Bassler BL Quorum sensing across bacterial and viral domains. PLoS Pathog. 2021; 17:e1009074. 10.1371/journal.ppat.1009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Erez Z, Steinberger-Levy I, Shamir M et al. Communication between viruses guides lysis-lysogeny decisions. Nature. 2017; 541:488–93. 10.1038/nature21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Santoriello FJ, Bassler BL The LuxO-OpaR quorum-sensing cascade differentially controls Vibriophage VP882 lysis-lysogeny decision making in liquid and on surfaces. PLoS Genet. 2024; 20:e1011243. 10.1371/journal.pgen.1011243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Suckow G, Seitz P, Blokesch M Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J Bacteriol. 2011; 193:4914–24. 10.1128/JB.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Smith HO, Tomb JF, Dougherty BA et al. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae rd genome. Science. 1995; 269:538–40. 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 177. Stein DC Transformation of Neisseria gonorrhoeae: physical requirements of the transforming DNA. Can J Microbiol. 1991; 37:345–9. 10.1139/m91-056. [DOI] [PubMed] [Google Scholar]
- 178. Achtman M, Schwuchow S, Helmuth R et al. Cell-cell interactions in conjugating Escherichia coli: Con− mutants and stabilization of mating aggregates. Molec Gen Genet. 1978; 164:171–83. 10.1007/BF00267382. [DOI] [Google Scholar]
- 179. Anthony KG, Sherburne C, Sherburne R et al. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol. 1994; 13:939–53. 10.1111/j.1365-2958.1994.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 180. Haudiquet M, Buffet A, Rendueles O et al. Interplay between the cell envelope and mobile genetic elements shapes gene flow in populations of the nosocomial pathogen Klebsiella pneumoniae. PLoS Biol. 2021; 19:e3001276. 10.1371/journal.pbio.3001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Wang S, Ding Q, Zhang Y et al. Evolution of virulence, fitness, and carbapenem resistance transmission in ST23 hypervirulent Klebsiella pneumoniae with the capsular polysaccharide synthesis gene wcaJ inserted via insertion sequence elements. Microbiol Spectr. 2022; 11:e04478-22. 10.1128/spectrum.02400-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Yong M, Chen Y, Oo G et al. Dominant carbapenemase-encoding plasmids in clinical enterobacterales isolates and hypervirulent Klebsiella pneumoniae, Singapore. Emerg Infect Dis. 2022; 28:1578–88. 10.3201/eid2808.212542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Haudiquet M, Le Bris J, Nucci A et al. Capsules and their traits shape phage susceptibility and plasmid conjugation efficiency. Nat Commun. 2024; 15:2032. 10.1038/s41467-024-46147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Johnson CM, Grossman AD Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol. 2014; 93:1284–301. 10.1111/mmi.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Johnson CM, Grossman AD The composition of the cell envelope affects conjugation in Bacillus subtilis. J Bacteriol. 2016; 198:1241–9. 10.1128/JB.01044-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Harden MM, Anderson ME, Grossman AD CRISPR interference screen reveals a role for cell wall teichoic acids in conjugation in Bacillus subtilis. Mol Microbiol. 2022; 117:1366–83. 10.1111/mmi.14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Ouyang R, Ongenae V, Muok A et al. Phage fibers and spikes: a nanoscale Swiss army knife for host infection. Curr Opin Microbiol. 2024; 77:102429. 10.1016/j.mib.2024.102429. [DOI] [PubMed] [Google Scholar]
- 188. de Sousa JAM, Buffet A, Haudiquet M et al. Modular prophage interactions driven by capsule serotype select for capsule loss under phage predation. ISME J. 2020; 14:2980–96. 10.1038/s41396-020-0726-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Manning PA, Morelli G, Achtman M traG protein of the F sex factor of Escherichia coli K-12 and its role in conjugation. Proc Natl Acad Sci USA. 1981; 78:7487–91. 10.1073/pnas.78.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Klimke WA, Frost LS Genetic analysis of the role of the transfer gene, traN, of the F and R100-1 plasmids in mating pair stabilization during conjugation. J Bacteriol. 1998; 180:4036–43. 10.1128/JB.180.16.4036-4043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Klimke WA, Rypien CD, Klinger B et al. The mating pair stabilization protein, TraN, of the F plasmid is an outer-membrane protein with two regions that are important for its function in conjugation. Microbiology. 2005; 151:3527–40. 10.1099/mic.0.28025-0. [DOI] [PubMed] [Google Scholar]
- 192. Low WW, Wong JLC, Beltran LC et al. Mating pair stabilization mediates bacterial conjugation species specificity. Nat Microbiol. 2022; 7:1016–27. 10.1038/s41564-022-01146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Ried G, Henning U A unique amino acid substitution in the outer membrane protein OmpA causes conjugation deficiency in Escherichia coli K-12. FEBS Lett. 1987; 223:387–90. 10.1016/0014-5793(87)80324-1. [DOI] [PubMed] [Google Scholar]
- 194. Komano T, Kim SR, Yoshida T et al. DNA rearrangement of the shufflon determines recipient specificity in liquid mating of IncI1 plasmid R64. J Mol Biol. 1994; 243:6–9. 10.1006/jmbi.1994.1625. [DOI] [PubMed] [Google Scholar]
- 195. Komano T Mating variation by DNA inversions of shufflon in plasmid R64. Adv Biophys. 1995; 31:181–93. 10.1016/0065-227X(95)99391-2. [DOI] [PubMed] [Google Scholar]
- 196. Allard N, Collette A, Paquette J et al. Systematic investigation of recipient cell genetic requirements reveals important surface receptors for conjugative transfer of IncI2 plasmids. Commun Biol. 2023; 6:1172. 10.1038/s42003-023-05534-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Ishiwa A, Komano T PilV adhesins of Plasmid R64 thin pili specifically bind to the lipopolysaccharides of recipient cells. J Mol Biol. 2004; 343:615–25. 10.1016/j.jmb.2004.08.059. [DOI] [PubMed] [Google Scholar]
- 198. Komano T Shufflons: multiple inversion systems and integrons. Annu Rev Genet. 1999; 33:171–91. 10.1146/annurev.genet.33.1.171. [DOI] [PubMed] [Google Scholar]
- 199. Komano T, Kim SR, Nisioka T Distribution of shufflon among IncI plasmids. J Bacteriol. 1987; 169:5317–9. 10.1128/jb.169.11.5317-5319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Komano T, Fujitani S, Funayama N et al. Physical and genetic analyses of IncI2 plasmid R721: evidence for the presence of shufflon. Plasmid. 1990; 23:248–51. 10.1016/0147-619X(90)90057-J. [DOI] [PubMed] [Google Scholar]
