Abstract
The sulfolipid sulfoquinovosyldiacylglycerol is one of the three nonphosphorous glycolipids that provide the bulk of the structural lipids in photosynthetic membranes of seed plants. Unlike the galactolipids, sulfolipid is anionic at physiological pH because of its 6-deoxy-6-sulfonate-glucose (sulfoquinovose) head group. The biosynthesis of this lipid proceeds in two steps: first, the assembly of UDP-sulfoquinovose from UDP-glucose and sulfite, and second, the transfer of the sulfoquinovose moiety from UDP-sulfoquinovose to diacylglycerol. The first reaction is catalyzed by the SQD1 protein in Arabidopsis. Here we describe the identification of the SQD2 gene of Arabidopsis. We propose that this gene encodes the sulfoquinovosyltransferase catalyzing the second step of sulfolipid biosynthesis. Expression of SQD1 and SQD2 in Escherichia coli reconstituted plant sulfolipid biosynthesis in this bacterium. Insertion of a transfer DNA into this gene in Arabidopsis led to complete lack of sulfolipid in the respective sqd2 mutant. This mutant showed reduced growth under phosphate-limited growth conditions. The results support the hypothesis that sulfolipid can function as a substitute of anionic phospholipids under phosphate-limited growth conditions. Along with phosphatidylglycerol, sulfolipid contributes to maintaining a negatively charged lipid–water interface, which presumably is required for proper function of photosynthetic membranes.
The photosynthetic membranes of seed plants are rich in nonphosphorous glycolipids, which include the galactolipids mono- and digalactosyldiacylglycerol and the sulfolipid sulfoquinovosyldiacylglycerol. This lipid is unusual, because its polar head group consists of sulfoquinovose, a 6-deoxy-6-sulfonate-glucose. Much has been speculated about the possible function of this lipid in photosynthesis (1, 2), but in recent years the isolation of sulfolipid-deficient bacterial mutants (3, 4) and a mutant of the unicellular algae Chlamydomonas has provided some novel clues (5). It has become apparent from these mutants that there is no essential role for sulfolipid in photosynthetic bacteria with anoxygenic or oxygenic photosynthesis or in eukaryotic cells containing chloroplasts, because all mutants were capable of photoautotrophic growth. The effects on photosynthesis are subtle under normal growth conditions, at best. However, if bacterial sulfolipid-deficient null mutants are starved for phosphate, they cease growth much earlier than the respective wild type (3, 4). Thus, sulfolipid is of conditional importance in these bacteria. Similar to bacteria, the ratio of nonphosphorous glycolipids to phospholipids drastically increases in Arabidopsis after phosphate deprivation (6, 7). In particular, the relative amount of sulfolipid rises several fold because of an active process based on the increased expression of at least one of the sulfolipid genes, SQD1 (6). Therefore, it seems likely that sulfolipid is of conditional importance also in Arabidopsis and other plants.
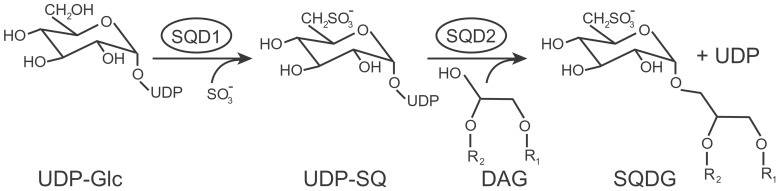
The unique step in sulfolipid biosynthesis is the synthesis of UDP-sulfoquinovose, the sulfolipid head group donor (Fig. 1), catalyzed by the SQD1 protein in Arabidopsis (8). The crystal structure for this protein is known, and a reaction mechanism has been proposed (9, 10). On the contrary, although the activity of the sulfolipid synthase catalyzing the last step of sulfolipid biosynthesis has been fairly well characterized in spinach chloroplast extracts (11), the protein had not been isolated, and the respective gene remained unidentified. Orthologs of the Arabidopsis SQD1 gene are highly conserved in different groups of bacteria and even in plants (12). However, two unrelated classes of genes encode the UDP-sulfoquinovose-dependent glycosyltransferases responsible for the final assembly of sulfolipid in the bacteria of the α group (13) and cyanobacteria (14). Because it seemed possible that plants inherited their sulfolipid synthase from the postulated cyanobacterial ancestor of chloroplasts, we searched for an ortholog of the cyanobacterial sqdX gene in the fully sequenced Arabidopsis genome. As a result of this genomic approach, the correct Arabidopsis ortholog of sqdX, designated SQD2, was identified, characterized, and exploited to answer open questions about sulfolipid biosynthesis and function in seed plants.
Figure 1.
The pathway for sulfolipid biosynthesis in Arabidopsis. Two enzymes, SQD1 and SQD2, are specific to this process and catalyze the reactions as indicated. DAG, diacylglycerol; R, fatty acyl groups; SQDG, sulfoquinovosyldiacylglycerol; UDP-Glc, UDP-glucose; UDP-SQ, UDP-sulfoquinovose.
Materials and Methods
Plant Material and Plant Growth Experiments.
Surface-sterilized seeds of Arabidopsis (Arabidopsis thaliana, ecotype Wassilewska) were germinated on 0.8% (wt/vol) agar-solidified MS medium (15) supplemented with 1% (wt/vol) sucrose. The seedlings were kept on agar for 10 days before the transfer to pots containing a soil mixture as described (16) except that sphagnum was replaced with Bactomix (Michigan Peat, Houston, TX). Plants were grown in growth chambers (AR-75L, Percival Scientific, Boone, IA) under light of a photosynthetic photon-flux density of 80 μmol of photons⋅m−2⋅s−1. A 14-h light/10-h dark regime was applied. The day/night temperature was controlled at 22/18°C. For growth under phosphate limitation, sterile Arabidopsis medium (16) was used but at half concentration and containing 0.8% agarose, 1% sucrose, 20 mM Mes (pH 6.0), and different concentrations of KH2PO4 as indicated. Plants were grown for 8 days on 0.8% (wt/vol) agar-solidified MS medium supplemented with 1% (wt/vol) sucrose and then transferred to the agar plates containing the phosphate-limited medium. For quantitative growth experiments, sucrose was left out of the medium after the transfer. Plant culture dishes for each experiment were placed on a single shelf of a CU-36L5 chamber (Percival Scientific). The dishes were covered with two layers of cheese cloth to diffuse the light and maintain a uniform photon-flux density of 80 μmol of photons⋅m−2⋅s−1. The dishes were rotated carefully over the shelf to ensure uniformity. At each time point, eight mutant and wild-type plants on matching dish positions were harvested, and their aerial parts were weighed. At least three independent biological repeats of the experiment were obtained with similar results.
