Abstract
Polyunsaturated fatty acids (PUFAs) are important membrane components and precursors of signaling molecules. To investigate the roles of these fatty acids in growth, development, and neurological function in an animal system, we isolated Caenorhabditis elegans mutants deficient in PUFA synthesis by direct analysis of fatty acid composition. C. elegans possesses all the desaturase and elongase activities to synthesize arachidonic acid and eicosapentaenoic acid from saturated fatty acid precursors. In our screen we identified mutants with defects in each fatty acid desaturation and elongation step of the PUFA biosynthetic pathway. The fatty acid compositions of the mutants reveal the substrate preferences of the desaturase and elongase enzymes and clearly demarcate the steps of this pathway. The mutants show that C. elegans does not require n3 or Δ5-unsaturated PUFAs for normal development under laboratory conditions. However, mutants with more severe PUFA deficiencies display growth and neurological defects. The mutants provide tools for investigating the roles of PUFAs in membrane biology and cell function in this animal model.
Polyunsaturated fatty acid (PUFA) components of phospholipids are necessary to create a fluid membrane environment. In mammals, specific PUFA composition affects many cellular processes including modulation of ion channels (1, 2), endocytosis/exocytosis (3), and activities of membrane-associated enzymes that are sensitive to the biophysical properties of lipid membranes (4). In addition, certain classes of PUFAs are precursors of powerful eicosanoid effectors such as prostaglandins and leukotrienes (5). Disruptions in proper PUFA intake and metabolism are associated with certain human disease states such as coronary artery disease, hypertension, diabetes, inflammatory disorders, and cancer (6). In most cases the mechanisms through which fatty acid composition affects these conditions are not well understood.
Mammals require the essential fatty acids linoleic acid (18:2n6) and linolenic acid (18:3n3) in their diet. Desaturation and elongation of these fatty acids produce the C20 PUFAs that become membrane components and precursors for eicosanoids. Plants produce these essential n6 and n3 PUFAs by desaturating oleic acid (18:1n9) at C12 and C15 positions to produce 18:2n6 and 18:3n3 but rarely possess the enzymes necessary to desaturate and elongate them further into C20 PUFAs. The free-living nematode Caenorhabditis elegans synthesizes a wide range of PUFAs including arachidonic acid (20:4n6) and eicosapentaenoic acid (20:5n3) by using saturated fatty acids obtained from their Escherichia coli diet (7). To this end, C. elegans expresses the full range of desaturase activities found in plants (Δ12 and n3 desaturase) and animals (Δ5 and Δ6 desaturase) as well as n6 and n3 PUFA elongase activities found in animals. cDNAs corresponding to C. elegans n3, Δ12, Δ6, and Δ5 desaturase genes have been expressed in Arabidopsis and yeast to confirm their desaturase activity (8–11). Another C. elegans gene confers the ability to elongate γ-linolenic acid (18:3n6) and stearidonic acid (18:4n3) when expressed in yeast (12). This gene, along with seven other C. elegans homologues presumably belonging to the same gene family, contains regions of similarity both to the yeast protein Elo1p, necessary for the elongation of myristic acid (14:0) to palmitic acid (16:0; ref. 13), and to Elo2p and Elo3p, which are involved in elongation of very long chain fatty acids necessary for sphingolipid formation in yeast (14).
The C. elegans fatty acid desaturases described above belong to a family of diiron-oxo integral membrane proteins that require cytochrome b5 and NADH-cytochrome b5 reductase as cofactors (15). Their protein sequences contain conserved motifs consisting of three histidine-rich sequences (H-boxes) that are believed to be the residues that coordinate the diiron-oxo structure at the active sites (16) as well as two long stretches (>40 residues each) of hydrophobic residues that appear to anchor the protein into the lipid bilayer and situate the H-boxes into proper position in the active site. Site-directed mutagenesis studies reveal that each of the histidine residues is critical for enzyme function (17). It is these highly conserved features of the protein sequence that has allowed extensive cloning of genes encoding desaturases and related enzyme functions. In contrast, the fatty acid elongase enzymes are not well characterized. Fatty acid elongation is a four-step process initiated by the condensation of malonyl-CoA with a long chain acyl-CoA, yielding a 3-ketoacyl-CoA in which the acyl moiety has been elongated by two carbon atoms. In subsequent steps, the 3-ketoacyl-CoA is reduced to 3-hydroxyacyl-CoA, which is dehydrated to an enoyl-CoA, and further reduced to yield the elongated acyl-CoA. The condensation reaction is the rate-limiting and substrate-specific step (18). Studies on the mammalian LCE gene, related to the yeast ELO family, recently provided evidence that the enzymes of this gene family mediate the condensation step of the fatty acid elongation system (19).
To investigate the fatty acid desaturation/elongation pathway in vivo and generate tools for examining the roles of PUFAs in development and behavior in an animal system, we isolated C. elegans mutants defective in fatty acid desaturation and elongation. In this article we report the characterization of a series of mutants that clearly demarcate the pathway of PUFA biosynthesis in this animal and reveal the substrate preferences of desaturase/elongase enzymes.
