Abstract
The objective of the present study was to make use of efficient molecular marker systems to reveal genetic relationships in traditional and evolved Basmati (EB) and semidwarf non-Basmati (NB) rice varieties. A subset of three rice groups was analyzed by using 19 simple sequence repeat (SSR) loci and 12 inter-SSR-PCR primers. A total of 70 SSR alleles and 481 inter-SSR-PCR markers were revealed in 24 varieties from the three groups. The lowest genetic diversity was observed among the traditional Basmati varieties, whereas the EB varieties showed the highest genetic diversity by both the marker assays. The results indicated that the subset of aromatic rice varieties analyzed in the present study is probably derived from a single land race. The traditional Basmati (TB) and semidwarf NB rice varieties used in the present study were clearly delineated by both marker assays. A number of markers, which could unambiguously distinguish the TB varieties used in the present study from the evolved and NB rice varieties, were identified. The potential use of these markers in Basmati rice-breeding programs and authentication of TB varieties used in the present study are envisaged.
Rice is the staple food for more than half of the world's population. Its rich genetic diversity in the form of thousands of land races and progenitor species, besides its economic significance, has aroused unending interest among scientists for several decades. In the evolution of rice and its genetic differentiation into distinct varietal groups, consumer quality preferences have played a significant role besides agroecological factors. One such varietal group comprising the aromatic pulao/biryani rice of the Indian subcontinent known as “Basmati” is the highly priced rice in domestic as well as international markets. Originating in the foothills of the Himalayas, Basmati rice is characterized by the extralong slender grain, pleasant and distinct aroma, and soft and fluffy texture of the cooked rice. These unique features of Basmati, said to be the culmination of centuries of selection and cultivation by farmers, are well preserved and maintained in their purest form in the traditional Basmati (TB) varieties. Historical and archeological findings imply that varieties with such unique morphological and quality attributes are not present in traditional rice-growing areas anywhere in the world (1). A number of undesirable traits of Basmati, such as tall stature, low yield, sensitivity to photoperiod, and poor response to fertilizer application, prompted breeders to develop “elite” Basmati varieties by making use of the high-yielding semidwarf non-Basmati (NB) rice varieties. Such “elite” evolved lines of Basmati (EB), however, fall short of the quality features of traditional varieties. Difficulty in recovering desirable recombinants from crosses involving NB and TB varieties and reversion often to parental types in the backcross generations suggest that probably indica and Basmati types are phylogenetically divergent (2). A study of Asian rice varieties using isozyme markers clustered Basmati varieties in the group V gene pool, which is well separated from groups I and VI comprising indica and japonica types, respectively (3). Further evidence of the high degree of divergence of Basmati from other indica varieties comes from the high percentage of hybrid sterility (4). The difficulties experienced in evolving “elite” Basmati varieties combining all the desirable traits of TB and NB varieties have retained the preeminent status of TB varieties in the rice industry. Consequently, TB varieties command a considerable price advantage in the market over EB varieties. The adulteration of TB grains with EB and NB grains is reported to be common and thus hampers the Basmati rice export market. Hence, identifying the genuine Basmati variety from the other Basmati-like NB varieties is considered important from the viewpoint of trade.
Traditionally used morphological and chemical parameters have not been found to be discriminative enough, warranting more precise techniques. Several molecular techniques are available for detecting genetic differences within and among cultivars (5–8). Among these, simple sequence repeat (SSR) markers are efficient and cost-effective and detect a significantly higher degree of polymorphism in rice (9–11). They are ideal for genetic diversity studies and intensive genetic mapping (12–14). An alternative method to SSR, called inter-SSR (ISSR)–PCR (15), has also been used to fingerprint the rice varieties (16).
The well-characterized Basmati rice-specific molecular markers could serve as marker tags for Basmati varieties. If the markers are shown to be tightly linked to any of the distinct traits of Basmati, they could be used in marker-assisted selection programs. Such markers could be further verified on the fully sequenced rice genome with regard to their location and linkage to the gene(s) of interest.
In the present study, we automated the ISSR-PCR marker assay to enhance genetic informativeness and used it along with rice SSRs to analyze the genetic relationships of TB, EB, and NB varieties.
Materials and Methods
Rice Materials.
The certified Basmati rice materials used in this study were provided by the Ministry of Commerce, Government of India, and NB varieties by the Directorate of Rice Research, Hyderabad, India. Details of the rice varieties are given in Table 1. DNA was extracted from 5 g of grains from each of the varieties by using the Phytopure plant DNA extraction kit (Pharmacia Amersham Pharmacia Biotech).
Table 1.