- 201. Venturini C, Hassan KA, Roy Chowdhury P et al. Sequences of two related multiple antibiotic resistance virulence plasmids sharing a unique IS26-related molecular signature isolated from different Escherichia coli pathotypes from different hosts. PLoS One. 2013; 8:e78862. 10.1371/journal.pone.0078862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Cottell JL, Saw HT, Webber MA et al. Functional genomics to identify the factors contributing to successful persistence and global spread of an antibiotic resistance plasmid. BMC Microbiol. 2014; 14:168. 10.1186/1471-2180-14-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Kim SR, Komano T The plasmid R64 thin pilus identified as a type IV pilus. J Bacteriol. 1997; 179:3594–603. 10.1128/jb.179.11.3594-3603.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Brouwer MSM, Tagg KA, Mevius DJ et al. IncI shufflons: assembly issues in the next-generation sequencing era. Plasmid. 2015; 80:111–7. 10.1016/j.plasmid.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 205. Achtman M, Kennedy N, Skurray R Cell–cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc Natl Acad Sci USA. 1977; 74:5104–8. 10.1073/pnas.74.11.5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Gottesman ME, Weisberg RA Little lambda, who made thee?. Microbiol Mol Biol Rev. 2004; 68:796–813. 10.1128/MMBR.68.4.796-813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Nguyen TVP, Wu Y, Yao T et al. Coinfecting phages impede each other’s entry into the cell. Curr Biol. 2024; 34:2841–53. 10.1016/j.cub.2024.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Auchtung JM, Lee CA, Garrison KL et al. Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICE Bs1 of Bacillus subtilis. Mol Microbiol. 2007; 64:1515–28. 10.1111/j.1365-2958.2007.05748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Garcillan-Barcia MP, de la Cruz F Why is entry exclusion an essential feature of conjugative plasmids?. Plasmid. 2008; 60:1–18. 10.1016/j.plasmid.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 210. Achtman M, Manning PA, Edelbluth C et al. Export without proteolytic processing of inner and outer membrane proteins encoded by F sex factor tra cistrons in Escherichia coli minicells. Proc Natl Acad Sci USA. 1979; 76:4837–41. 10.1073/pnas.76.10.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Gunton JE, Ussher JER, Rooker MM et al. Entry exclusion in the IncHI1 plasmid R27 is mediated by EexA and EexB. Plasmid. 2008; 59:86–101. 10.1016/j.plasmid.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 212. Haase J, Kalkum M, Lanka E TrbK, a small cytoplasmic membrane lipoprotein, functions in entry exclusion of the IncP alpha plasmid RP4. J Bacteriol. 1996; 178:6720–9. 10.1128/jb.178.23.6720-6729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Hartskeerl R, Tommassen J, Hoekstra W Relationship between the proteins encoded by the exclusion determining locus of the IncI plasmid R144 and the cellular localization of these proteins in Escherichia coli K-12. Molec Gen Genet. 1985; 200:138–44. 10.1007/BF00383327. [DOI] [PubMed] [Google Scholar]
- 214. Pohlman RF, Genetti HD, Winans SC Entry exclusion of the IncN plasmid pKM101 is mediated by a single hydrophilic protein containing a lipid attachment motif. Plasmid. 1994; 31:158–65. 10.1006/plas.1994.1017. [DOI] [PubMed] [Google Scholar]
- 215. Kamruzzaman M, Mathers AJ, Iredell JR A novel plasmid entry exclusion system in pKPC_UVA01, a promiscuous conjugative plasmid carrying the bla KPC carbapenemase gene. Antimicrob Agents Chemother. 2022; 66:aac.02322–02321. 10.1128/aac.02322-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Audette GF, Manchak J, Beatty P et al. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology. 2007; 153:442–51. 10.1099/mic.0.2006/001917-0. [DOI] [PubMed] [Google Scholar]
- 217. Humbert M, Huguet KT, Coulombe F et al. Entry exclusion of conjugative plasmids of the IncA, IncC, and related untyped incompatibility groups. J Bacteriol. 2019; 201:10.1128/jb.00731-18. 10.1128/jb.00731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Marrero J, Waldor MK The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J Bacteriol. 2007; 189:3302–5. 10.1128/JB.01902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Marrero J, Waldor MK Determinants of entry exclusion within Eex and TraG are cytoplasmic. J Bacteriol. 2007; 189:6469–73. 10.1128/JB.00522-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Sakuma T, Tazumi S, Furuya N et al. ExcA proteins of IncI1 plasmid R64 and IncIgamma plasmid R621a recognize different segments of their cognate TraY proteins in entry exclusion. Plasmid. 2013; 69:138–45. 10.1016/j.plasmid.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 221. Avello M, Davis KP, Grossman AD Identification, characterization and benefits of an exclusion system in an integrative and conjugative element of Bacillus subtilis. Mol Microbiol. 2019; 112:1066–82. 10.1111/mmi.14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Minkley EG Purification and characterization of pro-TraTp, the signal sequence-containing precursor of a secreted protein encoded by the F sex factor. J Bacteriol. 1984; 158:464–73. 10.1128/jb.158.2.464-473.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Rivard N, Humbert M, Huguet KT et al. Surface exclusion of IncC conjugative plasmids and their relatives. PLoS Genet. 2024; 20:e1011442. 10.1371/journal.pgen.1011442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Harrison JL, Taylor IM, Platt K et al. Surface exclusion specificity of the TraT lipoprotein is determined by single alterations in a five-amino-acid region of the protein. Mol Microbiol. 1992; 6:2825–32. 10.1111/j.1365-2958.1992.tb01462.x. [DOI] [PubMed] [Google Scholar]
- 225. Minkley EG, Willetts NS Overproduction, purification and characterization of the F traT protein. Mol Gen Genet. 1984; 196:225–35. 10.1007/BF00328054. [DOI] [PubMed] [Google Scholar]
- 226. Riede I, Eschbach M-L Evidence that TraT interacts with OmpA of Escherichia coli. FEBS Lett. 1986; 205:241–5. 10.1016/0014-5793(86)80905-X. [DOI] [PubMed] [Google Scholar]