Materials and Methods
Nematodes were cultured and maintained on agar plates according to standard methods at 20°C (20). The C. elegans wild-type (WT) strain was N2 (Bristol), and the E. coli used for feeding nematode cultures was strain OP50.
Isolation of fat and elo Mutants.
Mutations that resulted in alteration of fatty acid composition were isolated after ethyl methanesulfonate mutagenesis in a screen that relied on gas chromatography (GC) analysis of nematode fatty acid composition. WT L4 hermaphrodites (F0) were mutagenized with 50 mM ethyl methanesulfonate in M9 buffer for 4 h at room temperature. Individual F1 self-progeny were transferred to separate 6-cm plates and allowed to produce F3. Within 1 day of the clearing of the bacterial lawn, most of the F2 + F3 progeny were washed off the plate, and their overall fatty acid composition was determined (see below). For samples that produced a fatty acid profile different from WT, 20 F3 larvae were recovered and plated individually on 6-cm plates. The nematodes were allowed to self-propagate until they produced two generations and were harvested within 1 day of clearing the bacterial lawn. Segregation ratios of different fatty acid profiles were noted, and those lines producing the most extreme fatty acid composition defects were tested for homozygosity by replating another generation and analyzing the total fatty acids. Homozygous strains were backcrossed to WT 2–5 × before being used in the experiments reported here. For growth comparisons, 10 2-fold stage embryos were transferred to a fresh culture plate and allowed to grow at 20°C for 72 h.
Fatty Acid Analysis.
To determine overall fatty acid composition of nematodes, several thousand larvae and adult worms was washed off a single 6-cm plate with water and placed into glass screw-capped centrifuge tubes. After centrifugation at 1,000 × g for 1 min, as much water as possible was removed with a Pasteur pipette and replaced with 1 ml of 2.5% H2SO4 in methanol to extract fatty acids from tissues and transmethylate them. The samples were capped and incubated at 80°C for 1 h. After the addition of 0.2 ml of hexane and 1.5 ml of H2O, the fatty acid methyl esters were extracted into the hexane layer by shaking and centrifuging the tubes at low speed. Samples (1-μl) of the organic phase were analyzed by GC using an Agilent 6890 series gas chromatographer equipped with a 30 × 0.25-mm SP-2380 column (Supelco), helium as the carrier gas at 1.4 ml/min, and a flame ionization detector. The gas chromatograph was programmed for an initial temperature of 120°C for 1 min followed by an increase of 10°C/min to 190°C followed by an increase of 2°C/min to 200°C. We found that as few as 50 adult nematodes are required for a reproducible GC signal, and that the fatty acid composition determined by the direct methylation of nematodes is indistinguishable from the fatty acid composition of total lipid extracted with chloroform/methanol (data not shown). 4,4-Dimethyloxazoline derivatives of fatty acid methyl esters were prepared and analyzed as described in ref. 11.
Sequencing of Mutant Alleles.
Total RNA was prepared from mixed-stage nematodes by using Trizol reagent (Life Technologies, Rockville, MD). Reverse transcription was performed with Superscript II reverse transcriptase (Life Technologies) followed by PCR using primers designed to amplify sequences corresponding to the ORF of the gene of interest. The PCR products were gel-purified, and fragments were sequenced directly by using the amplification primers and internal sequencing primers. Each detected mutation was confirmed from at least two independent reverse transcription reactions.
Results
Identification of C. elegans Mutants with Altered Fatty Acid Composition.
We isolated mutants without selection by direct analysis of fatty acid composition of nematode populations derived from single F1 progeny of ethyl methanesulfonate-mutagenized worms. Each F1 worm was grown on an individual plate and allowed to self-propagate until the bacterial lawn was cleared, usually for two generations (6–8 days). A sample of the resulting population was harvested within 1 day of the clearing of the E. coli lawn and prepared for GC analysis (see Materials and Methods). Single F3 progeny from populations exhibiting altered fatty acid compositions were allowed to propagate and were analyzed to confirm the fatty acid change and determine segregation patterns.
fat-1 Mutants Lack n3 PUFAs.
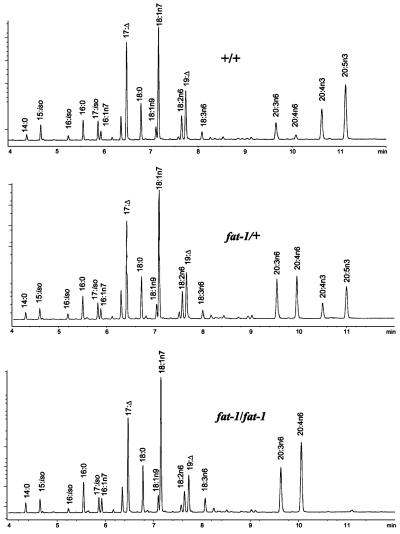
The GC analysis of populations produced from mutated F1 hermaphrodites revealed three independent lines that displayed a reduction in the n3 PUFAs 20:4n3 and 20:5n3. These lines contained ≈17% n3 PUFAs compared with 27% in WT controls. Concomitantly, the C20 n6 precursors (20:3n6 + 20:4n6) increased from 7.8% in WT to 13.7% in the mutant. Analysis of populations derived from single F3 generation worms revealed three segregating classes (Fig. 1): (i) a WT fatty acid composition, (ii) the originally identified reduced n3 composition, and (iii) a fatty acid profile consisting of no detectable n3 PUFAs and correspondingly increased levels of the C20 n6 fatty acids (20:3n6 + 20:4n6 = 35.5%). Populations derived from individuals of the first and third class always had fatty acid compositions identical to their parents, whereas the progeny of the second class fell into all three classes, which indicates that the reduction in n3 fatty acids was caused by a single nuclear mutation, and that members of the third class were homozygous for the mutant allele.