List of rice varieties (Oryza sativa) included in the study
| Name | Origin |
|---|---|
| TB | |
| Basmati 217 | Punjab (Indian subcontinent) |
| Basmati 370 | Punjab (Indian subcontinent) |
| Dehraduni (Type-3) | Uttar Pradesh |
| Ranbir Basmati | Jammu and Kashmir |
| Tarori (HBC-19) | Haryana |
| Basmati 386 | India |
| EB | |
| Basmati 385 (385) | TN1 × Basmati 370 |
| Super Basmati (SB) | Basmati 320 × IR 661 |
| Pusa Basmati (PB) | Pusa 150 × Karnal local |
| Kasturi (Kas) | CK 88-17-1-5 × Basmati 370 |
| Haryana Basmati (HB) | Sona × Basmati 370 |
| Mahi Sugantha (MS) | BK 79 × Basmati 370 |
| Haryana Gaurav (HG) | Mutant of Basmati 370 |
| Super (SU) | Equivalent to SB |
| Terricot (TER) | NA |
| Sharbati (SHA) | NA |
| CSR 30B (CSR) | Buraratha 4–10 × Pak Basmati? |
| Semidwarf NB | |
| IR 8 | Peta × Dee-gee-woo-gen |
| Jaya | TN 1 × T 141 |
| Taichung (N) 1 (TC) | NA |
| IR 22 | IR 8 × Tadukan |
| IR 20 | IR 262 × TKM 6 |
| IR BB5 | IR 24 × DZ 192 |
| PR 106 (PR) | IR 8 × Peta 5 × Bella patna |
Abbreviations indicated in parentheses.
DNA Markers and Laboratory Assay.
Two classes of markers were used in the present study: fluorescence-based ISSR-PCR and SSRs.
ISSR-PCR.
The ISSR-PCR method (15) was modified with a view to enhancing the speed and sensitivity of detection of markers. We designed and synthesized 12 5′ and 3′ anchored primers (Table 2). Amplification was performed in 10 mM Tris⋅HCl, pH 8.3 (50 mM KCl/1.5 mM MgCl2/0.01% gelatin/0.01% Triton X-100)/1 mM dNTPs/0.2 μM fluorescent dUTP (TAMARA, Perkin–Elmer)/0.3 unit of AmpliTaq Gold (Perkin–Elmer)/4 μM primer with 5 ng of genomic DNA per 5-μl reaction. Thermal cycling conditions were as follows: initial denaturation of 10 min at 94°C; 35 cycles of 30 s at 94°C; 30 s at 50°C; and 1 min at 72°C; and final extension of 10 min at 72°C. The PCR was performed on a Perkin–Elmer thermal cycler (9600). One microliter of PCR product was mixed with 1.5 μl of 6 × loading buffer (1:4 mixture of loading buffer and formamide; Sigma), and 0.4 μl of GENESCAN-1000 ROX-labeled molecular weight standard (red fluorescence) was included in the loading samples. The samples were denatured at 92°C for 1 min before loading onto an ABI 377 automated sequencer (Applied Biosystems) and electrophoresed on 5% polyacrylamide gel (Long Ranger, FMC) under denaturing conditions containing 7 M urea, in 1 × TBE buffer (90 mM Tris borate, pH 8.3, and 2 mM EDTA). Three replicate experiments were carried out to verify the reproducibility of markers.
Table 2.
List of ISSR primers, marker information, and diversity in three rice groups
| Primers | Sequences | Molecular weight range, bp | Total no. of markers | No. of markers
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TB
|
EB
|
NB
|
||||||||||
| M | P | D | M | P | D | M | P | D | ||||
| 5′ anchored | ||||||||||||
| Y (GA)7 | RAY RAT AY(GA)7 | 150–1,200 | 50 | 31 | 11 | 0.18 | 10 | 40 | 0.56 | 15 | 22 | 0.43 |
| R (CA)7 | GRT RCY GRT R (CA)7 | 150–1,500 | 45 | 20 | 9 | 0.27 | 14 | 27 | 0.51 | 24 | 18 | 0.23 |
| T (CA)7 | GYT ARY RYT(CA)7C | 180–1,200 | 38 | 17 | 6 | 0.16 | 6 | 30 | 0.68 | 16 | 9 | 0.21 |
| Y (GT)9 | CRT AY(GT)9 | 200–1,500 | 61 | 29 | 11 | 0.18 | 6 | 53 | 0.67 | 12 | 21 | 0.42 |
| R (TG)7 | RYA CRY RCA R(TG)7 | 160–1,400 | 42 | 23 | 6 | 0.15 | 14 | 27 | 0.49 | 14 | 16 | 0.29 |
| Y (TG)7 | YAY GYA CAY(TG)7T | 200–1,050 | 33 | 14 | 5 | 0.18 | 7 | 26 | 0.63 | 11 | 13 | 0.34 |
| T3(ATT)4 | RA TYT3(ATT)4 | 250–1,500 | 50 | 3 | 32 | 0.60 | 2 | 47 | 0.72 | 2 | 22 | 0.65 |
| RA(GCT)6 | AYA RA(GCT)6 | 180–760 | 30 | 20 | 4 | 0.07 | 11 | 18 | 0.37 | 13 | 7 | 0.24 |
| Y (ACC)7 | YYR AY(ACC)7A | 200–960 | 33 | 19 | 6 | 0.17 | 9 | 22 | 0.51 | 13 | 7 | 0.19 |
| (GACA)4 | RTY(GACA)4 | 315–1,400 | 18 | 7 | 7 | 0.30 | 4 | 14 | 0.59 | 5 | 10 | 0.60 |
| 3′ anchored | ||||||||||||
| (GA)8R | (GA)8RGY | 150–1,150 | 49 | 28 | 10 | 0.17 | 18 | 30 | 0.45 | 20 | 13 | 0.23 |
| (GT)8R | (GT)8RYR Y | 140–960 | 32 | 14 | 8 | 0.27 | 4 | 26 | 0.63 | 10 | 8 | 0.28 |
| Total | 481 | 0.2 ± 0.13 | 0.5 ± 0.10 | 0.3 ± 0.15 | ||||||||
M, monomorphic; P, polymorphic; D, diversity.
