Abstract
Recently, the FtsZ protein, which is known as a key component in bacterial cell division, was reported to be involved in mitochondrial division in algae. In yeast and animals, however, mitochondrial fission depends on the dynamin-like proteins Dnm1p and Drp1, respectively, whereas in green plants, no potential mitochondrial division genes have been identified. BLAST searches of the nuclear and mitochondrial genome sequences of Arabidopsis thaliana did not find any obvious homologue of the α-proteobacterial-type ftsZ genes. To determine whether mitochondrial division of higher plants depends on a dynamin-like protein, we cloned a cDNA for ADL2b, an Arabidopsis homologue of Dnm1p, and tested its subcellular localization and its dominant-negative effect on mitochondrial division. The fusion protein of green fluorescent protein and ADL2b was observed as punctate structures localized at the tips and at the constriction sites of mitochondria in live plant cells. Cells expressing dominant-negative mutant ADL2b proteins (K56A and T77F) showed a significant fusion, aggregation, and/or tubulation of mitochondria. We propose that mitochondrial division in higher plants is conducted by dynamin-like proteins similar to ADL2b in Arabidopsis. The evolutional points of loss of mitochondrial FtsZ and the functional acquisition of dynamin-like proteins in mitochondrial division are discussed.
Mitochondria and plastids are remnants of free-living prokaryotes that lost their autonomy during evolution by establishing an endosymbiosis with their host cells (reviewed in ref. 1). These two organelles divide to maintain themselves during cell proliferation (reviewed in ref. 2). Almost all prokaryotes divide by using the multimeric GTPase protein FtsZ. These organisms include the cyanobacteria and α-proteobacteria, which are the modern-day organisms that are most closely related to the progenitors of chloroplasts and mitochondria, respectively. In vivo, FtsZ assembles into a ring at the cell midpoint on the inner surface of the cytoplasmic membrane that constricts during bacterial cytokinesis (3).
In the case of plastid division, the cyanobacterial-type FtsZ and Min system genes, which regulate the correct position of the FtsZ ring, have been found in higher plants. Inhibition of the expression of chloroplast FtsZ genes affects plastid division, resulting in one or a few giant chloroplasts per cell (reviewed in ref. 4). Recently, FtsZ rings at the chloroplast midpoint have been revealed (5). These findings indicate that chloroplasts continue to have at least part of the original bacterial division apparatus.
Nuclear-encoded α-proteobacterial-type FtsZ genes were recently identified in the unicellular chromophyte alga Mallomonas splendens and the rhodophyte alga Cyanidioschizon merolae (6, 7). Moreover, immunofluorescence microscopy showed that the Mallomonas FtsZ localized at the constriction sites of dividing mitochondria (6). Therefore, at least in part, the FtsZ-based bacterial-type cell division apparatus seems to have been inherited by algal mitochondria, as was found to be the case with plastids.
On the other hand, no potential mitochondrial FtsZ has been identified in the complete genomes of the nematode (Caenorhabditis elegans) or yeast (Saccharomyces cerevisiae). In these organisms, mitochondrial fission has been found to be mediated by the oligomeric GTPase dynamin-like proteins, Drp1 and Dnm1p, respectively, which are localized on the mitochondrial outer membrane in punctate structures associated with sites of mitochondrial constriction and fission (8–11). It is well known that classic dynamins are required for endocytosis. Dynamin assembles into a multimeric spiral at the neck of clathrin-coated pits where it may pinch off vesicles from the plasma membrane (12).
In higher plants, to date, no mitochondrial fission components have been described. Because of the existence of an α-proteobacterial-type mitochondrial FtsZ (MtFtsZ) gene in red algae, which share a common ancestry with green algae and green plants (13, 14), it is possible that higher plants have mitochondrial FtsZ genes in their genomes. But no α-proteobacterial-type FtsZ has been identified in the nuclear or mitochondrial genome of Arabidopsis thaliana (15, 16).
In this study, we searched for and analyzed another candidate, Arabidopsis dynamin-like protein 2b (ADL2b), the protein that is most closely related to yeast Dnm1p. We also discuss the evolution of the mitochondrial division apparatus.
Materials and Methods
Plant Materials.
Bright yellow-2 (BY-2) tobacco (Nicotiana tabacum) cell suspension cultures were grown in modified Murashige and Skoog medium enriched with 0.2 mg/liter 2-4-D and were maintained as described by Nagata et al. (17). A. thaliana plants were grown in a growth chamber at 22°C with a short day photoperiod (10-h/14-h light/dark cycle).
Sequence Analysis and Comparisons.