- 227. Seddon C, David S, Wong JLC et al. Cryo-EM structure and evolutionary history of the conjugation surface exclusion protein TraT. Nat Commun. 2025; 16:659. 10.1038/s41467-025-55834-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Schmitt A, Hirt H, Järvå MA. et al. Enterococcal PrgA extends far outside the cell and provides surface exclusion to protect against unwanted conjugation. J Mol Biol. 2020; 432:5681–95. 10.1016/j.jmb.2020.08.018. [DOI] [PubMed] [Google Scholar]
- 229. Georjon H, Bernheim A The highly diverse antiphage defence systems of bacteria. Nat Rev Micro. 2023; 21:686–700. 10.1038/s41579-023-00934-x. [DOI] [PubMed] [Google Scholar]
- 230. Hampton HG, Watson BN, Fineran PC The arms race between bacteria and their phage foes. Nature. 2020; 577:327–36. 10.1038/s41586-019-1894-8. [DOI] [PubMed] [Google Scholar]
- 231. Arber W, Morse ML Host specificity of DNA produced by Escherichia coli. VI. Effects on bacterial conjugation. Genetics. 1965; 51:137–48. 10.1093/genetics/51.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Tesson F, Hervé A, Mordret E et al. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat Commun. 2022; 13:2561. 10.1038/s41467-022-30269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Ishikawa K, Fukuda E, Kobayashi I Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems. DNA Res. 2010; 17:325–42. 10.1093/dnares/dsq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Loenen WA, Raleigh EA The other face of restriction: modification-dependent enzymes. Nucleic Acids Res. 2014; 42:56–69. 10.1093/nar/gkt747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Dimitriu T, Szczelkun MD, Westra ER Various plasmid strategies limit the effect of bacterial restriction–modification systems against conjugation. Nucleic Acids Res. 2024; 52:12976–86. 10.1093/nar/gkae896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Oliveira PH, Touchon M, Rocha EP Regulation of genetic flux between bacteria by restriction–modification systems. Proc Natl Acad Sci USA. 2016; 113:5658–63. 10.1073/pnas.1603257113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Shaw LP, Rocha EP, MacLean RC Restriction–modification systems have shaped the evolution and distribution of plasmids across bacteria. Nucleic Acids Res. 2023; 51:6806–18. 10.1093/nar/gkad452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Wang R, Lou J, Li J A mobile restriction modification system consisting of methylases on the IncA/C plasmid. Mobile DNA. 2019; 10:26. 10.1186/s13100-019-0168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Fomenkov A, Sun Z, Murray IA et al. Plasmid replication-associated single-strand-specific methyltransferases. Nucleic Acids Res. 2020; 48:12858–73. 10.1093/nar/gkaa1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Shaw LP, Celis AP, Arroyo MA. et al. The leading region of many conjugative plasmids is depleted in restriction–modification targets. bioRxiv03 April 2025, preprint: not peer reviewed 10.1101/2025.04.03.647016. [DOI]
- 241. Naito Y, Naito T, Kobayashi I Selfish restriction modification genes: resistance of a resident R/M plasmid to displacement by an incompatible plasmid mediated by host killing. Biol Chem. 1998; 379:429–36. 10.1515/bchm.1998.379.4-5.429. [DOI] [PubMed] [Google Scholar]
- 242. Cooper TF, Heinemann JA Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc Natl Acad Sci USA. 2000; 97:12643–8. 10.1073/pnas.220077897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Mongold JA Theoretical implications for the evolution of postsegregational killing by bacterial plasmids. Am Nat. 1992; 139:677–89. 10.1086/285352. [DOI] [Google Scholar]
- 244. Westra ER, Buckling A, Fineran PC CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Micro. 2014; 12:317–26. 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 245. Shmakov SA, Sitnik V, Makarova KS et al. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. mBio. 2017; 8:mBio.01397–01317. 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Westra ER, Staals RHJ, Gort G et al. CRISPR-Cas systems preferentially target the leading regions of MOBF conjugative plasmids. RNA Biol. 2013; 10:749–61. 10.4161/rna.24202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Marraffini LA, Sontheimer EJ CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008; 322:1843–5. 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Foster K, Kalter J, Woodside W et al. The ribonuclease activity of Csm6 is required for anti-plasmid immunity by Type III-A CRISPR-Cas systems. RNA Biol. 2019; 16:449–60. 10.1080/15476286.2018.1493334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Rostøl JT, Marraffini LA Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR–Cas immunity. Nat Microbiol. 2019; 4:656–62. 10.1038/s41564-018-0353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Chou-Zheng L, Hatoum-Aslan A Critical roles for ‘housekeeping’ nucleases in type III CRISPR-Cas immunity. eLife. 2022; 11:e81897. 10.7554/eLife.81897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Garneau JE, Dupuis ME, Villion M et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010; 468:67–71. 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 252. Gophna U, Kristensen DM, Wolf YI et al. No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales. ISME J. 2015; 9:2021–7. 10.1038/ismej.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Liu Y, Botelho J, Iranzo J Timescale and genetic linkage explain the variable impact of defense systems on horizontal gene transfer. Genome Res. 2025; 35:268–78. 10.1101/gr.279300.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Kogay R, Wolf YI, Koonin EV Defence systems and horizontal gene transfer in bacteria. Environ Microbiol. 2024; 26:e16630. 10.1111/1462-2920.16630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Palmer KL, Gilmore MS Multidrug-resistant enterococci lack CRISPR-cas. mBio. 2010; 1:e00227. 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Touchon M, Rocha EPC The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One. 2010; 5:e11126. 10.1371/journal.pone.0011126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Aydin S, Personne Y, Newire E et al. Presence of type I-F CRISPR/Cas systems is associated with antimicrobial susceptibility in Escherichia coli. J Antimicrob Chemother. 2017; 72:2213–8. 10.1093/jac/dkx137. [DOI] [PubMed] [Google Scholar]