Selection of Primers and SSR Survey.
We selected 19 primer pairs (Table 3) from the list of 351 rice microsatellite loci displayed on the Cornell University Rice Genes web site (http://www.gramene.org/microsat/microsats.txt) for analysis. The primers for the selected loci were synthesized by Research Genetics (Huntsville, AL). Wherever possible, at least two loci with nonoverlapping alleles were multiplexed in the PCR reaction to increase the efficiency of genotyping.
Table 3.
Microsatellite marker information, allele distribution, and diversity in three rice groups
| Locus | Repeat motif | Total no. of alleles | Allele size range, bp | No. of alleles
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TB
|
EB
|
NB
|
||||||||||
| M | P | D | M | P | D | M | P | D | ||||
| RM 224 | (GA)13 | 4 | 130–155 | 1 | 1 | 0.49 | 0 | 3 | 0.74 | 0 | 4 | 0.91 |
| RM 16 | (GA)15 | 3 | 165–225 | 1 | 0 | 0.00 | 0 | 2 | 0.95 | 0 | 2 | 0.83 |
| RM 13 | (GA)16 | 6 | 135–158 | 0 | 2 | 0.62 | 0 | 4 | 0.91 | 0 | 4 | 0.86 |
| RM 252 | (GA)19 | 8 | 195–255 | 0 | 4 | 0.86 | 0 | 4 | 0.92 | 0 | 3 | 0.82 |
| RM 235 | (GA)24 | 3 | 102–138 | 0 | 2 | 0.62 | 0 | 2 | 0.74 | 0 | 3 | 0.82 |
| RM 234 | (GA)25 | 2 | 148–150 | 1 | 0 | 0.00 | 0 | 2 | 0.75 | 1 | 0 | 0.00 |
| RM 223 | (GA)25 | 4 | 170–180 | 1 | 0 | 0.00 | 0 | 4 | 0.88 | 1 | 0 | 0.00 |
| RM 1 | (GA)26 | 3 | 85–110 | 1 | 0 | 0.00 | 0 | 3 | 0.86 | 0 | 2 | 0.38 |
| RM 310 | (GT)19 | 5 | 85–110 | 0 | 2 | 0.27 | 0 | 4 | 0.92 | 0 | 3 | 0.88 |
| RM 302 | (GT)30(AT)8 | 4 | 136–205 | 1 | 0 | 0.00 | 0 | 4 | 0.93 | 0 | 2 | 0.62 |
| RM 160 | (GAA)23 | 2 | 100–105 | 1 | 0 | 0.00 | 0 | 2 | 0.71 | 1 | 0 | 0.00 |
| RM 330 | (CAT)5 | 5 | 160–225 | 3 | 0 | 0.00 | 1 | 3 | 0.42 | 1 | 3 | 0.26 |
| RM 72 | (TAT)5C(ATT)15 | 3 | 150–175 | 0 | 2 | 0.62 | 0 | 3 | 0.89 | 0 | 2 | 0.82 |
| RM 102 | (GGC)7(CG)6 | 3 | 435–445 | 2 | 0 | 0.00 | 0 | 3 | 0.58 | 1 | 2 | 0.42 |
| RM 171 | (GATG)5 | 3 | 325–346 | 0 | 2 | 0.49 | 0 | 2 | 0.77 | 0 | 2 | 0.55 |
| RM 163 | (GGAGA)4(GA)11C(GA)20 | 4 | 130–175 | 1 | 0 | 0.00 | 0 | 3 | 0.86 | 0 | 4 | 0.89 |
| RM 161 | (AG)20 | 3 | 163–180 | 1 | 1 | 0.49 | 0 | 2 | 0.74 | 1 | 0 | 0.00 |
| RM 136 | (AGG)7 | 3 | 100–124 | 1 | 0 | 0.00 | 0 | 2 | 0.71 | 1 | 1 | 0.49 |
| RM 238 | NA | 2 | 130–150 | 1 | 0 | 0.00 | 0 | 2 | 0.85 | 1 | 0 | 0.00 |
| Total no. of alleles | 70 | 16 | 16 | 0.2 ± 0.30 | 1 | 54 | 0.7 ± 0.13 | 8 | 37 | 0.5 ± 0.36 | ||
M, monomorphic; P, polymorphic; D, diversity.