Figure 1.
Mutations in the fat-1 gene result in the loss of n3 PUFA. GC traces of populations of worms of the following genotypes: +/+, WT; fat-1/+, segregating population consisting of WT, fat-1 heterozygotes, and fat-1 homozygotes; and fat-1/fat-1, fat-1 homozygotes. 17:Δ, 9,10-methylene hexadecanoic acid; 19:Δ, 11,12-methylene octadecanoic acid.
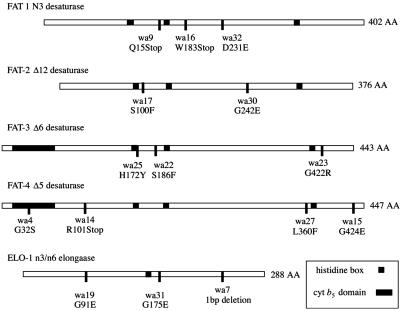
Because no n3 PUFAs were produced in the homozygous lines and the n6 precursors were increased, we hypothesized that the mutation affected the fat-1 locus, which encodes a gene product that has been shown to possess n3 desaturase activity (8, 21). To test this hypothesis, RNA from the mutant strains was isolated and the coding region of the fat-1 gene was amplified by reverse transcription–PCR and directly sequenced. We found that each of the three lines contained a mutation in the coding region of fat-1. Two alleles, wa9 and wa16, contained nucleotide changes that resulted in premature stops in the coding region (Fig. 2) and therefore are very probably null alleles. Despite the elimination of n3 PUFAs and the dramatic increase in n6 PUFAs, all three fat-1 alleles grow at the same rate as WT with no apparent defects in morphology, movement, or reproduction, indicating that under laboratory conditions, n3 fatty acids are not essential for these processes (Fig. 3B).
Figure 2.
Mutations in fatty acid desaturase and elongase ORFs. The predicted amino acid change for each mutant allele is noted.
Figure 3.
C. elegans mutants deficient in PUFA synthesis. Partial GC traces of populations of WT (A) and PUFA mutant (B–F) worms. The alleles used were as follows: fat-1(wa9), fat-2(wa17), fat-3(wa22), fat-4(wa14), and elo-1(wa7). For comparison of growth, 2-fold stage embryos from each genotype were placed onto culture plates, incubated at 20°C, and photographed after 72 h of growth. At this point WT, fat-1, fat-4, and elo-1 worms had reached adulthood and were just starting to lay eggs, whereas fat-3 had reached the L4 larval stage and fat-2 had grown only to the L2 larval stage. 19:Δ, 11,12-methylene octadecanoic acid.
fat-4 Mutants Contain No Δ5 Unsaturated Fatty Acids.
Several populations from our screen displayed a fatty acid profile with reduced 20:4n6 and 20:5n3 and increased levels of 20:3n6 and 20:4n3 that lack a double bond at the Δ5 position. The original segregating population contained 0.2% 20:4n6 and 12% 20:5n3 as compared with 1.7% 20:4n6 and 17.2% 20:5n3 in WT, and 7% 20:3n6 and 16.5.% 20:4n3 compared with 6% 20:3n6 and 9.8% 20:4n3 in WT. Analysis of progeny from the segregating population again revealed three classes: (i) WT, (ii) the original segregating fatty acid profile, and (iii) a population with no detectable 20:4n6 or 20:5n3 and 8.2% 20:3 and 24.8% 20:4n3 (Fig. 3E, Table 1). These data indicate that a single-gene mutation eliminates Δ5 unsaturation in each of these lines.
Table 1.
Fatty acid composition of WT and PUFA-deficient mutants
| WT | fat-1 | fat-2 | fat-3 | fat-4 | elo-1 | |
|---|---|---|---|---|---|---|
| Short/branch* | 11.6 | 11.0 | 12.9 | 12.9 | 13.0 | 11.8 |
| Cyclopropane† | 16.5 | 15.9 | 14.8 | 21.7 | 17.0 | 18.7 |
| 18:0 | 6.8 | 6.0 | 2.2 | 3.1 | 5.5 | 5.1 |
| 18:1n9 | 1.7 | 1.8 | 24.3 | 3.1 | 2.4 | 2.1 |
| 18:1n7 | 20.6 | 19.5 | 36.6 | 29.6 | 20.1 | 22.3 |
| 18:2n6 | 3.8 | 4.6 | 3.8 + 3.4‡ | 12.7 | 2.4 | 4.1 |
| 18:3n6 | 1.0 | 1.2 | n.d.§ | n.d. | 2.0 | 12.4 |
| 18:3n3 | 0.9 | n.d. | n.d. | 11.0 | 0.4 | 0.6 |
| 18:4n3 | n.d. | n.d. | n.d. | n.d. | n.d. | 6.3 |
| 20:3n6 | 4.4 | 13.8 | 0.8¶ | 1.0 + 1.6∥ | 8.2 | 1.1 |
| 20:4n6 | 1.7 | 21.7 | n.d. | n.d. | n.d. | n.d. |
| 20:4n3 | 8.7 | n.d. | n.d. | n.d. | 24.8 | 3.3 |
| 20:5n3 | 19.1 | n.d. | 1.9 | n.d. | n.d. | 7.8 |
| Total PUFAs | 39.6 | 41.3 | 9.9 | 26.3 | 39.9 | 31.5 |
Data are mol% and represent the average of at least eight independent determinations. SEM did not exceed 7%. Alleles used are as described in the Fig. 3 legend.