Mutant Isolation.
A transfer DNA (T-DNA) insertion into the SQD2 gene was isolated from the Arabidopsis knockout collection at the University of Wisconsin. The gene-specific primers used for the screening of insertions into the SQD2 gene were 5′-AATCTCTCTATACCTCCTCATTTGCTTCC-3′ and 5′-CACAAGCTCAATATAGGTTGACCCATAAG-3′, and the T-DNA-specific primer matching the left end of the T-DNA was 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′. PCR fragments obtained by using combinations of the T-DNA-specific primer and the two gene-specific primers were subcloned and sequenced at the Michigan State University genomics facility to determine the insertion point for the 5.5-kb T-DNA.
Cloning of the SQD2 cDNA.
The SQD2 ORF corresponding to the protein (GenBank accession no. CAB69850) predicted from bacterial artificial chromosome locus F7J8_200 (GenBank accession no. AL137189) was cloned by reverse transcription–PCR using the primers 5′-CGGGATCCCCATGACGACTCTTTCTTCTATA-3′ and 5′-AAGGTACCCTACACGTTACCTTCCGGTA-3′. Total leaf RNA was isolated from ≈20-day-old plants (Col-2) according to the method by Logemann et al. (17). Poly(A)+ mRNA was purified by using an oligotex kit from Qiagen (Chatsworth, CA) according to the instructions. Reverse transcription–PCRs were performed by using the ProSTAR HF single tube reverse transcription–PCR system from Stratagene. The PCR product was cloned into the ligation-ready Stratagene vector pPCR-Script Amp SK+ giving rise to pPCR-SQD2. The insert was sequenced and deposited at GenBank (accession no. AF454354).
Northern Blot Analysis.
Total RNA isolated as described above was separated by agarose gel electrophoresis and blotted onto Hybond N+ (Amersham Pharmacia) membranes by using standard procedures (17). Hybridization was done under standard conditions (18).
Complementation Analysis.
The insert of pPCR-SQD2 containing the full-length coding sequence including the transit peptide was amplified by PCR using the primers 5′-AAGGTACCATGACGACTCTTTCTTCTATA-3′ and 5′-AATCTAGACTACACGTTACCTTCCGGTA-3′. This fragment was cloned into the binary vector pBINAR-Hyg (19) by using the KpnI and XbaI sites. The resulting plasmid, pBINAR-Hyg/SQD2, was used to transform the sqd2 mutant by vacuum infiltration (20). Transgenic plants were selected in the presence of kanamycin (60 μg/ml) and hygromycin B (25 μg/ml) on MS medium lacking sucrose.
Expression of SQD1 and SQD2 in E. coli.
To express SQD2, the BamHI/KpnI fragment of pPCR-SQD2 containing the transit peptide-encoding sequence was cloned in-frame into the expression vector pQE32 from Qiagen, giving rise to pSQD2. The expression cassette encoding the SQD1 protein lacking the N-terminal 84-aa transit peptide was excised from pSQD1-TP (6) with XhoI/PvuII and ligated into pACYC184 (21) cut with SalI/EcoRV, giving rise to pSQD1, which is compatible with pSQD2 in E. coli. Both plasmids were transferred to E. coli host XL1-Blue (Stratagene) and selected for in the presence of 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. Expression was induced according to the Qiagen protocol.
Lipid and Fatty Acid Analysis.
Lipids were extracted from frozen leaves or E. coli pellets as described (22). Lipid extracts were analyzed as described (7), and lipids were visualized with iodine vapor or sugar-sensitive α-naphthol reagent (23) and identified by cochromatography with lipid extracts of known composition. For quantitative analysis, methyl esters were prepared and quantified by GLC (24). For the analysis of 35S-labeled lipids, four leaves of 3-week-old plants were cut midway through the petiole and transferred immediately petiole-down into a polypropylene microfuge tube containing 100 μl of an aqueous solution of carrier-free [35S]sulfate (3.7 MBq/ml; NEN). Lipids were extracted after 1 h of incubation and separated by two-dimensional TLC as described (23). Labeled compounds were visualized by autoradiography. Fast atom bombardment mass spectroscopy measurements of sulfolipid (25) were done at the Michigan State University mass-spectrometry facility.
Chlorophyll Fluorescence Measurements.
In vivo chlorophyll fluorescence was measured at room temperature with an FMS2 fluorometer (Hansatech Instruments, Pentney King's Lynn, U.K.) after 20 min of dark adaptation. The maximal variable fluorescence (Fv) was obtained by subtraction of the initial fluorescence (Fo) from the maximum fluorescence (Fm). The ratio of Fv/Fm served as a measure of the maximal photochemical efficiency of photosystem II. The effective quantum yield of photochemical energy conversion in photosystem II, ΦPSII, was determined as described by Härtel et al. (26). Calculation of ΦPSII was according to Genty et al. (27).
Results
Identification of an Arabidopsis SQD2 Candidate Gene.