PCR amplification was performed in a 5-μl volume containing 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.25 unit of AmpliTaq Gold (Perkin–Elmer), 50 μM of dNTPs, 0.2 μM fluorescent dUTP (TAMARA or R110 or R6G, Perkin–Elmer), and 0.35 μM of each primer with 5 ng of genomic DNA on a Thermal Cycler PTC 100 (MJ Research). The basic PCR program used to amplify the SSR DNA was as described in the Rice Genes web site. The sample preparation, loading, and electrophoretic conditions were as described under ISSR-PCR. When the allelic polymorphisms of any of the microsatellite loci revealed a >20-bp difference between the varieties, such loci were resolved on 3.5% MetaPhor agarose gels (FMC).
Evaluation of Polymorphisms and Data Analysis.
Polymorphic products from SSR and ISSR analyses were scored qualitatively for presence (+) or absence (−). The proportion of bands that were shared between any of the two varieties screened averaged over loci (SSRs) and primers (ISSR-PCR) were used as the measure of similarity. Genetic diversity (5) was calculated as follows:
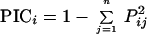 |
where Pij is the frequency of the jth allele for marker i and the summation extends over n alleles. The calculation was based on the number of alleles/locus in SSR and the number of bands/primer in the case of ISSR.
Cluster analysis was based on distance matrices by using the unweighted pair group method analysis (upgma) program in winboot software (17). The relationships between varieties were represented graphically in the form of dendrograms.
Results
Evaluation of ISSR-PCR Markers.
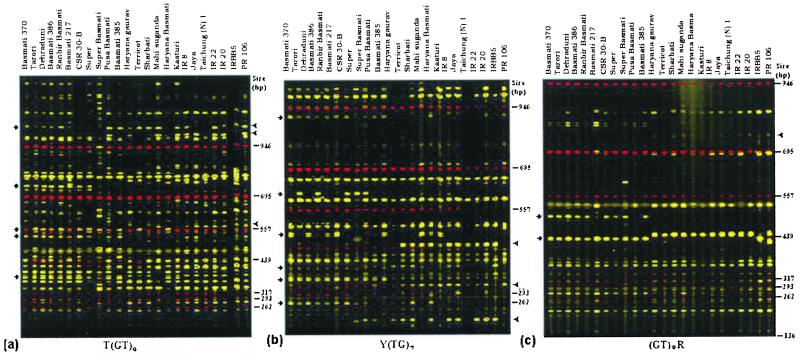
The fluorescence-based ISSR-PCR markers could be clearly resolved on an ABI automated sequencing gel. All 12 of the anchored SSR motifs used in the present study produced varying number of DNA fragments in a different size range (Table 2, Fig. 1). The average number of bands produced by ISSR primers with different repeat motifs was negatively correlated with the number of nucleotides in the repeat unit of the motif. For example, among the 5′ anchored primers, the dinucleotide-based primers produced more bands (43.8 ± 9.82) than tri- (37.6 ± 7.89) and tetranucleotide (18) primers. Although we have not used a similar number of primers in each repeat class of primers for such a comparison, the number of products amplified in different repeat length classes reflected the frequency of different repeat motifs distributed in the rice genome. The 12 primers produced a total of 481 PCR products, of which 389 (80.9%) were polymorphic in all 24 rice varieties. There appeared to be no correlation between the number of bands amplified and the degree of polymorphisms. For example, the primers R(TG)7 and Y(TG)7 generated 42 and 33 bands, respectively, of which 73.8 and 78.8% were polymorphic; on the other hand, 98% of the 50 bands amplified by T3(ATT)4 primer were polymorphic. Among the 12 primers, two 5′ anchored, T3(ATT)4 and T(GT)9 and one 3′ anchored primer (GT)8R, generated more than 90% scorable polymorphisms.
Figure 1.
Fluorescence-based ISSR profiles of TB, EB, and NB rice varieties for three representative primers: (a) T(GT)9; (b) Y(TG)7; and (c) (GT)8R. Arrows and arrowheads indicate the markers that differentiate Basmati and NB rice varieties, respectively.
Because our efforts were directed toward comparative genetic analysis of TB, EB, and NB rice varieties, we evaluated the polymorphisms in these three groups. The degree of polymorphisms differed substantially among the three groups. The TB varieties showed a very low level of polymorphism compared with EB and NB varieties. Of 340 bands scored in the TB varieties, only 115 (33.8%) were polymorphic (Table 2). On the other hand, 360 (77.4%) of 465 bands were polymorphic in the EB, and 166 (51.7%) of 321 bands were polymorphic in NB varieties. The primer T3(ATT)4 was found to produce the most polymorphic bands in all three rice groups: TB (91.4%), EB (92%), and NB (91.66), whereas RA(GCT)6 generated the least degree of polymorphisms (Table 2).