The complete coding sequence of ADL2b was determined from an expressed sequence tag (EST) clone (GenBank accession no. AV529344) obtained from Kazusa DNA Research Institute (Chiba, Japan). Sequencing was performed with an automated fluorescent laser sequencer ABI PRISM 3100 and a Big-Dye Terminator Cycle Sequencing kit (Perkin–Elmer Applied Biosystems, Foster City, CA). The sequencing primers were based on the A. thaliana genomic sequence. Sequence comparisons and phylogenetic tree analyses were performed by using the clustal w multiple alignment program (18) and the N-J method (19). To estimate the reliability of the branches of the tree, the bootstrap procedure in clustal w was used (1,000 replications). GenBank accession numbers for the proteins used in these analyses are given in the legend of Fig. 1.
Figure 1.
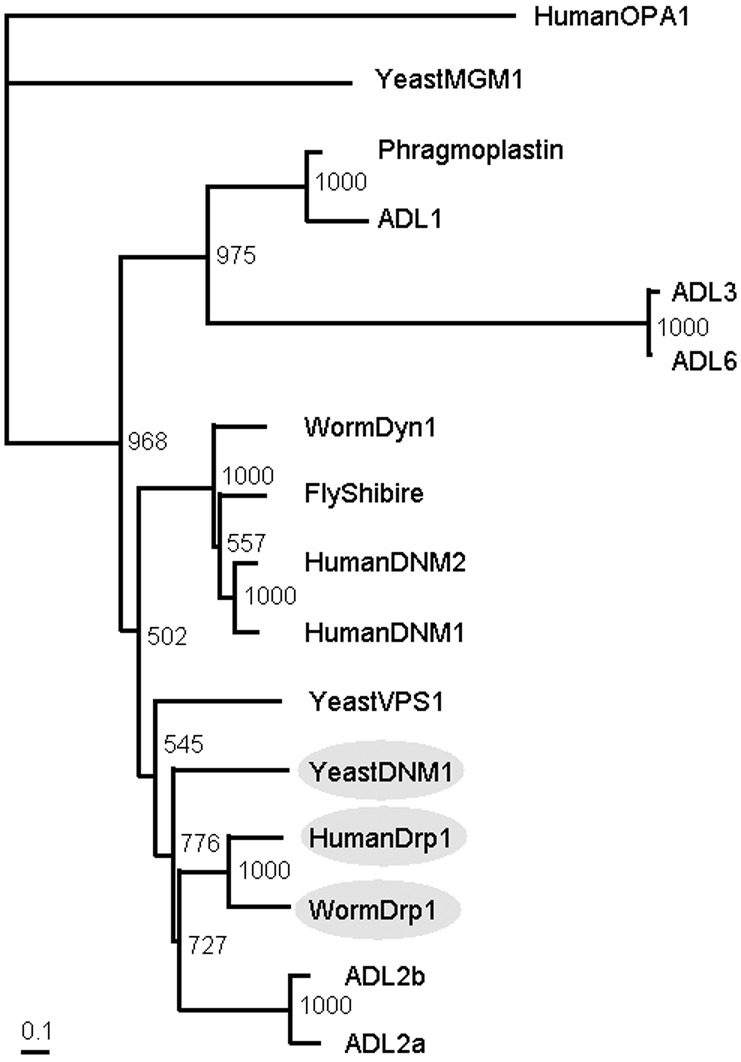
ADL2b is likely a member of the mitochondrial dividing dynamin-like protein family. Phylogenetic tree of dynamin and its related GTPase superfamily is shown. ADL2b is subgrouped with the yeast Dnm1p, nematode Drp1, and human Drp1 (gray ovals), all of which are mitochondria-dividing dynamin-like proteins. GenBank accession nos. for the proteins used in this analysis are as follows: WormDrp1, AF166274; HumanDrp1, AF000430; YeastDNM1, L40588; ADL1, L36939; Phragmoplastin, U25547; YeastMGM1, X62834; HumanOPA1, AB011139; ADL3, AB026987; ADL6, AF180732; WormDyn1, AF167982; FlyShibire, X59435; HumanDNM2, NM004945; Human DNM1, NM004408; and YeastVPS1, M33315. This phylogenetic tree was calculated by the N-J method (19). The bootstrap values based on 1,000 replications are shown.
Construction of Fusion Genes and Site-Directed Mutagenesis.