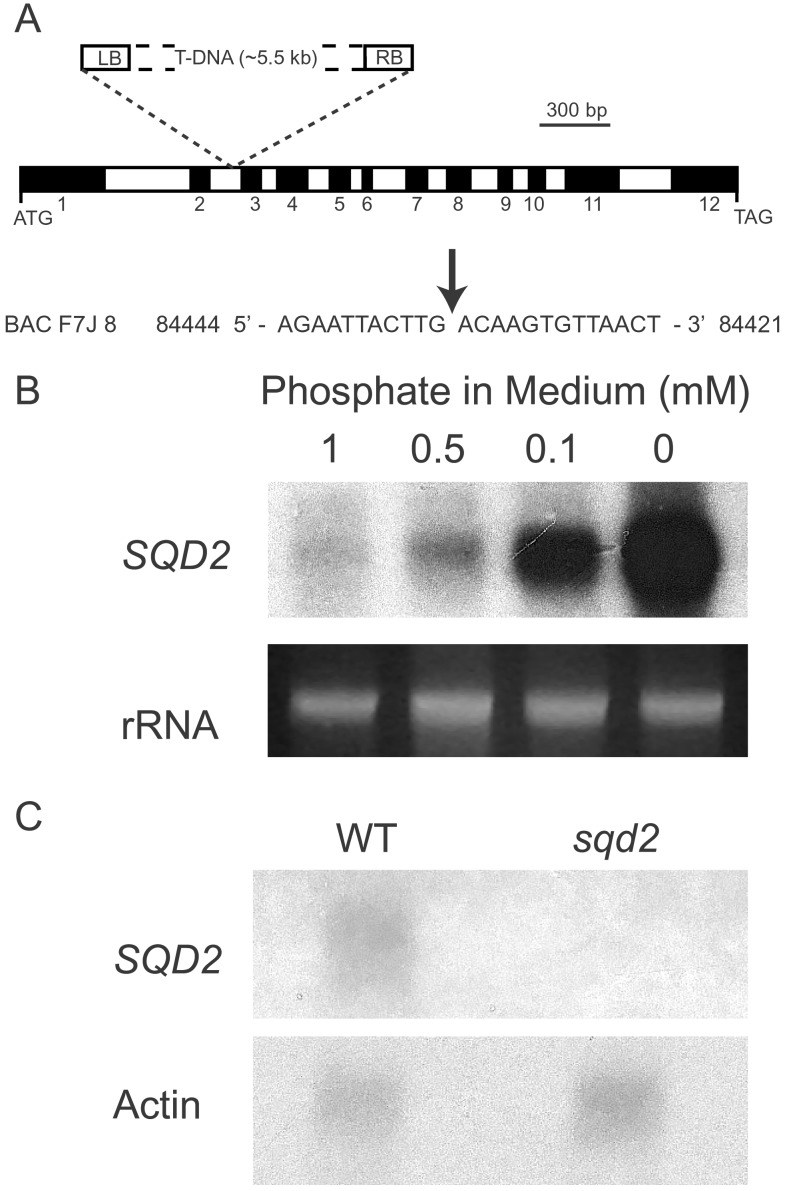
Taking advantage of the published Arabidopsis genome (28), we developed a set of criteria for the identification of an SQD2 candidate gene encoding the sulfolipid synthase of Arabidopsis. First, given the ancestral relationship between chloroplasts and cyanobacteria it seemed likely that SQD2 would be similar to the sqdX gene of the cyanobacterium Synechococcus strain sp. PCC7942, which was proposed to encode the sulfolipid synthase (14). Indeed we, and recently Berg et al. (29), identified Arabidopsis gene F7J8_200 (AT5g01220, GenBank accession no. AL137189.2) on chromosome 5 as a possible sqdX ortholog. The originally predicted protein (GenBank accession no. CAB69850) for this gene showed 37% identity over 370 amino acid residues with the cyanobacterial SqdX protein, sufficient to be considered an ortholog. A wild-type cDNA corresponding to gene F7J8_200 was isolated by reverse transcription–PCR and sequenced (GenBank accession no. AF454354). The ORF for this cDNA disagreed with the current protein predictions for the F7J8_200 gene (GenBank accession nos. CAB69850 and AAK76635), leading us to revise the exon-intron structure for F7J8_200 as shown in Fig. 2A. Exon 6 was added, and exons 7, 8, and 9 increased slightly in size. The revised protein sequence for the F7J8_200 ORF of 510 aa is 41% identical over 370 residues to the cyanobacterial SqdX protein. As a second criterion for the correct identification of an SQD2 candidate, a protein domain search using the PFAM database (30) revealed a glycosyltransferase group 1 motif (PFAM accession no. 00534) from amino acids 294 to 458 of the putative SQD2 protein encoded by the cDNA. According to the glycosyltransferase classification database CAZY (http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html; ref. 31), this protein is grouped in with glycosyltransferase family 4, which is characterized mechanistically by “retaining” glycosyltransferases. This classification would be in agreement with the α-anomeric proton configuration of the substrate and product of the sulfolipid synthase reaction. Third, sulfolipid synthase is an enzyme of the inner envelope in chloroplasts (32). As such, the translation product should contain a chloroplast transit peptide. According to an in silico analysis (33), the putative SQD2 protein contains an 83-aa transit peptide. Fourth, we expected that the expression of the candidate SQD2 gene would be induced as phosphate is depleted, because sulfolipid biosynthesis and the expression of SQD1 are induced under these conditions (6). As shown in Fig. 2B, the expression of the candidate SQD2 gene followed this prediction. With all four prediction criteria satisfied, a series of experiments was conducted to determine whether Arabidopsis gene F7J8_200 indeed encodes a sulfolipid synthase.
Figure 2.
Structure of the SQD2 gene carrying the T-DNA insertion (A), induction of SQD2 expression after phosphate deprivation (B), and lack of expression of SQD2 in the sqd2 mutant (C). (A) Exons are numbered as referred to in the text. A small portion of the bacterial artificial chromosome (BAC) F7J8 sequence is shown with nucleotide numbers referring to GenBank accession no. AL137189.2. The arrow indicates the T-DNA insertion point. (B) Plants were grown on agar-solidified medium for ≈20 days. As loading control, one of the rRNA bands stained with ethidium bromide is shown. (C) Plants were grown on soil for ≈25 days. An actin cDNA (GenBank accession no. M20016.1) was used as a probe for control purposes. WT, wild type.
Isolation of an sqd2 T-DNA Insertion Mutant.