Short/branched fatty acid are the total mol% of 14:0, C15:iso, C16:iso, 16:0, C17:iso, 16:1n7, and 17:0.
Cyclopropane fatty acids are the sum of 9,10-methylene hexadecanoic acid and 11,12-methylene octadecanoic acid.
fat-2 contains undetectable 18:2n6. Values given are for 18:2n9 and 18:2n7.
n.d., not detected (<0.3%).
¶ fat-2 contains undetectable 20:3n6. Value given is for 20:2n9.
∥ fat-3 contains undetectable 20:3n6. Values given are for 20:2n6 and 20:3n3.
In C. elegans, the fat-4 gene has been shown to encode Δ5 desaturase activity when expressed in yeast (11). We determined that the four lines lacking detectable Δ5-unsaturated fatty acid contained mutations in the coding region of the fat-4 gene (Fig. 2). One allele, wa14, contains a mutation that results in the insertion of a premature stop codon. Another allele, wa4, contains a point mutation in the highly conserved heme-binding domain of the cytochrome b5 region of FAT-4, changing the HPGG motif to HPSG. All four alleles contain no detectable Δ5 desaturated fatty acids, underscoring the functional importance of the mutated amino acids. Similar to the fat-1 mutants, fat-4 homozygotes are indistinguishable phenotypically from WT under laboratory conditions (Fig. 3E).
fat-3 Mutants Lack Δ6 Fatty Acids and Are Deficient in C20 Fatty Acids.
Three populations from our screen displayed a modest increase in 18:2n6 (9.1 vs. 5.4% in WT) and 18:3n3 (2 vs. 0.2% in WT). Homozygotes segregating from this population displayed a striking fatty acid composition, with only 1.0% 20:2n6 and 1.6% 20:3n3 and no other detectable C20 fatty acids (Fig. 3D, Table 1). The 18:2n6 increased to 12.7% and the 18:3n3 accumulated to 11.0%.
The fat-3 gene has been shown to confer Δ6 desaturase activity when expressed in yeast (10). To test whether the fatty acid composition change was caused by loss of activity of the Δ6 desaturase, we amplified and sequenced the fat-3 coding region from RNA isolated from homozygous mutant lines. We found that all three alleles contained mutations in the fat-3 coding region (Fig. 2). Unlike fat-1 and fat-4, however, none of the mutations results in a premature stop codon. However, one allele contains a mutation in a conserved histidine residue of the first H-box, which, in addition to the lack of detectable Δ6 fatty acids, suggests that these mutations are nulls.
The scarcity of C20 fatty acids in these lines indicates that Δ6 desaturation of C18 fatty acids is substantially a prerequisite for elongation to 20 carbons. The low levels of 20:2n6 and 20:3n3 that accumulate in these strains (Table 1) demonstrate that the efficiency of elongation of the non-Δ6-unsaturated precursors 18:2n6 and 18:3n3 is very low even in the absence of the preferred substrates.
Although fat-3 homozygotes are viable and fertile, they grow slowly, move sluggishly, and have a reduced brood size. In our growth comparisons, we found that after 3 days of growth at 20°C, staged embryos produced by fat-3 homozygous mothers had developed to L4 larval stage, whereas similar stage WT embryos had reached adulthood and were beginning to lay eggs (Fig. 3 A and D). The fat-3 animals required an additional day of development before commencing egg laying.
Complete Loss of Normal PUFAs in fat-2 Mutants.
The fatty acid composition of fat-2 mutants differs remarkably from that of WT. These mutants were detected in F1 segregating populations because of a noticeable increase in the amount of 18:1n9, which increases from 2.6 to 7%, whereas the amounts of 18:2n6 and C20 PUFAs are reduced slightly. Analysis of segregating populations revealed that the homozygous strain was incapable of producing the normal levels of C18 or C20 PUFAs (Fig. 3C, Table 1). In C. elegans, the fat-2 gene has been shown to encode a Δ12 desaturase (9). Because the Δ12 desaturase is the first step in the synthesis of PUFAs, mutants lacking this activity would be expected to be devoid of PUFAs. To confirm that the mutants devoid of PUFAs were defective in the Δ12 desaturase, we sequenced the amplification products of the fat-2 locus and found a point mutation in both mutant lines (Fig. 2). Detailed analysis was carried out on the wa17 allele, because this strain grows somewhat more robustly than wa30. It is likely that the wa17 strain is capable of carrying out a small amount of Δ12 desaturase activity (≈5% of WT activity), because we detected a small amount of 20:5n3 (1.9%) in these worms compared with 39.6% of Δ12 desaturated PUFAs in WT (Table 1).
