Abstract
Phosphoglucose isomerase (EC 5.3.1.9) catalyzes the second step in glycolysis, the reversible isomerization of d-glucose 6-phosphate to d-fructose 6-phosphate. The reaction mechanism involves acid-base catalysis with proton transfer and proceeds through a cis-enediol(ate) intermediate. 5-Phospho-d-arabinonohydroxamic acid (5PAH) is a synthetic small molecule that resembles the reaction intermediate, differing only in that it has a nitrogen atom in place of C1. Hence, 5PAH is the best inhibitor of the isomerization reaction reported to date with a Ki of 2 × 10−7 M. Here we report the crystal structure of rabbit phosphoglucose isomerase complexed with 5PAH at 1.9 Å resolution. The interaction of 5PAH with amino acid residues in the enzyme active site supports a model of the catalytic mechanism in which Glu-357 transfers a proton between C1 and C2 and Arg-272 helps stabilize the intermediate. It also suggests a mechanism for proton transfer between O1 and O2.
Phosphoglucose isomerase (PGI; EC 5.3.1.9) is a cytosolic glycolytic enzyme that catalyzes the reversible isomerization of d-glucose 6-phosphate (G6P) to d-fructose 6-phosphate (F6P). In addition to isomerase activity, PGI has been shown to display anomerase activity between pyranose anomers of G6P (1), between furanose anomers of F6P (2), and between those of d-mannose 6-phosphate (3), as well as C-2-epimerase activity on G6P (4). It also has roles in protein glycosylation, gluconeogenesis, and the pentose phosphate pathway (5). This central role in the metabolism of phosphorylated sugars explains the strong impact of PGI deficiency in humans (6, 7), as well as the interest as a therapeutic target in parasite metabolism (8). The PGI polypeptide chain, or perhaps a variation thereof, also has extracellular roles as neuroleukin, autocrine motility factor, and differentiation and maturation mediator (9–12). These proteins promote antibody secretion by mononuclear cells, tumor cell motility, and differentiation of leukemia cells and a promyelocytic cell line (HL-60 cells), respectively. More recently, PGI has also been identified as the antigen involved in rheumatoid arthritis of a mouse line (13), as a specific inhibitor toward myofibril-bound serine proteinase (14), and as the surface antigen involved in sperm agglutination (15). Because very little is known about how PGI is precisely involved in these various biological processes, this moonlighting protein (16) has been the subject of intense investigations.
Biochemical characterization of PGI began over 50 years ago and includes inhibitor studies (1, 17–25), labeling studies (26–32), solvent exchange (33, 34), mutagenesis (35, 36), and pH profile studies (20, 37). A proposed multistep catalytic mechanism includes a ring-opening step followed by an isomerization step. The isomerization step proceeds via general acid-base catalysis with proton transfer. In the G6P to F6P direction, abstraction of the proton from C2 by an active site amino acid residue yields a 1,2-cis-enediol(ate) intermediate. That same residue then stereospecifically transfers a proton back to the intermediate on the Re face of C1 (38) to produce F6P.
In recent years, several PGI crystal structures have helped to identify the location of the active site and amino acid residues likely to be involved in catalysis and substrate specificity. Published PGI crystal structures include a partial structure from pig (39), structures from Bacillus stearothermophilus [unliganded (40) or complexed with 5-phospho-d-arabinonate (5PAA) (41) or N-bromoacetylethanolamine phosphate (41)], from rabbit [complexed with 6-phospho-d-gluconate (42), 5PAA (43), or the cyclic form of a substrate, F6P (44)], and from human [complexed with β-mercaptoethanol and a sulfate ion (7)].