Genetic diversity was calculated for each of the ISSR primers in all three rice groups (Table 2). The difference in diversity values was as striking as the degree of polymorphism. The two primers, T3(ATT)4 and RA(GCT)6, that recorded the highest and lowest polymorphisms, respectively, in the three groups also recorded the highest and lowest diversity values. The 12 primers had an average diversity value of 0.23 ± 0.132 with a range of 0.07 to 0.6 for TB. The diversity values ranged from 0.37 to 0.72, averaging 0.57 ± 0.10 for the EB, and 0.19 to 0.65 with a mean of 0.34 ± 0.15 for the NB varieties. The difference was significant (P < 0.001 between TB and EB; P < 0.10 between TB and NB groups). Of the 12 ISSR-PCR primers, 5 revealed 12 PCR products, a combination of which could distinguish Basmati from NB varieties (data not shown) (Fig. 1). On the other hand, 21 specific PCR products generated by 9 ISSR-PCR primers could distinguish EB from NB varieties (data not shown) (Fig. 1). By making use of two to three informative primers, the TB from the EB and NB varieties could be unambiguously distinguished. For example, by using two primers, T(GT)9 and (GT)8R, one can make a decision whether a given variety is a TB or an EB rice variety.
Evaluation of SSR Polymorphisms.
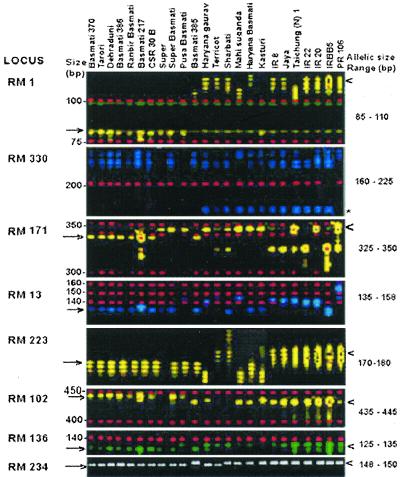
We also used 19 microsatellite loci for the genetic analysis of the three rice groups. Table 3 summarizes the total number of alleles detected and their size range across 24 rice varieties for each of the 19 microsatellite loci used. The number of alleles ranged from two to eight, with an average of 3.8 alleles. Only RM 252 detected a maximum of eight alleles. As could be seen from Table 3, there appears to be no correlation between the number of alleles detected and the number of SSR repeats in the SSR loci. For example, the microsatellite loci containing the (GA) repeat motifs varying from (GA)15 to (GA)25 did not show any correlation with the number of alleles they revealed. The allele number varied between the three rice groups (Table 3). More alleles were resolved in the EB (56) and NB (42) as compared with TB (28) varieties. Of 19 loci, only eight revealed polymorphisms in the TB varieties, whereas 14 were polymorphic in the NB varieties, and all of the loci were polymorphic in the EB varieties (Table 3, Fig. 2). The diversity values also varied from one locus to another and between the three rice groups (Table 3). An average diversity of 0.23 ± 0.3 was observed for the TB group, whereas EB and NB rice groups recorded much higher diversity values of 0.79 ± 0.13 and 0.50 ± 0.36, respectively. However, the extent of variation in average diversity is not as large as the difference in the number of alleles per locus, and it does not appear to correlate with the number of alleles. Across the three rice groups, the highest diversity of 0.87 was observed for RM 252 with eight alleles, whereas RM 72 and RM 16 with three alleles each displayed diversities of 0.70 and 0.76, respectively. The difference in diversity between the three groups was statistically significant (P < 0.001 between TB and EB; P < 0.05 between the TB and NB groups).
Figure 2.
Fluorescence-based SSR polymorphisms detected by eight representative SSR loci in TB, EB, and NB rice varieties. Arrows and arrowheads indicate the markers that differentiate Basmati and NB rice varieties, respectively. The asterisk indicates the duplication of locus 330 only in NB and some of the EB varieties. The first seven loci are genescan images, and the last locus (RM 234) is a 3.5% Metaphor agarose run ethidium bromide-stained image.
We scored the SSR alleles, which showed a preponderance in Basmati varieties. Of 70 alleles, 9 were found only in TB and in some of the EB varieties. These alleles were absent in all seven NB varieties analyzed in the present study (Table 4). Further analysis of seven more NB varieties confirmed that these nine alleles are confined only to the TB varieties analyzed in the present study (data not shown). Of 19 loci, one locus RM 330 was found to be duplicated only in NB varieties (except PR 106), and none of the TB varieties showed any such duplication for this locus (Fig. 2). None of the 19 SSR loci could distinguish the TB from the EB varieties independently. However, a combination of polymorphic loci with different Basmati-specific alleles enabled discrimination of the traditional from the evolved ones except CSR 30B. For example, RM 171 locus in combination with RM 238 or RM 16 or RM 302 could discriminate all traditional ones from the evolved ones (except CSR 30B) (Table 5).