In fat-2 homozygous worms, 18:1n9 accumulates nearly 10-fold above that found in WT to 24.3%. The animals are not completely devoid of PUFAs, however. In the absence of the Δ12 desaturase, small peaks of unusual 18:2 and 20:2 were identified. Analysis of 4,4-dimethyloxazoline derivatives of these fatty acids by GC mass spectroscopy indicated the presence of 18:2n9 (18:2Δ6,9), 18:2n7(Δ8,11), and 20:2n9(Δ8,11) (Fig. 3C). The mass spectra of 18:2n9 contained the diagnostic 166/167 and 194/206 fragments indicative of double bonds at C6 and C9 (22). The mass spectra of 18:2n7 and 20:2n9 revealed the characteristic fragments at 182/194 and 222/234 that are diagnostic of double bonds at C8 and C11.
The loss of normal PUFAs in fat-2 homozygotes causes many defects in this strain including slow growth, abnormal body shape, sluggish movement, cuticle defects, and reduced brood size. After 3 days of growth, 2-fold embryos produced by fat-2 homozygotes developed to the L2 larval stage, whereas similar staged WT embryos reached adulthood and already were laying eggs (Fig. 3 A and C). We found that 7 days at 20°C were required before the 2-fold fat-2 embryos reached adulthood.
elo-1 Worms Are Deficient in Elongation of n3 and n6 C18 Fatty Acids.
One class of mutants displayed a 10-fold increase in 18:3n6 (8.3% compared with 0.8% in WT). Segregating populations revealed that the homozygotes accumulate 12.4% 18:3n6 as well as 6.3% 18:4n3, a fatty acid normally not detected in WT (Fig. 3F, Table 1). The amounts of C20 PUFAs are reduced to only 12.2% of total fatty acids compared with 33.9% in WT. However, the substantial increase in C18 PUFAs means that the proportion of total PUFAs in this strain was 31.5%, similar to fat-3 mutants. This strain, however, did not exhibit the slow growth or movement defects observed in fat-3 mutants.
The elo-1 gene product (F56H11.4) expressed in yeast confers the ability to elongate C18 PUFA to C20 PUFAs (12). We amplified by reverse transcription–PCR and directly sequenced the elo-1 alleles in three homozygous strains that all displayed nearly identical fatty acid composition and found that all three strains carried a mutation in the elo-1 gene (Fig. 2). The mutant lesion in one allele, wa7, is a 1-bp deletion that causes a frameshift that affects the 50 C-terminal amino acids of the protein including the C-terminal lysines that act as an endoplasmic reticulum retention/recycling signal (23). The other two strains carrying the wa19 and wa31 alleles contained point mutations in the elo-1 gene. The persistence of n3 and n6 elongated C20 fatty acids in all strains carrying homozygous mutations in elo-1 indicates that at least one additional n6 and n3 PUFA elongase activity is likely to be present.
Discussion
Genetic studies of lipid composition have been crucial for elucidating the biochemical pathways of lipid synthesis in plants, yeast, and bacteria (24–27). Mutants deficient in fatty acid desaturase activities have provided a resource to study the role of unsaturated fatty acids in cellular and physiological processes as diverse as mitochondrial segregation in yeast (28), temperature acclimation in cyanobacteria (29), skin defects and cholesterol synthesis in mice (30, 31), and chloroplast development, pollen formation, pathogen defense, and photosynthesis in Arabidopsis (32–35). Because laboratory-cultured C. elegans consume E. coli, all the PUFAs are synthesized de novo or by desaturation and elongation of the bacterial saturated fatty acid precursors. The fatty acid desaturase and elongase activities encoded by fat-1, fat-2, fat-3, fat-4, and elo-1 have been characterized previously by heterologous expression in yeast (36). We have identified mutants that confirm the desaturase/elongase activities of these previously characterized gene products and demonstrate in vivo substrate preferences. They show that each desaturase step is carried out exclusively by one gene product, whereas the n3/n6 PUFA elongation is likely to be carried out by two or more isozymes. Finally, the mutants establish a link between changes in fatty acid composition and altered growth rate in these animals.
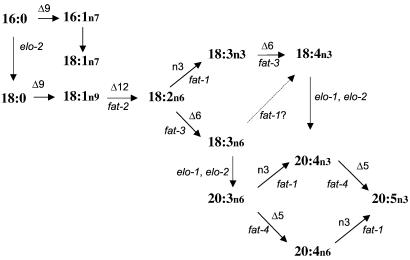
The series of C. elegans mutants described here represent each step of the biosynthetic pathway of PUFAs from 18:1n9 to 20:5n3 (Fig. 4). This pathway differs from the mammalian pathway because the nematodes can convert 18:1n9 into PUFAs because of the FAT-2 Δ12 desaturase activity that is absent in mammals. In addition, C. elegans can convert a range of C18 and C20 n6 PUFAs into n3 PUFAs by the FAT-1 n3 desaturase activity, whereas in mammals the absence of n3 desaturase activity results in parallel pathways for n6 and n3 PUFAs. Thus the ratio of n6/n3 PUFAs in mammalian tissues is determined largely by the dietary intake of n6 and n3 PUFAs.