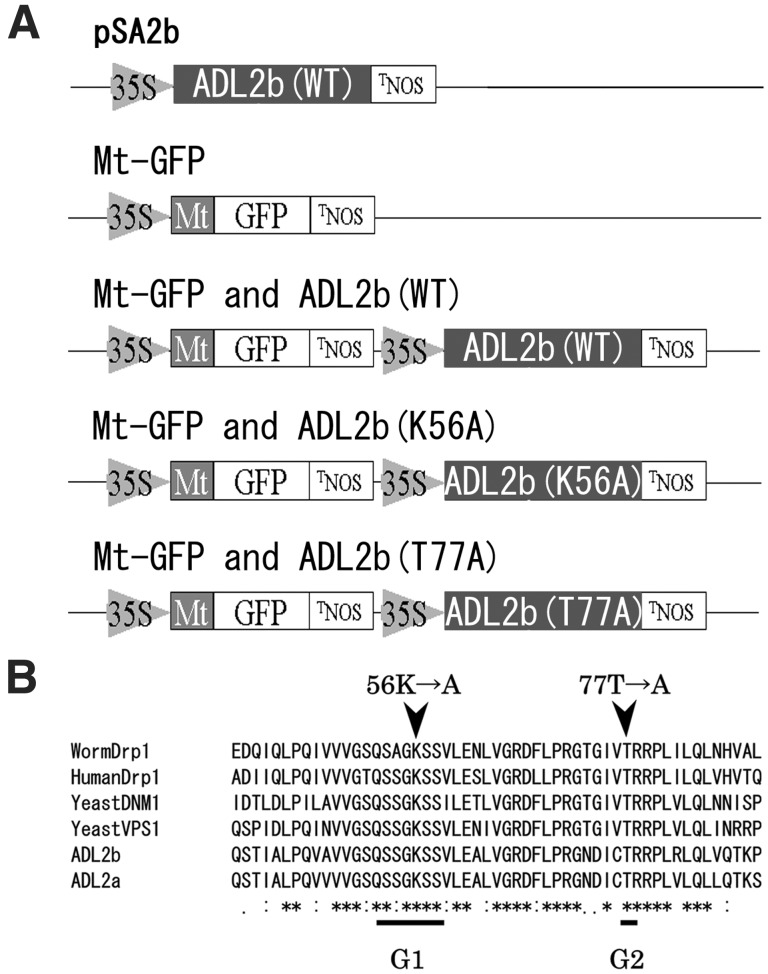
Before the construction of all plasmids containing ADL2b, we constructed the transient ADL2b expression vector pSA2b. The DNA fragment containing the ADL2b ORF was cut from the ADL2b EST plasmid (AV529344) and subcloned into blunt-ended NcoI and NotI sites of the S65TGFP expression vector pTH2, resulting in a gene encoding ADL2b in place of green fluorescent (GFP) that was driven by the cauliflower mosaic virus (CaMV) 35S promoter (pSA2b in Fig. 3).
Figure 3.
Schematic drawings of the plasmids and G1 and G2 motifs of dynamin-like proteins. (A) Schematic drawings of pSA2b (the top construct, see Materials and Methods) and the plasmids used to transform plant cells in Fig. 4. The ORFs of GFP (S65T) cDNA (GFP), its in-frame fusions with mitochondrial targeting presequence of ATPase delta subunit cDNA (Mt), and ADL2b wild type (WT), ADL2b K56A mutant (K56A), or ADL2b T77A mutant (T77A) cDNA were transiently coexpressed under the control of the CaMV 35S promoter (35S) and the terminator of nopaline synthase (TNOS). (B) G1 and G2 motifs, the targeted sites of the in vitro mutagenesis, in the GTPase domains of representative dynamin family members. Alignment of a part of short GTPase core segment from Arabidopsis ADL2a and ADL2b, yeast (S. cerevisiae) DNM1, human Drp1, nematode (C. elegans) Drp1, and yeast (S. cerevisiae) Vps1. Underlined segments show G1 and G2 motifs in the GTPase domains of dynamin family members (22). The site-directed mutated amino acids are shown at the top. Alignment was performed by using the CLUSTAL W multiple alignment program (18). See legend of Fig. 1 for accession nos.
A fusion protein in which GFP was fused to the NH2 terminus of ADL2b was constructed by using a 5′ SalI site just after the initiator ATG codon in pSA2b. The S65TGFP ORF was cut out and ligated into the SalI site of pSA2b, placing the GFP tag at the N terminus of the ADL2b.
Plasmid pWS (kindly provided by W. Sakamoto, Okayama Univ.) contains a mitochondrial-targeting form of GFP (Mt-GFP) that is located downstream of the CaMV 35S promoter (Fig. 3). (Mitochondrial targeting was accomplished by adding a targeting signal for the ATPase δ-subunit). Mt-GFP was cut out of pWS by digestion with EcoRI and HindIII. The fragment containing CaMV35Spro∷MtGFP∷NOSpolyA was purified, blunt-ended, and cloned into the blunt-ended SphI site of pSA2b (Fig. 3). The direction of the DNA fragment containing the MtGFP gene was checked by PCR and sequencing. Then, the point mutants K56A and T77A were generated with a QuickChange XL-Site Directed Mutagenesis kit (Stratagene). The mutagenesis was checked by sequencing. Each of the constructed plasmids was checked by sequencing.