- 258. Botelho J, Cazares A, Schulenburg H The ESKAPE mobilome contributes to the spread of antimicrobial resistance and CRISPR-mediated conflict between mobile genetic elements. Nucleic Acids Res. 2023; 51:236–52. 10.1093/nar/gkac1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Crowley VM, Catching A, Taylor HN et al. A type IV-A CRISPR-Cas system in Pseudomonas aeruginosa mediates RNA-guided plasmid interference In vivo. CRISPR J. 2019; 2:434–40. 10.1089/crispr.2019.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Faure G, Shmakov SA, Yan WX et al. CRISPR-Cas in mobile genetic elements: counter-defence and beyond. Nat Rev Micro. 2019; 17:513–25. 10.1038/s41579-019-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Newire E, Aydin A, Juma S et al. Identification of a type IV-A CRISPR-Cas system located exclusively on IncHI1B/IncFIB plasmids in Enterobacteriaceae. Front Microbiol. 2020; 11:fmicb.2020.01937. 10.3389/fmicb.2020.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Pinilla-Redondo R, Mayo-Muñoz D, Russel J et al. Type IV CRISPR–Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res. 2020; 48:2000–12. 10.1093/nar/gkz1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Bernheim A, Bikard D, Touchon M et al. Atypical organizations and epistatic interactions of CRISPRs and cas clusters in genomes and their mobile genetic elements. Nucleic Acids Res. 2020; 48:748–60. 10.1093/nar/gkz1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Benz F, Camara-Wilpert S, Russel J et al. Type IV-A3 CRISPR-Cas systems drive inter-plasmid conflicts by acquiring spacers in trans. Cell Host Microbe. 2024; 32:875–86. 10.1016/j.chom.2024.04.016. [DOI] [PubMed] [Google Scholar]
- 265. Sanchez-Londono M, Rust S, Hernández-Tamayo R et al. Visualization of type IV-A1 CRISPR-mediated repression of gene expression and plasmid replication. Nucleic Acids Res. 2024; 52:12592–603. 10.1093/nar/gkae879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Guo X, Sanchez-Londono M, Gomes-Filho JV et al. Characterization of the self-targeting type IV CRISPR interference system in Pseudomonas oleovorans. Nat Microbiol. 2022; 7:1870–8. 10.1038/s41564-022-01229-2. [DOI] [PubMed] [Google Scholar]
- 267. Cui N, Zhang J-T, Liu Y et al. Type IV-A CRISPR-csf complex: assembly, dsDNA targeting, and CasDinG recruitment. Mol Cell. 2023; 83:2493–508. 10.1016/j.molcel.2023.05.036. [DOI] [PubMed] [Google Scholar]
- 268. Koonin EV, Makarova KS CRISPR in mobile genetic elements: counter-defense, inter-element competition and RNA-guided transposition. BMC Biol. 2024; 22:295. 10.1186/s12915-024-02090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Wu WY, Mohanraju P, Liao C et al. The miniature CRISPR-Cas12m effector binds DNA to block transcription. Mol Cell. 2022; 82:4487–502. 10.1016/j.molcel.2022.11.003. [DOI] [PubMed] [Google Scholar]
- 270. Swarts DC, Jore MM, Westra ER et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014; 507:258–61. 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Weill F-X, Domman D, Njamkepo E et al. Genomic history of the seventh pandemic of cholera in Africa. Science. 2017; 358:785–9. 10.1126/science.aad5901. [DOI] [PubMed] [Google Scholar]
- 272. Amaro C, Aznar R, Garay E et al. R plasmids in environmental Vibrio cholerae non-O1 strains. Appl Environ Microb. 1988; 54:2771–6. 10.1128/aem.54.11.2771-2776.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Newland JW, Voll MJ, McNicol LA Serology and plasmid carriage in Vibrio cholerae. Can J Microbiol. 1984; 30:1149–56. 10.1139/m84-180. [DOI] [PubMed] [Google Scholar]
- 274. Jaskólska M, Adams DW, Blokesch M Two defence systems eliminate plasmids from seventh pandemic Vibrio cholerae. Nature. 2022; 604:323–9. 10.1038/s41586-022-04546-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Robins WP, Meader BT, Toska J et al. DdmABC-dependent death triggered by viral palindromic DNA sequences. Cell Rep. 2024; 43:114450. 10.1016/j.celrep.2024.114450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Panas MW, Jain P, Yang H et al. Noncanonical SMC protein in Mycobacterium smegmatis restricts maintenance of Mycobacterium fortuitum plasmids. Proc Natl Acad Sci USA. 2014; 111:13264–71. 10.1073/pnas.1414207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Doron S, Melamed S, Ofir G et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science. 2018; 359:eaar4120. 10.1126/science.aar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Deep A, Gu Y, Gao Y-Q et al. The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA. Mol Cell. 2022; 82:4145–59. 10.1016/j.molcel.2022.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Weiß M, Giacomelli G, Assaya MB et al. The MksG nuclease is the executing part of the bacterial plasmid defense system MksBEFG. Nucleic Acids Res. 2023; 51:3288–306. 10.1093/nar/gkad130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Pradhan B, Deep A, König J et al. Loop-extrusion-mediated plasmid DNA cleavage by the bacterial SMC Wadjet complex. Mol Cell. 2025; 85:107–16. 10.1016/j.molcel.2024.11.002. [DOI] [PubMed] [Google Scholar]
- 281. Roisné-Hamelin F, Liu HW, Taschner M et al. Structural basis for plasmid restriction by SMC JET nuclease. Mol Cell. 2024; 84:883–96. 10.1016/j.molcel.2024.01.009. [DOI] [PubMed] [Google Scholar]
- 282. Baca CF, Marraffini LA Nucleic acid recognition during prokaryotic immunity. Mol Cell. 2025; 85:309–22. 10.1016/j.molcel.2024.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Bravo JPK, Ramos DA, Fregoso Ocampo R et al. Plasmid targeting and destruction by the DdmDE bacterial defence system. Nature. 2024; 630:961–7. 10.1038/s41586-024-07515-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Loeff L, Adams DW, Chanez C et al. Molecular mechanism of plasmid elimination by the DdmDE defense system. Science. 2024; 385:188–94. 10.1126/science.adq0534. [DOI] [PubMed] [Google Scholar]
- 285. Yang X-Y, Shen Z, Wang C et al. DdmDE eliminates plasmid invasion by DNA-guided DNA targeting. Cell. 2024; 187:5253–66. 10.1016/j.cell.2024.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Zongo PD, Cabanel N, Royer G et al. An antiplasmid system drives antibiotic resistance gene integration in carbapenemase-producing Escherichia coli lineages. Nat Commun. 2024; 15:4093. 10.1038/s41467-024-48219-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. DeWeirdt PC, Mahoney EM, Laub MT DefensePredictor: a machine learning model to discover novel prokaryotic immune systems. bioRxiv8 January 2025, preprint: not peer reviewed 10.1101/2025.01.08.631726. [DOI]