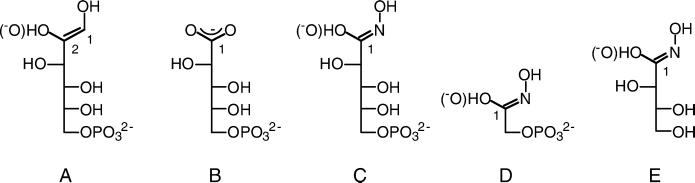
Although 5PAA is a good mimic of the cis-enediol(ate) intermediate (Fig. 1A) and has a Ki of 2 × 10−6 M at pH 8 (19, 45), it lacks an atom corresponding to O1 of the intermediate. 5PAA also has a carboxylate group in place of C2, C1, and O2 (Fig. 1B). Recently, a better mimic of the cis-enediol(ate) intermediate, 5PAH (Fig. 1C), has been synthesized (45). 5PAH is both the best reported inhibitor of the isomerization reaction, with a Ki of 2 × 10−7 M (pH 8), and a good structural and chemical mimic of the cis-enediol(ate) intermediate. 5PAH is the same length and configuration as the substrates and intermediate and differs only by the presence of a nitrogen atom in place of C1. This substitution of a nitrogen atom in place of C1 results in a planar hydroxamic acid moiety, possibly in the hydroximic or hydroximate form within the active site, in place of the cis-enediol(ate) at C1 and C2 and is stable enough in the conditions reported herein for structural studies of the enzyme/inhibitor complex. Similar hydroxamic acid ligands have been used successfully in biochemical and structural studies of two other sugar isomerases, triosephosphate isomerase [TIM (46)] and xylose isomerase [XYL (47)]. A crystal structure of PGI with bound 5PAH would result in an improved model of how the cis-enediol(ate) intermediate binds in the active site and provide the most accurate distances between the reactive intermediate and catalytic amino acid residues. We report here the x-ray crystal structure of rabbit PGI with bound 5PAH solved at 1.9 Å resolution.
Figure 1.
Catalytic intermediate and inhibitor molecules mentioned in the text. (A) The 1,2-cis-enediol(ate) intermediate of the isomerization reaction, (B) 5PAA, (C) 5PAH, (D) PGH, and (E) THA. Hydroxamic acids C–E are presented as hydroximic acid (or hydroximate) form.
Experimental Procedures
Crystallization of PGI.
Rabbit skeletal muscle PGI was purchased from Sigma and transferred into crystallization buffer as described (42). 5PAH was prepared as described (25). Crystals were grown by the hanging drop vapor-diffusion method at room temperature. The drops contained an equal mixture of protein solution (15 mg/ml PGI/50 mM KCl/10 mM imidazole, pH 7.5) and reservoir solution (12–14% PEG 8000/250 mM magnesium acetate/100 mM sodium cacodylate, pH 6.5) and were allowed to equilibrate with 1 ml of reservoir solution. The crystals grew as colorless hexagonal rods ≈0.3 × 0.3 × 1.0 mm in size.
Data Collection and Structure Refinement.
Before data collection, the crystals were soaked for 5 min in 16% PEG 8000, 250 mM Mg acetate, 100 mM Na cacodylate (pH 6.5), 20% glycerol, and 5 mM 5PAH and then flash frozen at −180°C. A diffraction data set to 1.9 Å resolution was collected from a single crystal of the enzyme/inhibitor complex by using monochromatic x-rays at APS beamline 14C (BioCARS) at Argonne National Laboratories. The data set was processed with the programs denzo and scalepack (48).
Refinement consisted of alternating rounds of computational refinement with cns (49) and manual fitting of the model to electron density maps with the program o (50). The initial model for refinement was a previously reported PGI structure (Protein Data Bank ID code 1DQR) from which all ordered water molecules and inhibitors were removed. Ten percent of the data set was set aside to compute Rfree (51). Simulated annealing was used to remove model bias. Data between 12 and 1.9 Å were used in positional and individual B factor refinement. Ordered water molecules were added based on electron density maps with coefficients |Fo − Fc|, together with criteria based on geometry and refined B factors. The geometry of the final model was checked with the programs procheck (52) and cns.
Results and Discussion
The crystal structure of rabbit PGI with bound 5PAH has been solved at 1.9 Å resolution with an R factor of 22.0 and an Rfree of 25.4 (Table 1). The shape of the protein in the PGI/5PAH complex is similar to the previously reported structures of rabbit and human PGI. The protein is a homodimer with 557 residues per subunit. Our model includes amino acid residues 1–556. Because of the absence of clear electron density corresponding to side chains for Lys-555 and Ile-556, the residues were modeled as alanines in both subunits.
Table 1.