To obtain corroborating evidence that the gene F7J8_200 encodes a protein essential for sulfolipid biosynthesis in Arabidopsis, the isolation of a T-DNA insertion mutant from the University of Wisconsin (Madison, WI) collection (34) was pursued. One homozygous mutant line that lacked sulfolipid was isolated. By sequencing PCR products obtained by using gene-specific and T-DNA-specific primers, it was determined that the T-DNA was inserted after base pair 84,434 of the F7J8 bacterial artificial chromosome sequence (GenBank accession no. AL137189.2), which places the insertion point into the second intron of the predicted F7J8_200 gene as shown in Fig. 2A. A comparative Northern analysis between the wild type and the homozygous sqd2 mutant (Fig. 2C) suggested complete lack of expression. Homozygosity was determined at the molecular level by using combinations of gene-specific and T-DNA-specific primers to generate PCR products diagnostic for mutant and wild-type chromosomes. After a cross between the homozygous mutant and the wild type, genetic segregation was determined in the F2 generation. Approximately one quarter of the plants were kanamycin-sensitive (93 of 379 total), and approximately one third of the kanamycin-resistant plants (31 of 100) were sulfolipid-deficient (see below). The observed segregation ratios agreed with a single T-DNA insertion into the F7J8_200 locus, with kanamycin resistance scoring as a dominant marker and sulfolipid deficiency as a recessive marker. To ensure that the observed T-DNA insertion indeed was causing the lipid phenotype, the cDNA derived from F7J8_200 was ligated into a binary expression vector and transferred into the homozygous mutant (sulfolipid-deficient, kanamycin-resistant) by Agrobacterium-mediated transformation. A hygromycin B (transgene T-DNA) and kanamycin-resistant (Ko-T-DNA) transgenic line was recovered. This line produced sulfolipid (data not shown) and showed wild-type growth characteristics (see below), thereby demonstrating complementation. Therefore, disruption of the F7J8_200 locus was responsible for the sulfolipid lipid deficiency in the sqd2 mutant and growth impairment as discussed below. This locus was designated SULFOQUINOVOSYLDIACYLGLYCEROL 2 (SQD2).
The sqd2 Mutant Is Completely Devoid of Sulfolipid.
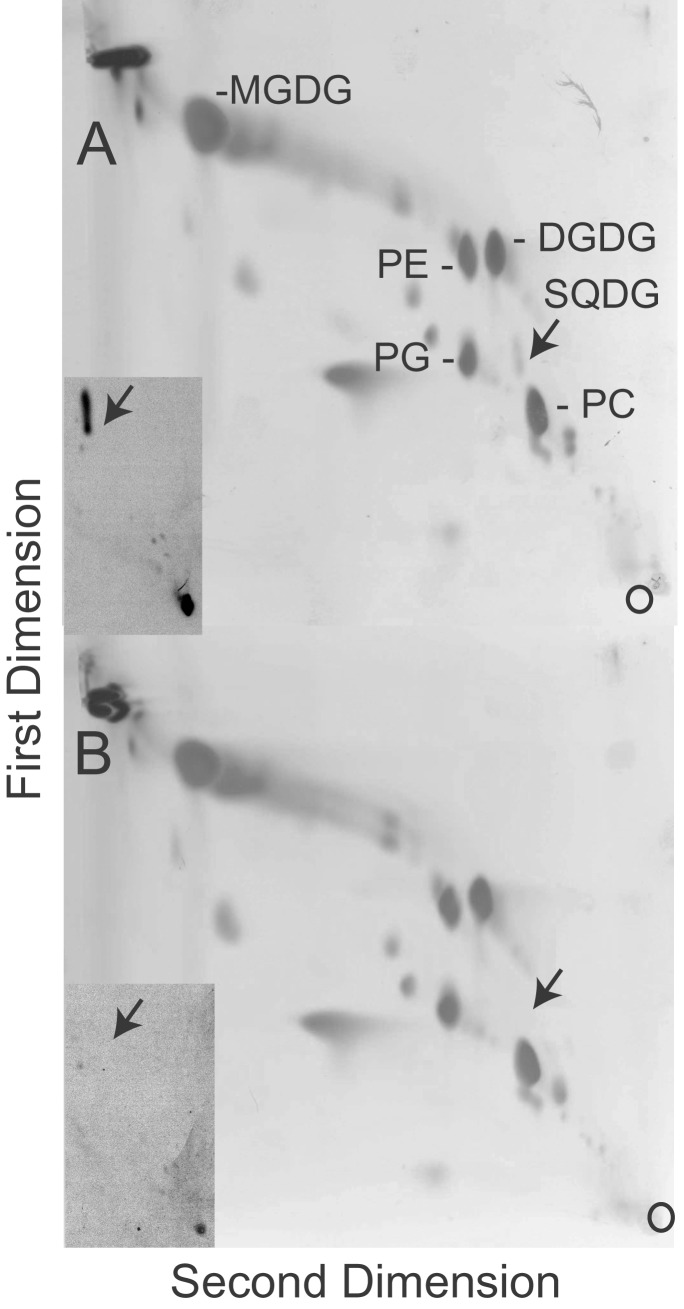
Lipids extracted from the sqd2 mutant and wild type were separated by two-dimensional TLC. No sulfolipid was detected in the mutant extracts by iodine staining (Fig. 3) or highly sensitive isotope labeling (Fig. 3 Inserts). On the basis of this result, it seems that sulfolipid biosynthesis is inactivated completely in this mutant because of an sqd2 null allele. To investigate compensatory effects in the sqd2 mutant, the leaf lipid composition was compared for extracts from mutant and wild-type (Wassilewska) plants grown in the presence and absence of phosphate (Table 1). Within the experimental error and besides the complete lack of sulfolipid, the leaf composition of the mutant showed no drastic differences compared with the wild type. Given that the relative amount of sulfolipid under phosphate-sufficient conditions does not exceed 2 mol%, this result was not surprising. However, under phosphate-deficient growth conditions, under which relative sulfolipid amounts increased and phosphatidylglycerol amounts decreased in the wild type, phosphatidylglycerol did not decrease in the sulfolipid-deficient Arabidopsis mutant. The same has been observed for sulfolipid-deficient bacterial mutants (3, 4) and was interpreted as a requirement to maintain the overall amount of anionic lipids in the thylakoid membrane. Taking sulfolipid and phosphatidylglycerol out of consideration, the glycolipid-to-phospholipid ratio increased in similar ways in the sqd2 mutant and wild type in response to phosphate starvation (Table 1).
Figure 3.
Two-dimensional separation of lipids extracted from wild-type (A) and sqd2 mutant (B) leaves. The lipids were visualized by iodine. (Inserts) Autoradiographs in the area of the sulfolipid spots. Leaves were incubated with [35S]sulfate to label sulfolipid. Lipids were separated as described for A and B. The small circles in the lower-right corners indicate the origins, and the arrows point toward the position of the sulfolipid. DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; SQDG, sulfoquinovosyldiacylglycerol.