Table 4.
SSR loci that distinguish Basmati and NB rice varieties
| SSR loci | Allele size, bp | TB | NB |
|---|---|---|---|
| RM 163 | 162 | − | + (only PR, TC) |
| RM 171 | 338 | + | − |
| RM 161 | 180 | + | − |
| RM 72 | 175 | + | − |
| RM 1 | 85 | + | − |
| RM 13 | 135 | + | − |
| RM 102 | 448 | + | − |
| RM 16 | 225 | − | + (only Jaya) |
| RM 16 | 165 | + | − |
| RM 330 | 160 | − | + (except IR 8, PR) |
| RM 302 | 140 | + | − |
| RM 252 | 255 | − | + (only IR20) |
| RM 252 | 228 | − | + (only TC) |
| RM 234 | 148 | + | − |
| RM 223 | 200 | + | − |
| RM 136 | 125 | + | − |
Table 5.
SSR loci that distinguish TB and EB rice varieties
| Locus | Allele size, bp | TB | EB |
|---|---|---|---|
| RM 163 | 140 | − | + (except 385, SB, SU) |
| RM 171 | 345 | − | + (except 385, TER) |
| RM 171 | 338 | + | − (except 385 CSR) |
| RM 72 | 175 | + | − (except 385, SB) |
| RM 238 | 140 | + | − (except SU, PB, SB) |
| RM 102 | 448 | + | − (except PB, SB) |
| RM 16 | 165 | + | − (except SB, HB) |
| RM 302 | 140 | + | − (except SU, PB, SB) |
| RM 330 | 180 | − | + (except 385, PB, SB, SU) |
| RM 136 | 124 | − | + (except 385, SU, SB) |
Cluster Analyses.
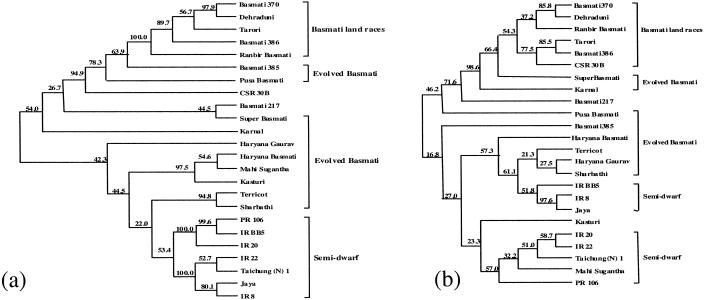
The ISSR-PCR and SSR profiles were used to determine the genetic similarity matrices, which were then used to construct dendrograms. Both methods separated the TB from the NB varieties (Fig. 3). The EB varieties got clustered in between, depending on the degree of genetic similarity to the two groups. In ISSR-PCR analysis, all of the TB varieties, except Basmati 217, displayed 95% similarity among themselves. Genetic similarity estimates obtained by SSR analysis also revealed similar results. Both marker assays included the land race Basmati 217 along with the EB varieties. As revealed by ISSR analysis, among the EB varieties, CSR, SB, 385, PB, and SU showed higher genetic similarity to the TB varieties as compared with the other EB varieties, which showed a higher similarity to NB varieties (Fig. 3).
Figure 3.
Dendrograms derived from an unweighted pair group method analysis (upgma) cluster analysis by using Nei and Li Coefficients based on (a) ISSR markers and (b) SSR markers. Numbers on the nodes indicate the number of times a particular branch was recorded per 100 bootstrap replications after 1,000 replications.
Discussion
The TB and EB varieties included in the present study probably represent a major component of the Basmati gene pool of the Indian subcontinent. In addition to the Basmati varieties, we also included in our study many semidwarf NB varieties, some of which have been used for development of EB varieties. We used two molecular marker assays, fluorescence-based ISSR-PCR and SSR; the former is an improvised version of the ISSR-PCR method developed earlier (15).
The number of bands produced across 24 rice varieties by different anchored SSR motifs is consistent with published reports on microsatellite frequency in the rice genome. Among dinucleotide repeats, (GA)n and (CA)n are the most abundant in the rice genome (9, 12, 14). Both repeat classes were amenable to fluorescence-based ISSR-PCR analysis of the rice genome, as both were equally polymorphic. Of the two 3′ anchored primers, one with the (GA)n and another with the (GT)n motif, (GA)n produced more bands, probably because of its greater abundance in the rice genome as reported earlier (9, 14). The two (GT)n-based primers, one anchored at the 5′ and the other at the 3′ end, amplified 61 and 31 bands, respectively. Although comparison of only two primers may not allow us to make definitive conclusions, taken together with earlier inferences (16), such a difference may be due to the lack of selective nucleotides at the 3′ end of the 5′ anchored primers. On the other hand, 5′ and 3′ anchored (GA)n repeats did not reveal such a difference, probably because 5′ anchored primers impose selection for a long stretch of SSRs, whereas amplification with the 3′ anchored primer would not impose selection for repeat length. Because (GA)n motifs are reported to be longer in the rice genome as compared with (GT)n motifs, both 5′ and 3′ anchored (GA)n primers produce a similar number of bands. On the other hand, the lack of length advantage in the (GT)n motif probably results in a difference between 5′ and 3′ end anchored (GT)n primers.