- 288. Mordret E, Hervé A, Vaysset H. et al. Protein and genomic language models chart a vast landscape of antiphage defenses. bioRxiv8 January 2025, preprint: not peer reviewed 10.1101/2025.01.08.631966. [DOI]
- 289. Russell AB, Peterson SB, Mougous JD Type VI secretion system effectors: poisons with a purpose. Nat Rev Micro. 2014; 12:137–48. 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Ho BT, Basler M, Mekalanos JJ Type 6 secretion system–mediated immunity to Type 4 secretion system–mediated gene transfer. Science. 2013; 342:250–3. 10.1126/science.1243745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Di Venanzio G, Moon KH, Weber BS et al. Multidrug-resistant plasmids repress chromosomally encoded T6SS to enable their dissemination. Proc Natl Acad Sci USA. 2019; 116:1378–83. 10.1073/pnas.1812557116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Getino M, de la Cruz F Natural and artificial strategies to control the conjugative transmission of plasmids. Microbiol Spectr. 2018; 6:10.1128/microbiolspec.mtbp-0015-2016 10.1128/microbiolspec.MTBP-0015-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Cascales E, Atmakuri K, Liu Z et al. Agrobacterium tumefaciens oncogenic suppressors inhibit T-DNA and VirE2 protein substrate binding to the VirD4 coupling protein. Mol Microbiol. 2005; 58:565–79. 10.1111/j.1365-2958.2005.04852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Getino M, Palencia-Gándara C, Garcillán-Barcia MP et al. PifC and Osa, plasmid weapons against rival conjugative coupling proteins. Front Microbiol. 2017; 8:fmicb.2017.02260. 10.3389/fmicb.2017.02260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Maindola P, Raina R, Goyal P et al. Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids. Nat Commun. 2014; 5:5322. 10.1038/ncomms6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Beamud B, Benz F, Bikard D Going viral: the role of mobile genetic elements in bacterial immunity. Cell Host Microbe. 2024; 32:804–19. 10.1016/j.chom.2024.05.017. [DOI] [PubMed] [Google Scholar]
- 297. Niault T, van Houte S, Westra E et al. Evolution and ecology of anti-defence systems in phages and plasmids. Curr Biol. 2025; 35:R32–44. 10.1016/j.cub.2024.11.033. [DOI] [PubMed] [Google Scholar]
- 298. Yan Y, Zheng J, Zhang X et al. dbAPIS: a database of anti-prokaryotic immune system genes. Nucleic Acids Res. 2024; 52:D419–25. 10.1093/nar/gkad932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Tesson F, Huiting E, Wei L et al. Exploring the diversity of anti-defense systems across prokaryotes, phages and mobile genetic elements. Nucleic Acids Res. 2025; 53:gkae1171. 10.1093/nar/gkae1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Wilkins BM Plasmid promiscuity: meeting the challenge of DNA immigration control. Environ Microbiol. 2002; 4:495–500. 10.1046/j.1462-2920.2002.00332.x. [DOI] [PubMed] [Google Scholar]
- 301. González-Montes L, del Campo I, Garcillán-Barcia MP et al. ArdC, a ssDNA-binding protein with a metalloprotease domain, overpasses the recipient hsdRMS restriction system broadening conjugation host range. PLoS Genet. 2020; 16:e1008750. 10.1371/journal.pgen.1008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. McMahon SA, Roberts GA, Johnson KA et al. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res. 2009; 37:4887–97. 10.1093/nar/gkp478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Skutel M, Yanovskaya D, Demkina A et al. RecA-dependent or independent recombination of plasmid DNA generates a conflict with the host EcoKI immunity by launching restriction alleviation. Nucleic Acids Res. 2024; 52:5195–208. 10.1093/nar/gkae243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Iida S, Streiff MB, Bickle TA et al. Two DNA antirestriction systems of bacteriophage P1, darA, and darB: characterization of darA-phages. Virology. 1987; 157:156–66. 10.1016/0042-6822(87)90324-2. [DOI] [PubMed] [Google Scholar]
- 305. Bondy-Denomy J, Pawluk A, Maxwell KL et al. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013; 493:429–32. 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Mahendra C, Christie KA, Osuna BA et al. Broad-spectrum anti-CRISPR proteins facilitate horizontal gene transfer. Nat Microbiol. 2020; 5:620–9. 10.1038/s41564-020-0692-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Pinilla-Redondo R, Shehreen S, Marino ND et al. Discovery of multiple anti-CRISPRs highlights anti-defense gene clustering in mobile genetic elements. Nat Commun. 2020; 11:5652. 10.1038/s41467-020-19415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Davidson AR, Lu W-T, Stanley SY et al. Anti-CRISPRs: protein inhibitors of CRISPR-Cas systems. Annu Rev Biochem. 2020; 89:309–32. 10.1146/annurev-biochem-011420-111224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Roy D, Huguet KT, Grenier F et al. IncC conjugative plasmids and SXT/R391 elements repair double-strand breaks caused by CRISPR-Cas during conjugation. Nucleic Acids Res. 2020; 48:8815–27. 10.1093/nar/gkaa518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Masai H, Arai K Frpo: a novel single-stranded DNA promoter for transcription and for primer RNA synthesis of DNA replication. Cell. 1997; 89:897–907. 10.1016/S0092-8674(00)80275-5. [DOI] [PubMed] [Google Scholar]
- 311. Althorpe NJ, Chilley PM, Thomas AT et al. Transient transcriptional activation of the IncI1 plasmid anti-restriction gene (ardA) and SOS inhibition gene (psiB) early in conjugating recipient bacteria. Mol Microbiol. 1999; 31:133–42. 10.1046/j.1365-2958.1999.01153.x. [DOI] [PubMed] [Google Scholar]