Table 1.
Quantitative analysis of leaf lipid extracts from wild type (Wassilewska) and the sqd2 mutant of A. thaliana grown on medium with or without phosphate for 20 days*
| Lipid | 1 mM Phosphate in
medium
|
No Pi in medium
|
||
|---|---|---|---|---|
| Wild type (Wassilewska) | sqd2 | Wild type (Wassilewska) | sqd2 | |
| MGDG | 34.8 ± 0.9 | 32.2 ± 3.3 | 38.4 ± 0.7 | 37.7 ± 0.2 |
| PG | 10.5 ± 0.5 | 12.8 ± 1.1 | 3.2 ± 0.3 | 11.2 ± 0.3 |
| DGDG | 16.5 ± 1.0 | 16.6 ± 0.7 | 39.9 ± 2.0 | 37.5 ± 1.0 |
| SQDG | 1.3 ± 0.2 | n.d. | 5.7 ± 0.7 | n.d. |
| PI | 2.0 ± 0.2 | 2.8 ± 0.2 | 0.8 ± 0.4 | 1.7 ± 1.1 |
| PE | 15.1 ± 0.1 | 15.6 ± 1.1 | 4.1 ± 0.3 | 4.9 ± 0.2 |
| PC | 19.8 ± 1.2 | 20.2 ± 0.6 | 7.6 ± 0.9 | 6.9 ± 0.4 |
Values (mol%) represent means of three measurements. Lipid symbols are as described in the legends for Fig. 3; n.d., not detected; PI, phosphatidylinositol.
Expression of Arabidopsis SQD1 and SQD2 in E. coli.
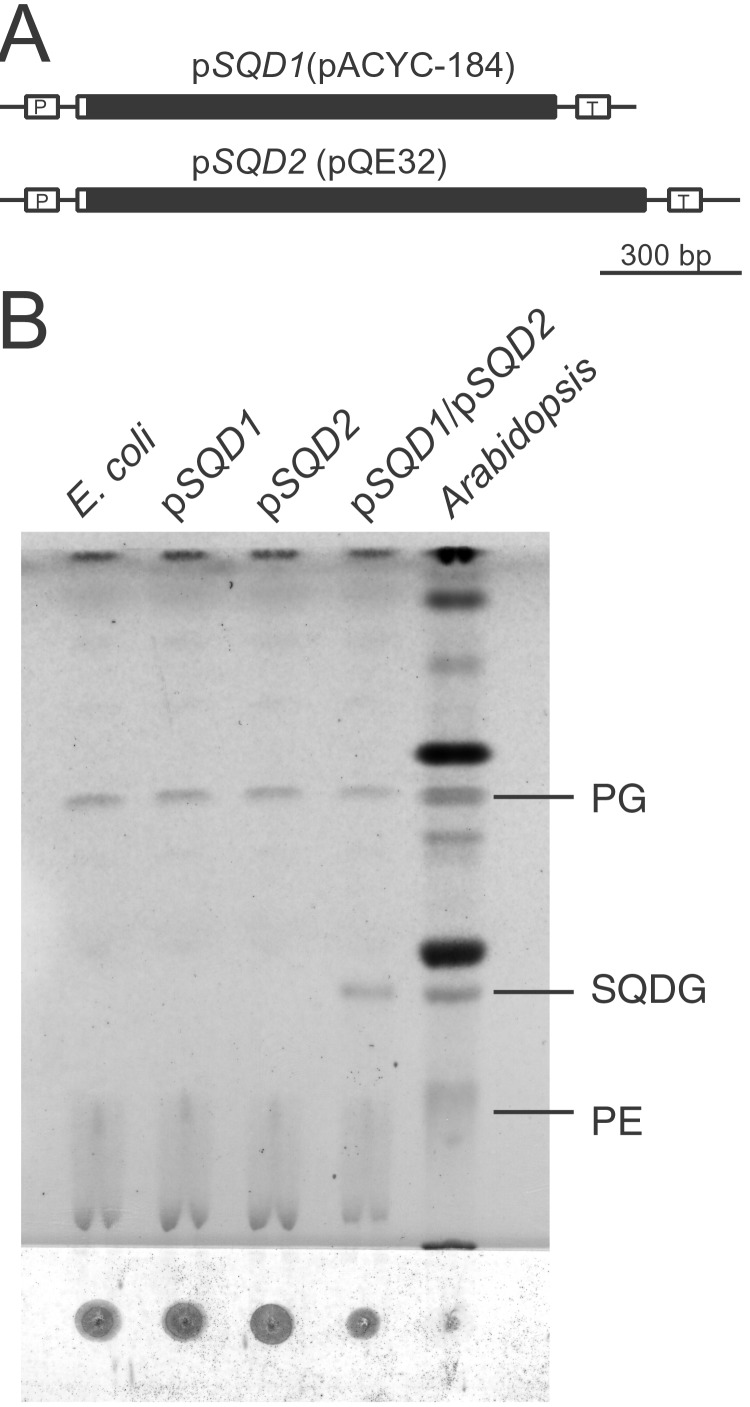
To demonstrate directly that SQD2 encodes the sulfolipid synthase and test whether the two proteins SQD1 and SQD2 of Arabidopsis represent the biosynthetic machinery specific to sulfolipid biosynthesis in plants, these two genes were coexpressed in E. coli. This bacterium normally does not contain sulfolipid. A truncated version of the SQD1 protein lacking the chloroplast transit peptide had been expressed previously in E. coli, and the purified recombinant protein was shown to catalyze the formation of UDP-sulfoquinovose from UDP-glucose and sulfite in vitro (8). To express both proteins, we followed a two-plasmid strategy as was used for the reconstitution of plant galactolipid biosynthesis in E. coli (22). Both constructs are shown in Fig. 4A. No new glycolipid bands were visible by α-naphthol staining in lipid extracts from E. coli cells containing only one of each of the two plasmids. However, when induced E. coli cells that contained both constructs were analyzed, a new band appeared, which cochromatographed with authentic plant sulfolipid from an Arabidopsis leaf lipid extract (Fig. 4B). This new lipid was isolated from induced E. coli cultures by using TLC purification and was analyzed by negative fast atom bombardment mass spectrometry. Two diagnostic molecular ion peaks (M-H) at 791 and 819 m/z were detected and agreed with the predicted M-H mass for sulfoquinovosyldiacylglycerol containing 16:0/16:1 and 16:0/18:1 fatty acids, respectively. Gas chromatographic analysis of fatty acid methylesters derived from the newly produced lipid revealed 16:0, 16:1, and 18:1 fatty acids as the predominant fatty acids. Taken together, all data were in agreement with the formation of authentic sulfolipid in E. coli after the expression of the Arabidopsis SQD1 and SQD2 cDNAs. These data also suggested that SQD2 indeed encodes the UDP-sulfoquinovosyl: diacylglycerol sulfoquinovosyl transferase from Arabidopsis.