Transformation.
Plasmid DNA was introduced into tobacco BY-2 cells and Arabidopsis leaves by particle bombardment with a helium-driven particle accelerator (PDS/1000; Bio-Rad) with all basic adjustments set according to the manufacturer's recommendations. The applied bombardment parameters were 1,100 psi (1 psi = 6.89 kPa) of bombardment pressure, 1.0 μm of gold particles, a distance of 12 cm from macrocarrier to leaf pieces and suspension cells, and a decompression vacuum of 28 in Hg. The bombarded samples were incubated at a constant 22°C under light conditions for Arabidopsis leaves or at a constant 27°C under dark conditions for tobacco suspension cells until microscopic observations.
Microscopic Observations.
All pieces of the bombarded samples were imaged vitally without fixation by using a fluorescence microscope (Nikon Eclipse-600 and a 100 × 1.4 numerical aperture objective) and a confocal laser scanning microscope system (MicroRadiance MR/AG-2, Bio-Rad). Transformed Arabidopsis leaves were placed on glass slides without any pretreatment. Transformed BY-2 cells were treated with 500 nM MitoTracker Red CMXRos (Molecular Probes) according to the manufacturer's manual. The samples were illuminated with an argon ion laser using 488 nm of light for GFP and a green HeNe laser using 543 nm of light for chlorophyll autofluorescence and MitoTracker. All images were obtained by superimposing several images on the same position except that the z axis position of the focal plane was varied by steps of 0.5 μm. All images were prepared by using Adobe Systems (Mountain View, CA) PHOTOSHOP 6.0.
Results
Identification of the ADL2b Gene, an Orthologue of DNM1 and Drp1.
By using the amino acid sequences of the yeast dynamin-like protein Dnm1p (GenBank accession no. L40588) (20), we searched the Arabidopsis genome and EST databases and identified two closely related genes, named ADL2a [reported as ADL2 (21), encoded on chromosome 4] and ADL2b (encoded on chromosome 2). We found two ESTs containing the complete coding sequence of ADL2a (GenBank accession nos. AV528354 and AV528645) and one EST containing the complete coding sequence of ADL2b (GenBank accession no. AV529344). Full-length ADL2a and ADL2b cDNAs were sequenced with primers based on the genomic sequence. The sequences are available in GenBank under accession nos. AB072373 and AB072374 (ADL2a) and AB072375 (ADL2b). Both genes were expressed in all of the tissues we examined (rosette leaf, cauline leaf, root, silique, stem, and flower), but the patterns of the expression were slightly different in each tissue (unpublished data).
The newly identified ADL2b cDNA encodes a 780-amino acid (87-kDa) protein with the characteristic arrangement of protein domains found in other dynamin family members, especially in the N-terminal GTPase domain (22). But ADL2b does not have a typical PH domain, which is found in classical dynamin I. Among published sequences, the amino acid sequence of ADL2b shows a higher similarity to the so-called mitochondria-dividing, dynamin-like proteins of yeast and animals, Dnm1p and/or Drp1s (39–41% amino acid identity) than to the conventional dynamins and other dynamin-related proteins (18–38% identity). Fig. 1 also shows that ADL2b (as well as ADL2a) is likely a member of the mitochondrial-dividing, dynamin-like protein family (gray ovals) in the phylogenetic tree of the dynamin and dynamin-related protein families. Furthermore, the homology among ADL2b, ADL2a, Dnm1p, and Drp1s extends to the C-terminal GTPase-effector domain (unpublished data), indicating a closer relationship among them. But ADL2b shows a slightly higher identity to Dnm1p and Drp1s (39–41%) than does ADL2a (37–39%). Based on the relatedness of the amino acid sequences, we tried to find a functional similarity between ADL2b and the mitochondria-dividing proteins Dnm1p and Drp1.
ADL2b Is Attached to the Constriction Site and the Tips of Mitochondria.
To localize ADL2b in live plant cells, we analyzed its subcellular localization with the aid of GFP. A derivative of GFP, which can be expressed in Arabidopsis and which has an S65T modification for lighter fluorescence (23), was fused to the N terminus of a full-length ADL2b clone and placed under the control of the CaMV 35S promoter for ubiquitous and high expression. The fusion gene named GFP-ADL2b was introduced into tobacco suspension cells, and its expression was observed by fluorescence and confocal laser scanning microscopy. Labrousse et al. (24) previously confirmed the validity of this approach by proper localization of the fusion protein of GFP and dynamin in transgenic worms.