- 312. Samuel B, Mittelman K, Croitoru SY et al. Diverse anti-defence systems are encoded in the leading region of plasmids. Nature. 2024; 635:186–92. 10.1038/s41586-024-07994-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Al Mamun AAM, Kishida K, Christie PJ Protein transfer through an F plasmid-encoded type IV secretion system suppresses the mating-induced SOS response. mBio. 2021; 12:10.1128/mbio.01629-21. 10.1128/mbio.01629-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Cury J, Oliveira PH, de la Cruz F et al. Host range and genetic plasticity explain the coexistence of integrative and extrachromosomal mobile genetic elements. Mol Biol Evol. 2018; 35:2230–9. 10.1093/molbev/msy123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Rodriguez-Beltran J, Hernandez-Beltran JCR, DelaFuente J et al. Multicopy plasmids allow bacteria to escape from fitness trade-offs during evolutionary innovation. Nat Ecol Evol. 2018; 2:873–81. 10.1038/s41559-018-0529-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Sastre-Dominguez J, DelaFuente J, Toribio-Celestino L et al. Plasmid-encoded insertion sequences promote rapid adaptation in clinical Enterobacteria. Nat Ecol Evol. 2024; 8:2097–112. 10.1038/s41559-024-02523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Che Y, Yang Y, Xu X et al. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc Natl Acad Sci USA. 2021; 118:e2008731118. 10.1073/pnas.2008731118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Wang X, Zhang H, Yu S et al. Inter-plasmid transfer of antibiotic resistance genes accelerates antibiotic resistance in bacterial pathogens. ISME J. 2024; 18:wrad032. 10.1093/ismejo/wrad032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Yao Y, Maddamsetti R, Weiss A et al. Intra- and interpopulation transposition of mobile genetic elements driven by antibiotic selection. Nat Ecol Evol. 2022; 6:555–64. 10.1038/s41559-022-01705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Nicoloff H, Hjort K, Andersson DI et al. Three concurrent mechanisms generate gene copy number variation and transient antibiotic heteroresistance. Nat Commun. 2024; 15:3981. 10.1038/s41467-024-48233-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. DelaFuente J, Toribio-Celestino L, Santos-Lopez A et al. Within-patient evolution of plasmid-mediated antimicrobial resistance. Nat Ecol Evol. 2022; 6:1980–91. 10.1038/s41559-022-01908-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Pesesky MW, Tilley R, Beck DAC Mosaic plasmids are abundant and unevenly distributed across prokaryotic taxa. Plasmid. 2019; 102:10–8. 10.1016/j.plasmid.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 323. Plutarch’s Lives. 1914; Boston, MA: Harvard University Press; 608. [Google Scholar]
- 324. Lim JY, Yoon JW, Hovde CJ A brief overview of Escherichia coli O157:H7 and its plasmid O157. J Microbiol Biotechnol. 2010; 20:5–14. 10.4014/jmb.0908.08007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Harrison PW, Lower RP, Kim NK et al. Introducing the bacterial ‘chromid’: not a chromosome, not a plasmid. Trends Microbiol. 2010; 18:141–8. 10.1016/j.tim.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 326. diCenzo GC, Finan TM The divided bacterial genome: structure, function, and evolution. Microbiol Mol Biol Rev. 2017; 81:10/1128/mmbr.00019-17. 10.1128/mmbr.00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. MacLean RC, Lood C, Wheatley RM Chromosomal capture of beneficial genes drives plasmids towards ecological redundancy. ISME j. 2025; 19:wraf091. 10.1093/ismejo/wraf091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Hanke DM, Wang Y, Dagan T Pseudogenes in plasmid genomes reveal past transitions in plasmid mobility. Nucleic Acids Res. 2024; 52:7049–62. 10.1093/nar/gkae430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Zhang X, Deatherage DE, Zheng H et al. Evolution of satellite plasmids can prolong the maintenance of newly acquired accessory genes in bacteria. Nat Commun. 2019; 10:5809. 10.1038/s41467-019-13709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Scott JR A defective P1 prophage with a chromosomal location. Virology. 1970; 40:144–51. 10.1016/0042-6822(70)90386-7. [DOI] [PubMed] [Google Scholar]
- 331. Billard-Pomares T, Fouteau S, Jacquet ME et al. Characterization of a P1-like bacteriophage encoding an SHV-2 extended-spectrum beta-lactamase from an Escherichia coli strain. Antimicrob Agents Chemother. 2014; 58:6550–7. 10.1128/AAC.03183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Kidgell C, Pickard D, Wain J et al. Characterisation and distribution of a cryptic Salmonella typhi plasmid pHCM2. Plasmid. 2002; 47:159–71. 10.1016/S0147-619X(02)00013-6. [DOI] [PubMed] [Google Scholar]
- 333. Octavia S, Sara J, Lan R Characterization of a large novel phage-like plasmid in Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett. 2015; 362:fnv044. 10.1093/femsle/fnv044. [DOI] [PubMed] [Google Scholar]
- 334. Casjens S, Palmer N, van Vugt R et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000; 35:490–516. 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 335. Lederberg J, Cavalli LL, Lederberg EM Sex compatibility in Escherichia coli. Genetics. 1952; 37:720–30. 10.1093/genetics/37.6.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Castañeda-Barba S, Top EM, Stalder T Plasmids, a molecular cornerstone of antimicrobial resistance in the one health era. Nat Rev Micro. 2024; 22:18–32. 10.1038/s41579-023-00926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Hughes VM, Datta N Conjugative plasmids in bacteria of the ‘pre-antibiotic’ era. Nature. 1983; 302:725–6. 10.1038/302725a0. [DOI] [PubMed] [Google Scholar]
- 338. Wang Y, Dagan T The evolution of antibiotic resistance islands occurs within the framework of plasmid lineages. Nat Commun. 2024; 15:4555. 10.1038/s41467-024-48352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Forster SC, Liu J, Kumar N et al. Strain-level characterization of broad host range mobile genetic elements transferring antibiotic resistance from the human microbiome. Nat Commun. 2022; 13:1445. 10.1038/s41467-022-29096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Brophy JAN, Triassi AJ, Adams BL et al. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat Microbiol. 2018; 3:1043–53. 10.1038/s41564-018-0216-5. [DOI] [PubMed] [Google Scholar]
- 341. Ronda C, Chen SP, Cabral V et al. Metagenomic engineering of the mammalian gut microbiome in situ. Nat Methods. 2019; 16:167–70. 10.1038/s41592-018-0301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342. Adams BL The next generation of synthetic biology chassis: moving synthetic biology from the laboratory to the field. ACS Synth Biol. 2016; 5:1328–30. 10.1021/acssynbio.6b00256. [DOI] [PubMed] [Google Scholar]