Figure 4.
Plasmid constructs used for the expression of the SQD1 and SQD2 genes in E. coli (A) and separation of lipids from different E. coli strains (B). (A) The used vector plasmid is indicated in brackets for each construct. The open boxes indicate the His tag, and the black boxes indicate plant sequences. P, promoter; T, terminator. (B) The four left lanes represent extracts from E. coli harboring no plasmid or plasmids as indicated. An Arabidopsis leaf lipid extract was included for lipid-identification purposes. The lipids were visualized by α-naphthol, which primarily stains glycolipids. Other lipids are visualized because of light charring. The abbreviations are as described in the Fig. 3 legend.
The Effect of Sulfolipid Deficiency on the sqd2 Mutant.
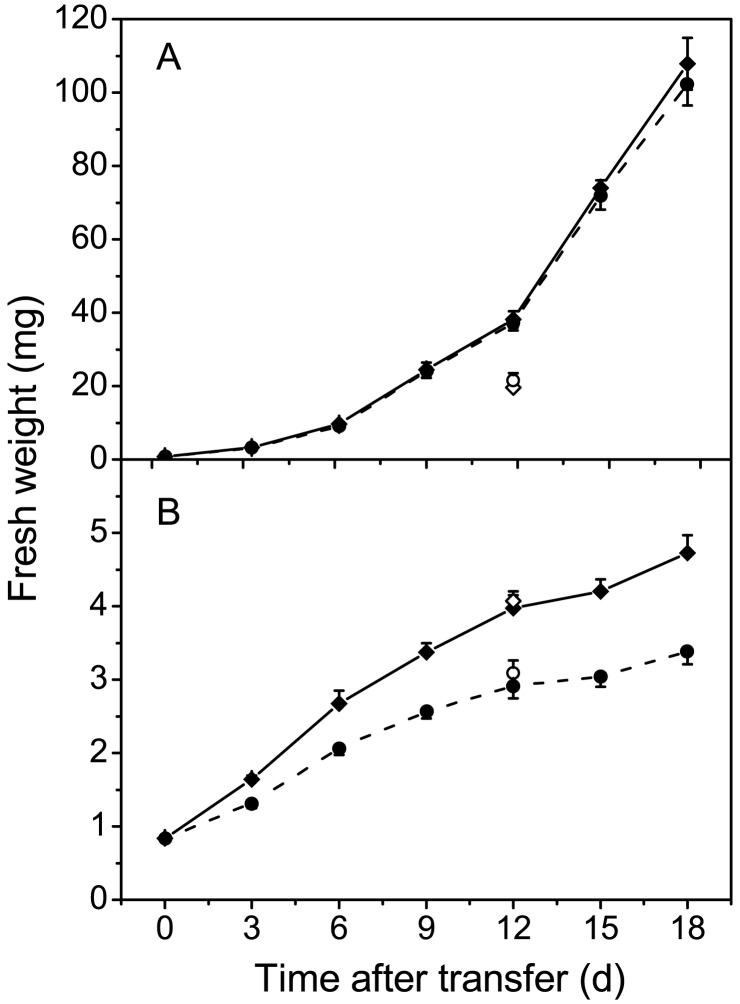
The sqd2 sulfolipid null mutant of Arabidopsis described here provides an excellent tool for the in vivo analysis of sulfolipid function in seed plants. Thus far, the molecular and genetic analyses of the mutant described above have not provided any evidence for multiple T-DNA insertions into the genome or other secondary mutations, which can occur in T-DNA-tagged mutant lines. Therefore, comparison of mutant and wild type should be a valid approach toward the elucidation of sulfolipid function. Comparing the growth of the mutant and wild type (Wassilewska) on agar plates in a controlled growth chamber environment on the basis of fresh weight did not reveal any difference when the plants were grown in the presence of phosphate-replete medium (Fig. 5A). However, the growth rate of the sqd2 mutant was reduced when plants were phosphate-depleted during growth on agar medium lacking phosphate (Fig. 5B). To eliminate the possibility that this subtle difference in growth was caused by a secondary mutation, we repeated one time point midway through the experiment comparing the homozygous mutant (sqd2) with the homozygous mutant line expressing the wild-type SQD2 cDNA (sqd2/SQD2 cDNA), lacking and containing sulfolipid, respectively. These two lines are nearly isogenic, but the second line contains an additional mutation caused by the insertion of an additional T-DNA. Again, no difference between the wild type and mutant was observed under normal growth conditions (Fig. 5A, open symbols), although, the onset of the exponential growth phase in this completely independent experiment was delayed, as is visible in the slight offset between the two data point sets (Fig. 5A, open and closed symbols). More importantly, the mutant lagged behind the complemented mutant line under phosphate-limited growth conditions (Fig. 5B, open symbols). Thus, secondary mutations as a cause for the observed growth difference between the mutant and wild type under phosphate-limiting conditions can be ruled out.
Figure 5.
Growth curves for wild type (filled diamonds, solid lines) and sqd2 mutant (filled circles, broken lines) on agar-solidified medium with 1 mM (A) or no (B) phosphate added. Eight plants were weighed for each time point. To rule out effects of secondary mutations, one time point (12 days) was repeated with an sqd2 mutant line transformed with the SQD2 cDNA (open diamonds) containing sulfolipid and the original sulfolipid-deficient sqd2 mutant (open circles). At least 15 plants are represented in each point in this experiment. Standard errors are indicated where they exceed the size of the symbols.