GFP-ADL2b was observed in punctate structures within tobacco suspension cells BY-2 (Fig. 2, in green). These spots were observed only in cells particle-bombarded with the GFP-ADL2b (unpublished data), indicating that they represent specific GFP fluorescence. The localization of fluorescence in detectable spots suggests that GFP-ADL2b molecules form aggregates. Visualization of mitochondria in these cells with MitoTracker, a vital mitochondrial fluorescent probe, indicated that many punctate structures were associated with the tips and constriction sites of mitochondria (Fig. 2, in red). In the superimposed image of Fig. 2 Right, the GFP signals were localized at constriction sites (arrowheads) and tips (arrows) of mitochondria. These results suggest that GFP-ADL2b is localized at the constriction sites of mitochondria that are about to divide and that GFP-ADL2b remained at the tips of mitochondria after division.
Figure 2.
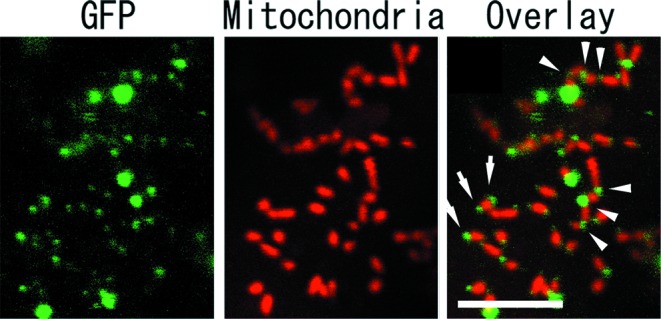
Localization of GFP-ADL2b with tips and constriction sites of mitochondria. BY-2 cells transiently expressing GFP-ADL2b with mitochondrial marker MitoTracker were examined by confocal laser scanning microscopy. High-magnification images of a part of a single BY-2 cell are shown. Left and Center are separate images obtained with the GFP and MitoTracker of Right images, respectively. GFP-ADL2b (green) was located in small punctate structures, many of which were localized in tips and constricted sites of mitochondria (red). GFP-ADL2b spots (arrowheads) were localized to places where the mitochondria were constricted. GFP signals (arrows) were localized at the tips of mitochondria. [Bar = 5 μm.]
ADL2b Mutants Cause a Dominant-Negative Effect on Plant Mitochondrial Fission.
The results described above suggest that ADL2b is involved in mitochondrial division. To test this hypothesis, we introduced wild-type and mutated ADL2b to tobacco suspension-cultured cells (BY-2) and Arabidopsis leaf epidermal cells, co-transformed with Mt-GFP as a mitochondrial marker. Schematic drawings of the plasmids used in this study and the mutation-target sites of ADL2b are shown in Fig. 3. In this experiment, unlike in the above experiment for the ADL2b localization (Fig. 2), GFP and ADL2b are not fused, and GFP is used to visualize the whole mitochondria in the transformed cells. (The Mt-GFP and MitoTracker completely matching images are shown in Fig. 4 Right).
Figure 4.
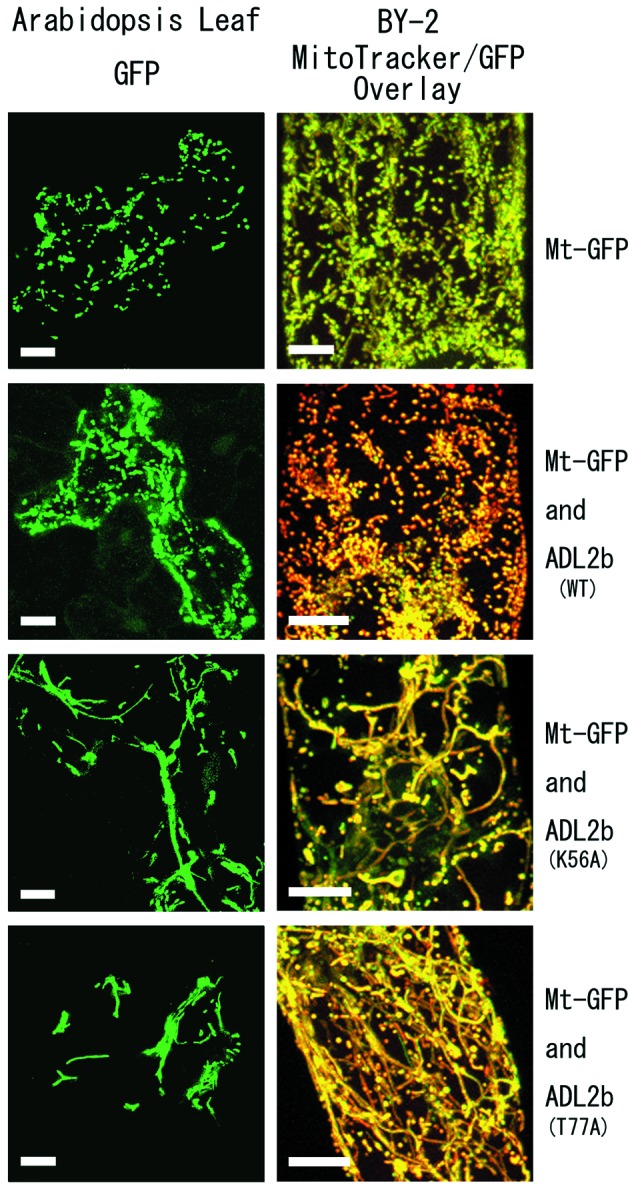
Mutagenized ADL2b causes dominant-negative effects on mitochondrial morphology. Live plant cells transiently expressing wild-type ADL2b or mutagenized ADL2b as well as Mt-GFP were examined by confocal laser scanning microscopy. (Left) Arabidopsis leaf epidermal cells expressing the vectors shown at the right end. The mitochondria were visualized only with Mt-GFP. (Right) High-magnification images of single BY-2 cells bombarded with the vector shown at the right end. The images show overlaid images of green Mt-GFP and red MitoTracker. Yellow signals indicate areas where the green and red signals completely match. In the upper four images, all cells with and without wild-type ADL2b show normal mitochondrial morphology, with small spherical or peanut-shaped particles. In the bottom four images, all cells bombarded with mutant ADL2b (K56A or T77A) show fusion of mitochondria (fewer and longer mitochondria). [Bars = 10 μm.]