- 343. Mell JC, Redfield RJ Natural competence and the evolution of DNA uptake specificity. J Bacteriol. 2014; 196:1471–83. 10.1128/JB.01293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Kittleson JT, DeLoache W, Cheng H-Y et al. Scalable plasmid transfer using engineered P1-based phagemids. ACS Synth Biol. 2012; 1:583–9. 10.1021/sb300054p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Encinas D, Garcillán-Barcia MP, Santos-Merino M et al. Plasmid conjugation from proteobacteria as evidence for the origin of xenologous genes in cyanobacteria. J Bacteriol. 2014; 196:1551–9. 10.1128/JB.01464-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. García-Gutiérrez C, Aparicio T, Torres-Sánchez L et al. Multifunctional SEVA shuttle vectors for actinomycetes and gram-negative bacteria. MicrobiologyOpen. 2020; 9:1135–49. 10.1002/mbo3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Samperio S, Guzmán-Herrador DL, May-Cuz R et al. Conjugative DNA transfer from E. coli to transformation-resistant lactobacilli. Front Microbiol. 2021; 12:fmicb.2021.606629. 10.3389/fmicb.2021.606629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Neil K, Allard N, Rodrigue S Molecular mechanisms influencing bacterial conjugation in the intestinal microbiota. Front Microbiol. 2021; 12:fmicb.2021.673260. 10.3389/fmicb.2021.673260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349. Brödel AK, Charpenay LH, Galtier M et al. In situ targeted base editing of bacteria in the mouse gut. Nature. 2024; 632:877–84. 10.1038/s41586-024-07681-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350. Robledo M, Álvarez B, Cuevas A et al. Targeted bacterial conjugation mediated by synthetic cell-to-cell adhesions. Nucleic Acids Res. 2022; 50:12938–50. 10.1093/nar/gkac1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Li YG, Kishida K, Ogawa-Kishida N et al. Ligand-displaying Escherichia coli cells and minicells for programmable delivery of toxic payloads via type IV secretion systems. mBio. 2023; 14:e02143–23. 10.1128/mbio.02143-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352. Alves J, Rand JD, Johnston ABE et al. Methylome-dependent transformation of emm1 group A streptococci. mBio. 2023; 14:e00798–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353. Vento JM, Durmusoglu D, Li T et al. A cell-free transcription-translation pipeline for recreating methylation patterns boosts DNA transformation in bacteria. Mol Cell. 2024; 84:2785–96. 10.1016/j.molcel.2024.06.003. [DOI] [PubMed] [Google Scholar]
- 354. Dorado-Morales P, Lambérioux M, Mazel D Unlocking the potential of microbiome editing: a review of conjugation-based delivery. Mol Microbiol. 2024; 122:273–83. 10.1111/mmi.15147. [DOI] [PubMed] [Google Scholar]
- 355. Benz F, Beamud B, Laurenceau R et al. CRISPR–Cas therapies targeting bacteria. Nat Rev Bioeng. 2025; 3:s44222-025-00311-8. 10.1038/s44222-025-00311-8. [DOI] [Google Scholar]
- 356. Citorik RJ, Mimee M, Lu TK Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol. 2014; 32:1141–5. 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357. Bikard D, Euler CW, Jiang W et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014; 32:1146–50. 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. Tao S, Chen H, Li N et al. Elimination of blaKPC−2-mediated carbapenem resistance in Escherichia coli by CRISPR-Cas9 system. BMC Microbiol. 2023; 23:310. 10.1186/s12866-023-03058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. Wu Z-Y, Huang Y-T, Chao W-C et al. Reversal of carbapenem-resistance in Shewanella algae by CRISPR/Cas9 genome editing. J Adv Res. 2019; 18:61–9. 10.1016/j.jare.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Okesanya OJ, Ahmed MM, Ogaya JB et al. Reinvigorating AMR resilience: leveraging CRISPR–Cas technology potentials to combat the 2024 WHO bacterial priority pathogens for enhanced global health security—A systematic review. Trop Med Health. 2025; 53:43. 10.1186/s41182-025-00728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. deSouza HCA, Panzenhagen P, Santos D et al. Unravelling the advances of CRISPR-Cas9 as a precise antimicrobial therapy: a systematic review. J Glob Antimicrob Resist. 2025; 42:51–60. 10.1016/j.jgar.2025.02.002. [DOI] [PubMed] [Google Scholar]
- 362. Neil K, Allard N, Roy P et al. High-efficiency delivery of CRISPR-Cas9 by engineered probiotics enables precise microbiome editing. Mol Syst Biol. 2021; 17:e10335. 10.15252/msb.202110335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Skovmand AB, Paaske NJ, Peñil-Celis A. et al. Exploring clinical class 1 integrons as valuable targets for the re-sensitization of multidrug resistant pathogenic bacteria using CRISPR-Cas. bioRxiv28 March 2025, preprint: not peer reviewed 10.1101/2025.03.28.645910. [DOI]
- 364. Zhou Y, Yang Y, Li X et al. Exploiting a conjugative endogenous CRISPR-Cas3 system to tackle multidrug-resistant Klebsiella pneumoniae. Ebiomedicine. 2023; 88:104445. 10.1016/j.ebiom.2023.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Lopez-Igual R, Bernal-Bayard J, Rodriguez-Paton A et al. Engineered toxin-intein antimicrobials can selectively target and kill antibiotic-resistant bacteria in mixed populations. Nat Biotechnol. 2019; 37:755–60. 10.1038/s41587-019-0105-3. [DOI] [PubMed] [Google Scholar]
- 366. Jalasvuori M, Friman V-P, Nieminen A et al. Bacteriophage selection against a plasmid-encoded sex apparatus leads to the loss of antibiotic-resistance plasmids. Biol Lett. 2011; 7:902–5. 10.1098/rsbl.2011.0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Parra B, Cockx B, Lutz VT et al. Isolation and characterization of novel plasmid-dependent phages infecting bacteria carrying diverse conjugative plasmids. Microbiol Spectr. 2023; 12:e02537–23. 10.1128/spectrum.02537-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Li J, García P, Ji X. et al. Male-specific bacteriophages and their potential on combating the spreading of T4SS-bearing antimicrobial resistance plasmids. Crit Rev Microbiol. 2024; 10:1–12. 10.1080/1040841X.2024.2400150. [DOI] [PubMed] [Google Scholar]
- 369. Draper O, César CE, Machón C et al. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc Natl Acad Sci USA. 2005; 102:16385–90. 10.1073/pnas.0506081102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Garcillán-Barcia MP, Jurado P, González-Pérez B et al. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol Microbiol. 2007; 63:404–16. 10.1111/j.1365-2958.2006.05523.x. [DOI] [PubMed] [Google Scholar]