Figure 4.
The pathway of PUFA synthesis in C. elegans. Horizontal or slanted arrows represent fatty acid desaturation. Vertical arrows represent fatty acid elongation. For the desaturation steps, the desaturase activity is noted above the arrow, and the C. elegans gene encoding this enzymatic activity is listed below the arrow.
In animals, the stearoyl-CoA Δ9 desaturases carry out the first desaturation of 16:0 and 18:0 to 16:1n7 and 18:1n9. Three C. elegans gene products with Δ9 desaturase activity, FAT-5, FAT-6, and FAT-7, have been characterized by expression in yeast (37). Our screen did not identify mutants in the fat-5, fat-6, and fat-7 desaturases. The activities of these enzymes are likely to overlap, rendering identification of mutants in segregating populations difficult. We also might have failed to identify lines in which homozygous mutants were inviable.
Clearly a critical step in the PUFA synthesis pathway is the Δ12 desaturation of 18:1n9 by FAT-2 to produce 18:2n6. The fat-2 mutants accumulate 18:1n9 levels that are 10-fold higher than WT. These mutants demonstrate that the Δ5 and n3 desaturases are unable to act on 18:1n9, whereas the Δ6 desaturase is able to form a small amount of 18:2n9 that is elongated to 20:2n9. In human fatty acid deficiency, Mead acid (20:3n9) is formed by a similar Δ6 desaturation of 18:1Δ9, elongation to 20:2n9, then desaturation by a Δ5 desaturase (38). The loss of PUFAs in fat-2 mutants result in severe growth, movement, and reproductive defects. Despite this, a fraction of the worms are viable and fertile, and this strain can be maintained as a homozygote.
18:2n6 is a substrate for two desaturase enzymes; the FAT-3 Δ6 desaturase produces 18:3n6, whereas the FAT-1 n3 desaturase produces 18:3n3. The latter fatty acid is present at less than 1% in WT worms but reveals itself as a pathway intermediate in the fat-3 mutants (Table 1). The fatty acid composition of fat-3 mutants demonstrates that deficiency in Δ6 desaturase activity has severe consequences on C20 PUFA formation. Despite the accumulation of 18:2n6, only a very small amount of 20:2n6 is produced, demonstrating that C18 fatty acids with a Δ6 double bond are the preferred substrate for elongation. In humans, the Δ6 desaturation step is thought to be the rate-limiting step in the conversion of the essential fatty acids to long-chain PUFAs, and reduced Δ6 desaturase activity has been associated with diabetes, aging, atopic dermatitis, premenstrual syndrome, and rheumatoid arthritis (39).
In addition to the reduction of PUFAs in fat-2 and fat-3 mutants, the levels of 18:0 in these mutants are 2–3-fold lower than WT, which may be indicative of an increase in Δ9 desaturase activity that would result if FAT-5, FAT-6, and/or FAT-7 expression was up-regulated by lack of C20 PUFAs. This observation suggests that the C. elegans stearoyl-CoA desaturases may be regulated by PUFAs as they are in mammals (40).
In WT worms the two products of the FAT-3 Δ6 desaturase, 18:3n6 and 18:4n3, appear to be elongated rapidly to 20:3n6 and 20:4n3. Although 18:4n3 does not accumulate in WT, it comprises 6.3% of fatty acids in elo-1 mutants (Fig. 3F, Table 1). The elo-1 mutants isolated in our screen displayed increased levels of 18:3n6 as well as 18:4n3 and reduced levels of C20 PUFAs. None, however, completely eliminated C20 n3 and n6 PUFAs. It is likely that at least one allele, elo-1(wa7) is a null allele, because a 1-bp deletion causes a frameshift affecting 50 C-terminal amino acids of the protein (Fig. 2). Recently, the technique of RNA inhibition was used to suppress the function of C. elegans ELO-2 (F11E6.5). These studies demonstrated that this protein is capable of elongating both 16:0 and C18 PUFAs (M. Kniazeva, M. Sieber, S. McCauley, K. Zhang, J.L.W., and M. Han, unpublished data). Suppression of elo-2 in the elo-1 mutants resulted in the elimination of C20 PUFAs. Therefore, ELO-1 and ELO-2 seem to act together to elongate C18n6 and C18n3 PUFAs.