Both mutations are thought to interfere with GTP hydrolysis activity of dynamin-like proteins (25, 26). The first mutation is K56A in the G1 consensus motif of the GTPase domain. The K56A mutation of ADL2b is analogous to inactivating mutations of dynamin, Dnm1p, and Drp1, which strongly interfere with mitochondrial division in transiently transformed cells (11, 27). ADL2b (K56A) molecules most likely exist in a nucleotide-free or GDP-bound form. The second mutation is T77A in the G2 motif, which has been shown to cause yeast Dnm1p and worm Drp1 to malfunction (11). This mutation most likely reduces the affinity for GTP and has been shown in other GTPases to inhibit interactions with GTPase-activating proteins. The dominant interfering effects of these two mutations were tested.
In cells bombarded with Mt-GFP only (without ADL2b) and in cells bombarded with Mt-GFP and with wild-type ADL2b, the mitochondria were typically dispersed with many small spherical and/or peanut-shaped particles (Fig. 4). We defined these types of mitochondria as “normal.” However, when we introduced the mutant genes ADL2b (K56A) and ADL2b (T77A), the mitochondria were found to be fewer and longer, or to have bulges and/or to occur in aggregates in both types of plant cells (Fig. 4). We defined these types of mitochondria as “abnormal.” Some mitochondrial abnormalities might be the results of variations in growth conditions, because wild-type leaf epidermal cells also have some variability in mitochondrial morphologies. The mitochondria in 4.2% of the wild-type epidermal cells did not have a normal shape (n = 160 cells, SD = 6.3%). However, leaves that were transformed with mutant ADL2bs had many more cells with abnormally shaped mitochondria [98.7% of cells with ADL2b (K56A) and 92.7% with ADL2b (T77A); SD = 1.4 and 6.8; n = 277 and 308]. These results indicate that the dysfunction of ADL2b prevents mitochondria from adopting their normal morphology in live plant cells, possibly because of the arrest of mitochondrial fission.
Discussion
Evolution of the Mitochondrial Division Apparatus.
Our results show that a dynamin-like protein, rather than an FtsZ protein of α-proteobacteria origin, functions in the mitochondrial fission of higher plants. In the course of evolution, the ancient mitochondria seem to have divided with the help of an α-proteobacterial-type FtsZ and other bacterial-type components (28). But now, in higher eukaryotes, the dynamin-like GTPases have replaced the prokaryotic FtsZ GTPases (29). If ancient mitochondria had an FtsZ GTPase and other bacterial-type division proteins, at what points in the various eukaryotic lineages would they have been lost? And at what points did dynamin-like proteins acquire a mitochondria-dividing function?