All (ATT)n and (GACA)4 anchored primers amplified a large number of bands in the present study, in contrast to earlier studies (16). Although (GACA)n repeats are fewer than the di- and trinucleotide repeats, they appear to be in good number in the rice genome, as reported earlier (18). Because the lengths of tri- and tetranucleotide repeats in the rice genome are mostly 5 to 8 and 5 to 6, respectively, the longer repeat motifs used by earlier studies (16) would have precluded amplification. Our results indicate that tri- and tetranucleotide-based ISSR-PCR markers could provide potential markers in the rice genome.
There has been a wide range of interest in the genetic differences between TB, EB, and NB rice varieties. The information available on genetic diversity and differences in the three groups is scanty except for the Asian rice varieties using isozyme markers (3). The results of the present study using fluorescence-based ISSR and SSR markers indicate that TB varieties have the least diversity compared with the EB and NB varieties. Besides, both marker assays showed that there are no significant differences between the TB varieties. We believe the high degree of genetic similarity among the TB varieties indicates that they are possibly the descendants of a single land race, and the minor genetic variation is maintained as a result of the selection and preference imposed by farmers for several years. This observation draws support from the historical relationship of Basmati varieties used in the present study. Most TB varieties classified under different names are likely to have been selected from the local variety such as Basmati 370, which was released for commercial cultivation in 1933 at the Rice Research Station, Kalashah Kaku (now in Pakistan). For example, the isozyme patterns of 60 of 65 Pakistani accessions described as Basmati matched the isozyme pattern of Basmati 370 and Type 3. Similarly, of the nine varieties from India, all except Karnal local were identical to the isozyme pattern of Basmati 370 and Type 3 (1). Among the 19 (19) and 5 (20) EB varieties released since 1965 for cultivation in India and Pakistan, respectively, 12 and 4 had Basmati 370 as one of the donor parents. Our studies, along with these reports, suggest that the TB varieties used in the present study could be considered the bulk of the narrow TB gene pool of the Indian subcontinent. A recent report on random amplified polymorphic DNA profiling of aromatic rices also shows a low level of genetic diversity (21). The variety Basmati 217, which we received as land race, showed only 75 and 66% similarities, whereas CSR 30B, which was received as EB, showed 82 and 96% similarities based on ISSR and SSR marker assays, respectively, to the five TB varieties. Considering the genetic diversity in the other EB varieties, it is unlikely that CSR 30B evolved from a direct cross between a Pakistani Basmati variety, and the salt-tolerant NB variety, Buraratha, has attained such a high level of genetic similarity to the TB varieties. However, further insights into its grain quality vis à vis TB and EB varieties and the pedigree details may be required for ascertaining its status. A reasonable explanation for the higher genetic distance of Basmati 270 from the other TB varieties is that it has differentiated from the rest of the TB varieties and probably represents a separate lineage. This argument is supported by the observation that, of the 70 SSR alleles among 24 varieties, 3 were unique to Basmati 217. The high level of genetic diversity and preponderance of NB alleles in most EB varieties indicate that most of them still retain a large genomic fraction of the NB varieties used in the breeding program.
Both marker assays clearly differentiate the Basmati and NB varieties as highlighted by isozyme and restriction fragment length polymorphism studies (3). The high level of genetic differentiation of Basmati and NB rice varieties suggests that the former might have possibly diverged a long time ago from NB varieties through conscious selection and patronage. The duplication event at locus RM 330 only in semidwarf NB varieties, including the old japonica varieties Taipai and Wu 10B, supports the divergence of aromatic varieties from the common ancestor. It would be interesting to study the other varieties in Group V, which embraces most of the long grain varieties (3), for a duplication event in this locus. The high-level genetic differentiation of the two groups may be the reason for the lack of finding desirable recombinants and introgression of useful genes in Basmati breeding programs (2), as reflected by the preponderance of NB alleles in most EB varieties. Some of the bands/alleles unique to the TB varieties used in the present study are not found at all in any of the EB varieties, either because of incompatible chromosome regions (coadapted gene complex) or because they are possibly linked to the negative traits of Basmati and are thus selected against. On the other hand, alleles from the NB varieties have survived in greater numbers in the EB varieties, possibly because they are in close proximity to the genes, which confer useful traits of NB varieties.