Using noninvasive chlorophyll fluorescence measurements, it was determined that the maximum quantum yields for photosystem II photochemistry (Fv/Fm) were very similar for the mutant and wild type (Table 2). The effective quantum yield of photochemical energy conversion in photosystem II (ΦPSII) was slightly but significantly reduced in the mutant grown under normal conditions (Table 2). Pigment content was not altered in the mutant (Table 2). Based on this basic analysis of photosynthesis, an essential function for sulfolipid in photosynthesis of seed plants can be ruled out. However, the current results do not exclude more subtle or conditional roles for sulfolipid in photosynthesis as were observed for Chlamydomonas and cyanobacteria (4, 5). It should be pointed out that chlorophyll fluorescence parameters were indistinguishable for the mutant and wild type under phosphate limitation (data not shown). Apparently, phosphate deficiency by itself has more pronounced effects on chlorophyll fluorescence (35) than sulfolipid deficiency.
Table 2.
| Wild type | sqd2 | |
|---|---|---|
| Chlorophyll a | 0.91 ± 0.06 | 0.93 ± 0.03 |
| Chlorophyll b | 0.29 ± 0.02 | 0.28 ± 0.02 |
| Chlorophyll a + b | 1.21 ± 0.07 | 1.21 ± 0.05 |
| Chlorophyll a/b | 3.14 ± 0.12 | 3.32 ± 0.14 |
| Carotenoids | 0.40 ± 0.02 | 0.42 ± 0.01 |
| Fv/Fm | 0.82 ± 0.01 | 0.80 ± 0.01 |
| ΦPSII | 0.77 ± 0.01 | 0.70 ± 0.01 |
Samples were taken from fully expanded leaves of 20–30-day-old plants grown on agar-solidified medium containing 1 mM phosphate. Pigment values represent means (± SE) of three independent determinations.
Actinic light for fluorescence measurements was 80 μmol photons⋅m−2⋅s−1, the same as used for raising the plants. Values represent means (± SE) of 10 independent determinations.
Discussion
More than 40 years after the discovery of the “plant sulfolipid” sulfoquinovosyldiacylglycerol by Benson et al. (36), the quest for the enzymes involved in the biosynthesis of sulfolipid in seed plants has come to an end. Only two Arabidopsis proteins, SQD1 and SQD2, are required for sulfolipid biosynthesis (Fig. 1) when simultaneously produced in E. coli. The first, SQD1, has been shown to catalyze the biosynthesis of UDP-sulfoquinovose from UDP-glucose and sulfite (8) in vitro. The successful reconstitution of sulfolipid biosynthesis in E. coli suggests that the proper in vivo sulfolipid donor is present in this bacterium but is not specific to organisms naturally capable of sulfolipid biosynthesis. Thus, these results support the hypothesis that sulfite as a product of the general sulfur assimilation pathway acts also as the in vivo sulfur donor. The second required plant enzyme, SQD2, is highly similar to glycosyltransferases (29), and it is proposed that this protein represents the sulfolipid synthase. Thus far, this enzyme has only been described in envelope preparations from spinach chloroplasts (11). The availability of an expression system for the production of SQD2 in E. coli provides the means to study the enzymatic reaction catalyzed by this protein in detail. The high similarity between the two Arabidopsis proteins SQD1 and SQD2 and the cyanobacterial sulfolipid proteins SqdB and SqdX (4, 14) and their ability to functionally reconstitute sulfolipid biosynthesis in E. coli suggests that these are true orthologs of SqdB and SqdX, respectively.
The isolation of the sulfolipid-deficient sqd2 null mutant of Arabidopsis allowed us to address another fundamental question about the role of sulfolipid in photosynthetic membranes of seed plants. Although the sqd2 mutant has not been studied yet in greater detail, our initial analysis of photosynthesis and growth suggests that sulfolipid does not have a crucial role in plant photosynthesis. The sqd2 mutant did not show any obvious signs of altered morphology or growth and photosynthetic parameters were affected only mildly under normal laboratory growth conditions. A similar conclusion can be drawn for bacterial mutants (3, 4) and a sulfolipid-deficient mutant in Chlamydomonas (5). Although the latter mutant appears to be impaired more severely in photosynthesis than the Arabidopsis sqd2 mutant or the bacterial mutants, effects caused by secondary mutations cannot be ruled out in this case. Unlike the Chlamydomonas mutant, the Arabidopsis sqd2 mutant and its nearly isogenic complementation line should provide the ideal material to probe further for specific roles of sulfolipid in chloroplastic photosynthesis.
If sulfolipid is not essential to photosynthesis, why did plants maintain this pathway during the course of evolution? One answer to this question is that sulfolipid is of importance under conditions under which phosphate becomes limiting for membrane biosynthesis. As in bacterial null mutants (3, 4), sulfolipid deficiency in the Arabidopsis sqd2 mutant led to growth impairment under phosphate-limiting growth conditions (Fig. 5). Although the effect was subtle, it was measurable clearly under highly controlled laboratory conditions, and one would assume that sulfolipid deficiency may be of even greater disadvantage under naturally occurring conditions. Remarkably, the wild type is able, within limitations, to maintain the relative amount of total anionic thylakoid lipids by reciprocally adjusting the sulfolipid and phosphatidylglycerol contents as phosphate availability changes (6). The lipid profiles for the bacterial and Arabidopsis sulfolipid-deficient null mutants share a common feature: phosphatidylglycerol amounts are not decreasing under phosphate-limiting conditions. Therefore, sulfolipid-deficient null mutants exhaust their phosphate reserves sooner than the corresponding wild types, resulting in reduced growth under these conditions. The fact that this effect occurs in bacteria and plants suggests that sulfolipid/phosphatidylglycerol substitution may be ubiquitous to photosynthetic organisms containing these lipids. It should be pointed out, although, that sulfolipid cannot substitute phosphatidylglycerol fully, which unlike sulfolipid is present prominently in the crystal structure of photosystem I (37) and may have specific functions beyond sulfolipid. However, photosynthetic membranes generally seem to need a certain amount of anionic lipids for proper function, and these can be provided by either phosphatidylglycerol or sulfolipid. One component of the underlying regulatory process involves changes in the transcription of the SQD1 (6) and SQD2 genes (Fig. 2) in response to phosphate availability. It will be interesting to see whether these lipid genes are controlled by a more general phosphate-sensing regulatory pathway or a regulatory pathway sensing biophysical properties of membranes such as the ratio of anionic to neutral lipids.