The loss of the MtFtsZ gene may not have occurred in the common ancestor of the animals, fungi, and plants because some red algae have MtFtsZ (Fig. 5). In the phylogenetic tree of life, red algae are now thought to share a common ancestry with green plants, outside of the lineage of animals and fungi (13, 14). In view of the above results, the loss of MtFtsZ may have occurred independently in the lineage of animals and fungi, and in the lineage of higher green plants after their divergence from the Rhodophyta (Fig. 5).
Figure 5.
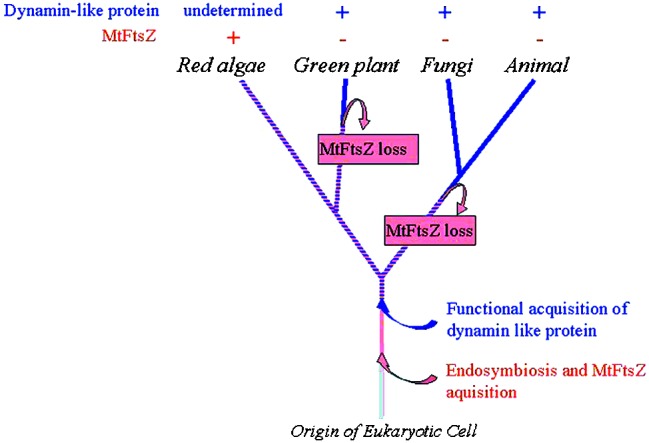
Tentative phylogenetic relationship showing parallel losses of MtFtsZ and the functional acquisition of a dynamin-like protein involved in mitochondrial division. This model assumes that (i) parallel losses of MtFtsZ occurred at least on the branch to higher plants and in the group of animals and fungi, and (ii) a functional acquisition of dynamin-like protein on mitochondrial division has occurred at the point before the divergence of these four groups. If this is correct, the common ancestor of green plants and red algae after the divergence of animals and fungi would have used both FtsZ and a dynamin-like protein in their mitochondrial division. It is possible that some presently living red algae have both proteins.
On the other hand, the functional acquisition of the dynamin-like proteins in the mitochondrial division apparatus might have occurred in the common ancestor of the animals, fungi, and plants. The higher degree of functional and sequence similarity among Dnm1p, Drp1, and ADL2b supports this scenario. In addition, other mitochondrial fission proteins such as Fis1/Mdv2 are shared among animals, fungi, and plants. Fis1/Mdv2 is reported to be a mitochondrial membrane-anchored protein that helps to attach Dnm1p to the mitochondrial membrane in yeast (30).
It is possible that some eukaryotes use both FtsZ and a dynamin-like protein in their mitochondrial division, although no such organisms have been found yet (28, 31). It is proposed that the FtsZ could pull from the inside of the mitochondrial inner membrane and a dynamin-like protein could squeeze from the outside of the outer membrane (31). If this is correct, then it would be possible for the two proteins to be present simultaneously, because they would not interfere with one another. Taken together, the above results suggest that the common ancestor of Rhodophyta and green plants used not only MtFtsZ but also other GTPase dynamin-like proteins in their mitochondrial division, as shown in Fig. 5. It is even possible that some modern Rhodophyta use both of these proteins for their mitochondrial division. Electron microscopy observations showed the presence of outer and inner mitochondrial dividing rings (MD-rings) in the rhodophyte C. merolae (2), which is known to have an MtFtsZ gene (7). It is interesting to consider the possibility that the outer MD-ring is made of a dynamin-like protein and that the inner ring is made of FtsZ. Further studies are needed to determine whether the ADL2b forms ring structures on the outside of the outer membrane that constrict the mitochondria during the division process.
Two ADL2 Genes in the Arabidopsis Genome.
There are two ADL2 genes (ADL2a and ADL2b) in the Arabidopsis genome. In this report, we characterized ADL2b and showed that it is concerned with mitochondrial fission. Kang et al. (21) reported that the N-terminal 35-aa residues of ADL2a (originally described as ADL2) were sufficient to direct the attached GFP into chloroplasts in tobacco protoplasts. In addition, it was recently reported that ADL2a, which is located in membrane-bound compartments within chloroplasts, assembles into a high-molecular weight complex (32). The strong identify in amino acids (77%) displayed between ADL2a and ADL2b raises the question of whether both proteins function in the chloroplasts and mitochondria, or in just one or the other. However, recent studies in our laboratory suggest that ADL2b does not have an N-terminal signal peptide for chloroplasts. One of our next goals is to determine whether ADL2a, like ADL2b, is able to locate to mitochondria and whether it is involved in mitochondrial fission.