The high resolution of the fluorescence-based ISSR-PCR assay described in the present study provides a large number of highly reproducible markers with as little as 5 ng of template DNA per PCR reaction without using radioactive isotopes or chemicals involved in silver staining. By varying SSR motifs and their anchors, a large number of markers could be generated that can be used for further saturation of the rice genome. We believe that, because the bulk of the global Basmati rice trade and breeding programs center around the TB varieties analyzed in the present study, the combination of SSR and ISSR markers can be used to identify these varieties from NB as well as EB varieties. These markers will also allow the determination of adulteration of this set of traditional Basmati varieties. It is hazardous to venture to the conclusion that the markers observed in the present study could be universally applied to all aromatic rice varieties. Further studies on geographically random samples of aromatic rice varieties would probably give a more complete and less discontinuous picture with regard to the allelic/marker association. Nevertheless, the markers specific to the TB varieties used in the present study should be further pursued to look for allelic association, if any, with the Basmati phenotype. Such a study would provide markers that would help eliminate unnecessary chromosome regions in the early stages of backcrossing, thus helping breeders shorten breeding cycles by rapid incorporation of Basmati traits into breeding lines.
Acknowledgments
Financial support to J.N. by Department of Biotechnology, New Delhi, is gratefully acknowledged.
Abbreviations
- TB
traditional Basmati
- EB
evolved Basmati
- NB
semidwarf non-Basmati
- SSR
simple sequence repeat
- ISSR
inter-SSR
References
- 1.Singh V P. In: Aromatic Rices. Singh R K, Singh U S, Khush G S, editors. New Delhi: Oxford and India Book House; 2000. pp. 135–153. [Google Scholar]
- 2.Khush G S, Paule C M, De La Cruz N M. Proc. Workshop on Chemical Aspects of Rice Grain Quality. Manila, The Philippines: Int. Rice Res. Inst.; 1979. pp. 21–31. [Google Scholar]
- 3.Glaszmann J C. Theor Appl Genet. 1987;74:21–30. doi: 10.1007/BF00290078. [DOI] [PubMed] [Google Scholar]
- 4.Engle L M, Chang T T, Ramirez D A. Philipp Agric. 1969;53:289–307. [Google Scholar]
- 5.Botstein D R, White R L, Skolnick M, Davis R W. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 6.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingy S V. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber J L, May P E. Am J Hum Genet. 1989;44:388–396. [PMC free article] [PubMed] [Google Scholar]
- 8.Vos P, Hogers R, Bleekr M, Reijans M, Zabeau M. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu K S, Tanksley S D. Mol Gen Genet. 1993;241:225–235. doi: 10.1007/BF00280220. [DOI] [PubMed] [Google Scholar]
- 10.Panaud O, Chen X, McCouch S R. Genome. 1995;38:1170–1176. doi: 10.1139/g95-155. [DOI] [PubMed] [Google Scholar]
- 11.Yang G P, Saghai Maroof M A, Xu C G, Zhang Q, Biyashev R M. Mol Gen Genet. 1994;245:187–194. doi: 10.1007/BF00283266. [DOI] [PubMed] [Google Scholar]
- 12.Cho Y G, Ishii T, Temnykh S, Chen X, Lipovich L, McCouch S R, Park W D, Ayres N, Cartinhour S. Theor Appl Genet. 2000;100:713–722. [Google Scholar]
- 13.Akagi H, Yokozeki Y, Inagaki A, Fujimura T. Theor Appl Genet. 1996;93:1071–1077. doi: 10.1007/BF00230127. [DOI] [PubMed] [Google Scholar]
- 14.Temnykh S, Park W D, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho Y G, Ishii T, McCouch S R. Theor Appl Genet. 2000;100:698–712. [Google Scholar]
- 15.Zietkiewicz E, Rafalski A, Labuda D. Genomics. 1994;20:176–183. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]
- 16.Blair M W, Panaud O, McCouch S R. Theor Appl Genet. 1999;98:780–792. [Google Scholar]
- 17.Yap I, Nelson R J. IRRI Discussion Paper Series No. 14. Manila, The Philippines: Int. Rice Res. Inst.; 1996. [Google Scholar]
- 18.Ramakrishna W, Lagu M D, Gupta V S, Ranjekar P K. Theor Appl Genet. 1994;88:402–406. doi: 10.1007/BF00223651. [DOI] [PubMed] [Google Scholar]
- 19.Kumar S, Rani N S, Krishniah K. Proc. Workshop on Problems and Prospects of Aromatic Rices in India. 1996. (Sept. 21–Oct. 10, 1996, Int. Rice Res. Inst., Manila, The Philippines), pp. 21–29. [Google Scholar]
- 20.Khan M G. Proc. Workshop on Problems and Prospects of Aromatic Rices in India. 1996. (Sept. 21–Oct. 10, 1996, Int. Rice Res. Inst., Manila, The Philippines), pp. 96–101. [Google Scholar]
- 21.Ray Choudhury P, Kohli S, Srinivasan K, Mohapatra T, Sharma R P. Euphytica. 2001;118:243–251. [Google Scholar]