Acknowledgments
Financial support was provided in part by the U.S. Department of Energy and the Michigan State University Center for Novel Plant Products. We also acknowledge the Arabidopsis Stock Center at Ohio State University for providing the seeds for this study and the Arabidopsis Knock-Out facility at the University of Wisconsin (Madison, WI) for enabling us to isolate the sqd2 mutant.
Abbreviation
- T-DNA
transfer DNA
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF454354).
References
- 1.Barber J, Gounaris K. Photosynth Res. 1986;9:239–249. doi: 10.1007/BF00029747. [DOI] [PubMed] [Google Scholar]
- 2.Benning C. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:53–75. doi: 10.1146/annurev.arplant.49.1.53. [DOI] [PubMed] [Google Scholar]
- 3.Benning C, Beatty J T, Prince R C, Somerville C R. Proc Natl Acad Sci USA. 1993;90:1561–1565. doi: 10.1073/pnas.90.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Güler S, Seeliger A, Härtel H, Renger G, Benning C. J Biol Chem. 1996;271:7501–7507. doi: 10.1074/jbc.271.13.7501. [DOI] [PubMed] [Google Scholar]
- 5.Sato N, Sonoike K, Tsuzuki M, Kawaguchi A. Eur J Biochem. 1995;234:16–23. doi: 10.1111/j.1432-1033.1995.016_c.x. [DOI] [PubMed] [Google Scholar]
- 6.Essigmann B, Güler S, Narang R A, Linke D, Benning C. Proc Natl Acad Sci USA. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Härtel H, Dörmann P, Benning C. Proc Natl Acad Sci USA. 2000;97:10649–10654. doi: 10.1073/pnas.180320497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanda S, Leustek T, Theisen M, Garavito M, Benning C. J Biol Chem. 2001;276:3941–3946. doi: 10.1074/jbc.M008200200. [DOI] [PubMed] [Google Scholar]
- 9.Essigmann B, Hespenheide B M, Kuhn L A, Benning C. Arch Biochem Biophys. 1999;369:30–41. doi: 10.1006/abbi.1999.1344. [DOI] [PubMed] [Google Scholar]
- 10.Mulichak A M, Theisen M J, Essigmann B, Benning C, Garavito R M. Proc Natl Acad Sci USA. 1999;96:13097–13102. doi: 10.1073/pnas.96.23.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seifert U, Heinz E. Bot Acta. 1992;105:197–205. [Google Scholar]
- 12.Weissenmayer B, Geiger O, Benning C. Mol Plant–Microbe Interact. 2000;13:666–672. doi: 10.1094/MPMI.2000.13.6.666. [DOI] [PubMed] [Google Scholar]
- 13.Rossak M, Tietje C, Heinz E, Benning C. J Biol Chem. 1995;270:25792–25797. doi: 10.1074/jbc.270.43.25792. [DOI] [PubMed] [Google Scholar]
- 14.Güler S, Essigmann B, Benning C. J Bacteriol. 2000;182:543–545. doi: 10.1128/jb.182.2.543-545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 16.Estelle M A, Somerville C. Mol Gen Genet. 1987;206:200–206. [Google Scholar]
- 17.Logemann J, Schell J, Willmitzer L. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Dörmann P, Benning C. Plant J. 1998;13:641–652. doi: 10.1046/j.1365-313x.1998.00067.x. [DOI] [PubMed] [Google Scholar]
- 20.Bechtold N, Pelletier G. In: Arabidopsis Protocols. Martinez-Zapater J, Salinas J, editors. Totowa, NJ: Humana Press; 1998. pp. 259–266. [Google Scholar]
- 21.Chang A C, Cohen S N. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dörmann P, Balbo I, Benning C. Science. 1999;284:2181–2184. doi: 10.1126/science.284.5423.2181. [DOI] [PubMed] [Google Scholar]
- 23.Benning C, Huang Z H, Gage D A. Arch Biochem Biophys. 1995;317:103–111. doi: 10.1006/abbi.1995.1141. [DOI] [PubMed] [Google Scholar]
- 24.Rossak M, Schäfer A, Xu N, Gage D A, Benning C. Arch Biochem Biophys. 1997;340:219–230. doi: 10.1006/abbi.1997.9931. [DOI] [PubMed] [Google Scholar]
- 25.Gage D A, Huang Z H, Benning C. Lipids. 1992;27:632–636. doi: 10.1007/BF02536123. [DOI] [PubMed] [Google Scholar]
- 26.Härtel H, Reinhardt I, Grimm B. J Photochem Photobiol B. 1998;43:136–145. [Google Scholar]
- 27.Genty B, Briantais J M, Baker N R. Biochem Biophys Acta. 1989;990:87–92. [Google Scholar]
- 28.The Arabidopsis Genome Initiative. Nature (London) 2001;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 29.Berg S, Edman M, Li L, Wikstrom M, Wieslander A. J Biol Chem. 2001;276:22056–22063. doi: 10.1074/jbc.M102576200. [DOI] [PubMed] [Google Scholar]
- 30.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henrissat B, Coutinho P M, Davies G J. Plant Mol Biol. 2001;47:55–72. [PubMed] [Google Scholar]
- 32.Tietje C, Heinz E. Planta. 1998;206:72–78. [Google Scholar]
- 33.Emanuelsson O, Nielsen H, von Heijne G. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sussman M R, Amasino R M, Young J C, Krysan P J, Austin-Phillips S. Plant Physiol. 2000;124:1465–1467. doi: 10.1104/pp.124.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Härtel H, Essigmann B, Lokstein H, Hoffmann-Benning S, Peters-Kottig M, Benning C. Biochim Biophys Acta. 1998;1415:205–218. doi: 10.1016/s0005-2736(98)00197-7. [DOI] [PubMed] [Google Scholar]
- 36.Benson A A, Daniel H, Wiser R. Proc Natl Acad Sci USA. 1959;45:1582–1587. doi: 10.1073/pnas.45.11.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jordan P, Fromme P, Witt H, Klukas O, Saenger W, Krauss N. Nature (London) 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]