The FAT-4 Δ5 desaturase acts on 20:3n6 to produce arachidonic acid (20:4n6). We also detected the formation of 20:4n3 after supplementation of 20:3n6 in fat-2 and fat-3 mutants, demonstrating that the FAT-1 n3 desaturase can use 20:3n6 as a substrate (our unpublished observations). 20:4n3 is desaturated to 20:5n3 by the FAT-4 Δ5 desaturase, and 20:4n6 is desaturated to 20:5n3 by the FAT-1 n3 desaturase. Mammals produce 22:5n6, 22:5n3, and 22:6n3 by further elongation and desaturation of 20:4n6 and 20:5n3. C. elegans lacks this elongation activity, and the pathway terminates with 20:5n3 as the most abundant PUFA. Interestingly, the human ELO1 homologue, HELO1, has been shown by expression in yeast to elongate 18:3n6, 18:4n3, 20:4n6, and 20:5n3 (41). Thus, this human elongase produces C20 PUFAs from C18 Δ6 desaturated substrates, similar to the C. elegans activity, yet it also can convert 20:4n6 and 20:5n3 to 22:4n6 and 22:5n3, an activity absent in worms. A 5-bp deletion in another human ELO homologue, ELOVL4, has been shown to be associated with human autosomal dominant macular degeneration (42). This gene is expressed exclusively in photoreceptor cells in the retina, a region rich in C22 PUFA and sphingolipids.
Two of the mutants, fat-1 and fat-4, affect specific classes of C20 PUFAs. Both of these genes are represented by null mutations, and therefore we are certain that no enzymatic activity remains in the homozygotes. Despite this absence of activity, the worms develop and behave normally. In both mutants the amounts of remaining C20 PUFAs are increased such that fat-1, fat-4, and WT all contain ≈40% C20 PUFAs, although the composition of specific fatty acids varies considerably. Thus, the fat-4 mutant contains no arachidonic acid, but the fat-1 mutant contains 10-fold more arachidonic acid in its membranes than WT.
In humans, specific eicosanoid classes are derived from distinct C20 PUFA precursors (39). For example, series 1, 2, and 3 of prostaglandins are derived from 20:3n6, 20:4n6, and 20:5n3, respectively. Prostaglandins produced from certain precursors have distinct and sometimes opposing cellular effects to those of their counterparts derived from other precursors (43). Because large differences in the proportions of 20:3n6, 20:4n6, and 20:5n3 in our mutants have no obvious phenotypic consequences, we infer that these processes are less important in C. elegans. In addition, the elo-1 mutants demonstrate that moderate reduction in C20 PUFAs does not impair growth or movement.
The mutants described in this study provide tools to examine the mechanisms through which the physical properties of fatty acid molecules impact the cell biology and physiology of animals. The fat-2 and fat-3 strains grow slowly and exhibit considerable embryonic lethality as well as body shape and behavioral abnormalities. Our continuing studies of the developmental and behavioral defects in these strains promise to provide insights into the cellular processes most sensitive to changes in the unsaturation of membrane fatty acids.
Acknowledgments
We thank Eric Phillips and Shannon Casey for technical assistance and Jim Wallis for critical reading of the manuscript. Funding was provided by National Institutes of Health Grant R01 GM62521, National Science Foundation Postdoctoral Fellowship DBI-9804195 (to J.L.W.), and the Agricultural Research Center at Washington State University.
Abbreviations
- PUFA
polyunsaturated fatty acid
- X:YnZ
fatty acid chain of X carbon atoms and Y methylene-interrupted cis double bonds (Z indicates the position of the terminal double bond relative to the methyl end of the molecule)
- WT
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Chyb S, Raghu P, Hardie R. Nature (London) 1999;397:255–259. doi: 10.1038/16703. [DOI] [PubMed] [Google Scholar]
- 2.Xiao Y F, Ke Q, Wang S Y, Auktor K, Yang Y, Wang G K, Morgan J P, Leaf A. Proc Natl Acad Sci USA. 2001;98:3606–3611. doi: 10.1073/pnas.061003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt A, Wolde M, Thiele C, Fest W, Kratzin H, Podtelejnikov A V, Witke W, Huttner W B, Soling H D. Nature (London) 1999;401:133–141. doi: 10.1038/43613. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg E M, Zidovetzki R. Biophys J. 1997;73:2603–2614. doi: 10.1016/S0006-3495(97)78290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funk C D. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- 6.Simopoulos A P. Am J Clin Nutr. 1999;70:560S–569S. doi: 10.1093/ajcn/70.3.560s. [DOI] [PubMed] [Google Scholar]
- 7.Hutzell P A, Krusberg L R. Comp Biochem Physiol B Biochem Mol Biol. 1982;73:517–520. [Google Scholar]