Acknowledgments
We thank Prof. A. Hirai, Dr. M. Nakazono, and Dr. W. Sakamoto for helpful discussions during the course of this study. We thank Drs. Y. Niwa and W. Sakamoto for kindly providing the enhanced GFP expression vector and the ATPase delta presequence-GFP (Mt-GFP) expression vector, respectively. This research was supported by a research fellowship of the Japan Society for the Promotion of Science for young scientists and by grants-in-aid from the Basic Research for Innovative Biosciences.
Abbreviations
- CaMV
cauliflower mosaic virus
- GFP
green fluorescent protein
- Mt-GFP
mitochondrial matrix-targeted GFP
- MtFtsZ
mitochondrial FtsZ
- EST
expressed sequence tag
- BY-2
bright yellow-2
Footnotes
References
- 1.Gray M W. Int Rev Cytol. 1993;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- 2.Kuroiwa T, Kuroiwa H, Sakai A, Takahashi H, Toda K, Itoh R. Int Rev Cytol. 1998;181:1–41. doi: 10.1016/s0074-7696(08)60415-5. [DOI] [PubMed] [Google Scholar]
- 3.Bi E, Lutkenhaus J. Nature (London) 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 4.Osteryoung K W, McAndrew R S. Annu Rev Plant Phys Plant Mol Biol. 2001;52:315–333. doi: 10.1146/annurev.arplant.52.1.315. [DOI] [PubMed] [Google Scholar]
- 5.Vitha S, McAndrew R S, Osteryoung K W. J Cell Biol. 2001;153:111–119. doi: 10.1083/jcb.153.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beech P L, Nheu T, Schultz T, Herbert S, Lithgow T, Gilson P R, McFadden G I. Science. 2000;287:1276–1279. doi: 10.1126/science.287.5456.1276. [DOI] [PubMed] [Google Scholar]
- 7.Takahara M, Takahashi H, Matsunaga S, Miyagishima S, Takano H, Sakai A, Kawano S, Kuroiwa T. Mol Gen Genet. 2000;264:452–460. doi: 10.1007/s004380000307. [DOI] [PubMed] [Google Scholar]
- 8.Bleazard W, McCaffery J M, King E J, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw J M. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sesaki H, Jensen R E. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smirnova E, Shurland D L, Ryazantsev S N, van der Bliek A M. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labrousse A M, Zappaterra M D, Rube D A, van der Bliek A M. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 12.Takei K, McPherson P S, Schmid S L, Camilli P D. Nature (London) 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 13.Moreira D, Guyader H L, Philippe H. Nature (London) 2000;405:69–72. doi: 10.1038/35011054. [DOI] [PubMed] [Google Scholar]
- 14.Burger G, Diane S L, Gray M W, Lang F. Plant Cell. 1999;11:1675–1694. doi: 10.1105/tpc.11.9.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 16.Unseld M, Marienfeld J R, Brandt P, Brennicke A. Nature (London) Genetics. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 17.Nagata T, Nemoto Y, Hasezawa S. Int Rev Cytol. 1992;132:1–30. [Google Scholar]
- 18.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.Gammie A E, Kurihara L J, Vallee R B, Rose M D. J Cell Biol. 1995;130:553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang S G, Jin J B, Piao H L, Pih K T, Jang H J, Lim J H, Hwang I. Plant Mol Biol. 1998;38:437–447. doi: 10.1023/a:1006099718761. [DOI] [PubMed] [Google Scholar]
- 22.van der Bliek A M. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- 23.Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 24.Labrousse A M, Shurland D L, van der Bliek A M. Mol Biol Cell. 1998;9:3227–3239. doi: 10.1091/mbc.9.11.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bourne H R, Sanders D A, McCormick F. Nature (London) 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 26.van der Bliek A M, Redelmeier T E, Damke H, Tisdale E J, Meyerowitz E M, Schmid S L. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitts K R, Yoon Y, Krueger E W, McNiven M A. Mol Biol Cell. 1999;10:4403–4417. doi: 10.1091/mbc.10.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilson P L, Beech P L. Res Microbiol. 2001;152:3–10. doi: 10.1016/s0923-2508(00)01162-1. [DOI] [PubMed] [Google Scholar]
- 29.Erickson H P. J Cell Biol. 2000;148:1103–1105. doi: 10.1083/jcb.148.6.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mozdy A D, McCaffery J M, Shaw J M. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margolin W. Curr Biol. 2000;10:328–330. doi: 10.1016/s0960-9822(00)00458-9. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y W, Park D S, Park S C, Kim S H, Cheong G W, Hwang I. Plant Physiol. 2001;127:1243–1255. [PMC free article] [PubMed] [Google Scholar]