- 371. Guzmán-Herrador DL, Fernández-Gómez A, Depardieu F et al. Delivery of functional Cas:DNA nucleoprotein complexes into recipient bacteria through a type IV secretion system. Proc Natl Acad Sci USA. 2024; 121:e2408509121. 10.1073/pnas.2408509121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Lin A, Jimenez J, Derr J et al. Inhibition of bacterial conjugation by phage M13 and its protein g3p: quantitative analysis and model. PLoS One. 2011; 6:e19991. 10.1371/journal.pone.0019991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Oyedemi BOM, Shinde V, Shinde K et al. Novel R-plasmid conjugal transfer inhibitory and antibacterial activities of phenolic compounds from Mallotus philippensis (Lam.) Mull. Arg. J Glob Antimicrob Resist. 2016; 5:15–21. 10.1016/j.jgar.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 374. Palencia-Gándara C, Getino M, Moyano G et al. Conjugation inhibitors effectively prevent plasmid transmission in natural environments. mBio. 2021; 12:10.1128/mbio.01277-21. 10.1128/mbio.01277-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375. Shaffer CL, Good JAD, Kumar S et al. Peptidomimetic small molecules disrupt type IV secretion system activity in diverse bacterial pathogens. mBio. 2016; 7:10.1128/mbio.00221-16. 10.1128/mbio.00221-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Cabezón E, de la Cruz F, Arechaga I Conjugation inhibitors and their potential use to prevent dissemination of antibiotic resistance genes in bacteria. Front Microbiol. 2017; 8:fmicb.2017.02329. 10.3389/fmicb.2017.02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377. Prangishvili D, Albers SV, Holz I et al. Conjugation in archaea: frequent occurrence of conjugative plasmids in Sulfolobus. Plasmid. 1998; 40:190–202. 10.1006/plas.1998.1363. [DOI] [PubMed] [Google Scholar]
- 378. Stedman KM, She Q, Phan H et al. pING family of conjugative plasmids from the extremely thermophilic archaeon Sulfolobus islandicus: insights into recombination and conjugation in Crenarchaeota. J Bacteriol. 2000; 182:7014–20. 10.1128/JB.182.24.7014-7020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379. Beltran LC, Cvirkaite-Krupovic V, Miller J et al. Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery. Nat Commun. 2023; 14:666. 10.1038/s41467-023-36349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380. Catchpole RJ, Barbe V, Magdelenat G et al. A self-transmissible plasmid from a hyperthermophile that facilitates genetic modification of diverse Archaea. Nat Microbiol. 2023; 8:1339–47. 10.1038/s41564-023-01387-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381. Blesa A, Berenguer J.. Villa TG, Viñas M Alternative ways to exchange DNA: unconventional conjugation among bacteria. Horizontal Gene Transfer: Breaking Borders between Living Kingdoms. 2019; Cham: Springer International Publishing; 77–96. 10.1007/978-3-030-21862-1. [DOI] [Google Scholar]
- 382. Ummels R, Abdallah AM, Kuiper V et al. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio. 2014; 5:10.1128/mbio.01744-14. 10.1128/mbio.01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383. Thoma L, Muth G Conjugative DNA transfer in streptomyces by TraB: is one protein enough?. FEMS Microbiol Lett. 2012; 337:81–8. 10.1111/1574-6968.12031. [DOI] [PubMed] [Google Scholar]
- 384. Reuther J, Gekeler C, Tiffert Y et al. Unique conjugation mechanism in mycelial streptomycetes: a DNA-binding ATPase translocates unprocessed plasmid DNA at the hyphal tip. Mol Microbiol. 2006; 61:436–46. 10.1111/j.1365-2958.2006.05258.x. [DOI] [PubMed] [Google Scholar]
- 385. Kumar HD, Ueda K Conjugation in the cyanobacterium Anacystis nidulans. Mol Gen Genet. 1984; 195:356–7. 10.1007/BF00332771. [DOI] [Google Scholar]
- 386. Muro-Pastor AM, Kuritz T, Flores E et al. Transfer of a genetic marker from a megaplasmid of Anabaena sp. strain PCC 7120 to a megaplasmid of a different Anabaena strain. J Bacteriol. 1994; 176:1093–8. 10.1128/jb.176.4.1093-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Hug LA, Baker BJ, Anantharaman K et al. A new view of the tree of life. Nat Microbiol. 2016; 1:16048. 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 388. Quiñonero-Coronel M, del M, Cabello-Yeves PJ. et al. The Type IV Secretion System of patescibacteria is homologous to the bacterial monoderm conjugation machinery. Microbial Genomics. 2025; 11:001409. 10.1099/mgen.0.001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Erdmann S, Tschitschko B, Zhong L et al. A plasmid from an Antarctic haloarchaeon uses specialized membrane vesicles to disseminate and infect plasmid-free cells. Nat Microbiol. 2017; 2:1446–55. 10.1038/s41564-017-0009-2. [DOI] [PubMed] [Google Scholar]
- 390. Dougherty PE, Bernard C, Carstens AB. et al. Persistent virulent phages exist in bacterial isolates. bioRxiv1 January 2025, preprint: not peer reviewed 10.1101/2024.12.31.630880. [DOI]
- 391. Hay ID, Lithgow T Filamentous phages: masters of a microbial sharing economy. EMBO Rep. 2019; 20:e47427. 10.15252/embr.201847427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392. Lee CA, Babic A, Grossman AD Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol. 2010; 75:268–79. 10.1111/j.1365-2958.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393. Brito IL Examining horizontal gene transfer in microbial communities. Nat Rev Micro. 2021; 19:442–53. 10.1038/s41579-021-00534-7. [DOI] [PubMed] [Google Scholar]
- 394. Kent AG, Vill AC, Shi Q et al. Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial hi-C. Nat Commun. 2020; 11:4379. 10.1038/s41467-020-18164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395. Stalder T, Press MO, Sullivan S et al. Linking the resistome and plasmidome to the microbiome. ISME J. 2019; 13:2437–46. 10.1038/s41396-019-0446-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396. Fogarty EC, Schechter MS, Lolans K et al. A cryptic plasmid is among the most numerous genetic elements in the human gut. Cell. 2024; 187:1206–22. 10.1016/j.cell.2024.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397. Neil K, Allard N, Grenier F et al. Highly efficient gene transfer in the mouse gut microbiota is enabled by the Incl2 conjugative plasmid TP114. Commun Biol. 2020; 3:523. 10.1038/s42003-020-01253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398. León-Sampedro R, DelaFuente J, Díaz-Agero C et al. Pervasive transmission of a carbapenem resistance plasmid in the gut microbiota of hospitalized patients. Nat Microbiol. 2021; 6:606–16. 10.1038/s41564-021-00879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399. Grayson F, Loman L, Nonnenmacher T et al. Plasmid conjugation drives within–patient plasmid diversity. Microb Genomics. 2025; 11:001361. 10.1099/mgen.0.001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this research.