- 8.Spychalla J P, Kinney A J, Browse J. Proc Natl Acad Sci USA. 1997;94:1142–1147. doi: 10.1073/pnas.94.4.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peyou-Ndi M M, Watts J L, Browse J. Arch Biochem Biophys. 2000;376:399–408. doi: 10.1006/abbi.2000.1733. [DOI] [PubMed] [Google Scholar]
- 10.Napier J A, Hey S J, Lacey D J, Shewry P R. Biochem J. 1998;330:611–614. doi: 10.1042/bj3300611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watts J L, Browse J. Arch Biochem Biophys. 1999;362:175–182. doi: 10.1006/abbi.1998.1024. [DOI] [PubMed] [Google Scholar]
- 12.Beaudoin F, Michaelson L V, Hey S J, Lewis M J, Shewry P R, Sayanova O, Napier J A. Proc Natl Acad Sci USA. 2000;97:6421–6426. doi: 10.1073/pnas.110140197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toke D A, Martin C E. J Biol Chem. 1996;271:18413–18422. doi: 10.1074/jbc.271.31.18413. [DOI] [PubMed] [Google Scholar]
- 14.Oh C S, Toke D A, Mandala S, Martin C E. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 15.Strittmatter P, Spatz L, Corcoran D, Rogers M J, Setlow B, Redline R. Proc Natl Acad Sci USA. 1974;71:4565–4569. doi: 10.1073/pnas.71.11.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shanklin J, Cahoon E B. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- 17.Shanklin J, Whittle E, Fox B G. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 18.Cinti D L, Cook L, Nagi M N, Suneja S K. Prog Lipid Res. 1992;31:1–51. doi: 10.1016/0163-7827(92)90014-a. [DOI] [PubMed] [Google Scholar]
- 19.Moon Y A, Shah N A, Mohapatra S, Warrington J A, Horton J D. J Biol Chem. 2001;276:45358–45366. doi: 10.1074/jbc.M108413200. [DOI] [PubMed] [Google Scholar]
- 20.Wood W B. The Nematode Caenorhabditis elegans. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 21.Kang Z B, Ge Y, Chen Z, Cluette-Brown J, Laposata M, Leaf A, Kang J X. Proc Natl Acad Sci USA. 2001;98:4050–4054. doi: 10.1073/pnas.061040198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spitzer V. Prog Lipid Res. 1997;35:387–408. doi: 10.1016/s0163-7827(96)00011-2. [DOI] [PubMed] [Google Scholar]
- 23.Jackson M R, Nilsson T, Peterson P A. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sommerville C, Browse J. Science. 1991;252:80–87. doi: 10.1126/science.252.5002.80. [DOI] [PubMed] [Google Scholar]
- 25.Somerville C, Browse J. Trends Cell Biol. 1996;6:148–153. doi: 10.1016/0962-8924(96)10002-7. [DOI] [PubMed] [Google Scholar]
- 26.Carman G M, Henry S A. Annu Rev Biochem. 1989;58:635–669. doi: 10.1146/annurev.bi.58.070189.003223. [DOI] [PubMed] [Google Scholar]
- 27.Vanden Boom T, Cronan J E., Jr Annu Rev Microbiol. 1989;43:317–343. doi: 10.1146/annurev.mi.43.100189.001533. [DOI] [PubMed] [Google Scholar]
- 28.Stewart L C, Yaffe M P. J Cell Biol. 1991;115:1249–1257. doi: 10.1083/jcb.115.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wada H, Gombos Z, Murata N. Proc Natl Acad Sci USA. 1994;91:4273–4277. doi: 10.1073/pnas.91.10.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Y, Eilertsen K J, Ge L, Zhang L, Sundberg J P, Prouty S M, Stenn K S, Parimoo S. Nat Genet. 1999;23:268–270. doi: 10.1038/15446. [DOI] [PubMed] [Google Scholar]
- 31.Miyazaki M, Kim Y C, Gray-Keller M P, Attie A D, Ntambi J M. J Biol Chem. 2000;275:30132–30138. doi: 10.1074/jbc.M005488200. [DOI] [PubMed] [Google Scholar]
- 32.Wu J, Browse J. Plant Cell. 1995;7:17–27. doi: 10.1105/tpc.7.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Routaboul J M, Fischer S F, Browse J. Plant Physiol. 2000;124:1697–1705. doi: 10.1104/pp.124.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McConn M, Browse J. Plant J. 1998;15:521–530. doi: 10.1046/j.1365-313x.1998.00229.x. [DOI] [PubMed] [Google Scholar]
- 35.Vijayan P, Shockey J, Levesque C A, Cook R J, Browse J. Proc Natl Acad Sci USA. 1998;95:7209–7214. doi: 10.1073/pnas.95.12.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Napier J, Michaelson L. Lipids. 2001;36:761–766. doi: 10.1007/s11745-001-0782-9. [DOI] [PubMed] [Google Scholar]
- 37.Watts J L, Browse J. Biochem Biophys Res Commun. 2000;272:263–269. doi: 10.1006/bbrc.2000.2772. [DOI] [PubMed] [Google Scholar]
- 38.Horrobin D F. Prog Lipid Res. 1992;31:163–194. doi: 10.1016/0163-7827(92)90008-7. [DOI] [PubMed] [Google Scholar]
- 39.Fan Y, Chapkin R. J Nutr. 1998;128:1411–1414. doi: 10.1093/jn/128.9.1411. [DOI] [PubMed] [Google Scholar]
- 40.Ntambi J. J Lipid Res. 1999;40:1549–1558. [PubMed] [Google Scholar]
- 41.Leonard A, Bobik E, Dorado J, Kroeger P, Chuang L, Thurmond J, Parker-Barnes K, Das T, Huang Y, Mukerji P. Biochem J. 2000;350:765–770. [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K, Kniazeva M, Han M, Li W, Yu Z, Yang Z, Li Y, Metzker M L, Allikmets R, Zack D J, et al. Nat Genet. 2001;27:89–93. doi: 10.1038/83817. [DOI] [PubMed] [Google Scholar]
- 43.Dwang D. In: Fatty Acids in Foods and Their Health Implications. Chow C, editor. New York: Dekker; 1992. pp. 445–557. [Google Scholar]