Statistics for data collection and refinement
| Data collection | |
| Space group | C2221 |
| Cell dimensions, Å | a = 82.69, b = 115.97, c = 271.85 |
| Resolution limit, Å | 1.9 |
| Observed reflections | Total = 400188, Unique = 96961 (9158) |
| Completeness, % | 93.4 (89.0) |
| Redundancy | 4.1 |
| Rsym (% on I) | 5.7 (32.7) |
| Refinement | |
| Resolution range, Å | 12–1.9 (1.97–1.9) |
| R factor/Rfree, % | 22.0 (30.7)/25.4 (33.5) |
| Ordered water molecules | 802 |
| rms deviations from ideal geometry | |
| Bond lengths, Å | 0.007 |
| Bond angles, ° | 1.3 |
| Average B factor, Å2 | 44.4 |
Rsym = ∑|Ii − 〈I〉|/∑ Ii. R factor = ∑∥Fo − Fc∥/∑ Fo. Rfree is the equivalent of R factor but it is calculated for 10% randomly chosen set of reflections that were omitted from the refinement process. The numbers in parentheses refer to the last shell of data.
Interestingly, in subunit A of the PGI/5PAH complex, difference electron density maps with coefficients of |Fo-Fc| indicate that the helix containing residues 513–521 occupies two positions with approximately equal probability. One position corresponds to the position of the helix in the PGI/5PAA complex crystal structure (43), and the other corresponds to the position of the helix in the PGI/6-phospho-d-gluconate or PGI/F6P complex crystal structures (42, 44). Therefore, our model includes two copies of this helix in subunit A, and each was assigned 50% occupancy. In subunit B, the difference electron density maps indicate a single conformation of the helix, the one that corresponds to the position of the helix in the PGI/5PAA complex crystal structure.
Other than the partial occupancy of one helix, the PGI/5PAH dimer closely resembles the structure of the rabbit PGI/5PAA complex. The rms deviation for the two dimers calculated for all alpha carbon atoms is 0.28 Å. For the PGI/5PAH structure, a single conformation of the helix containing residues 513–521 was used in the calculation.
Active Site.
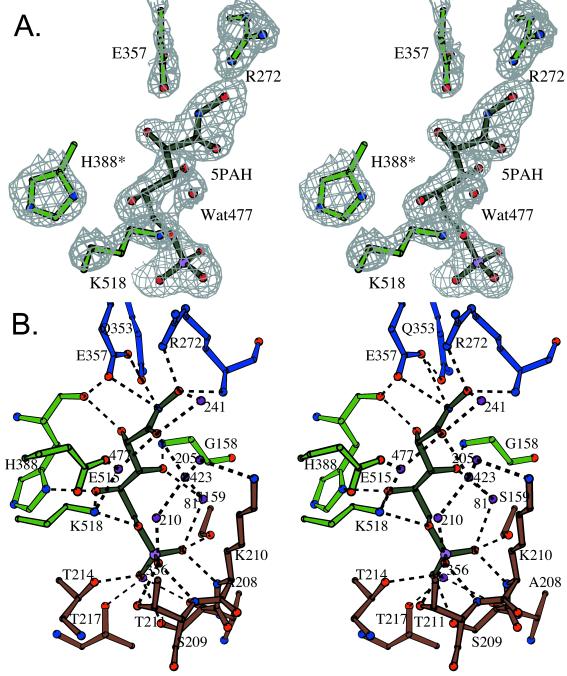
In subunit B of the new structure, in contrast to a structure described (43) in which 5PAH was hydrolyzed to 5PAA, the electron density maps show a complete, nonhydrolyzed 5PAH molecule (Fig. 2). This difference is probably because of the fact that previously, the 5PAH had been added to the crystallization mix weeks before data collection. The crystals for our new structure were grown without inhibitor, and the 5PAH was soaked into the crystals just before data collection.
Figure 2.
Active site (subunit B) of the PGI/5PAH complex. (A) An omit electron density map calculated with coefficients |Fo − Fc| is shown with a 1 σ contour level. The omit map was prepared by leaving out the inhibitor and surrounding amino acid residues. (B) A ball and stick model is shown including the inhibitor (dark green), ordered water molecules (purple), and amino acid residues interacting with the phosphate group (brown), with the hydroxyl groups of C2, C3, and C4 (light green) or with the hydroxamic acid part of 5PAH (dark blue). Interactions between the inhibitor and active site residues are shown as dashed lines. Figs. 2, 3, and 4 A and B were prepared with BOBSCRIPT (70).
In subunit A, the density for the 5PAH inhibitor is interrupted between the C1 and C2 positions and suggests that the 5PAH molecule bound in this subunit might also have two conformations, each partially occupied. For our analysis of the PGI/5PAH complex active site, we will refer to subunit B, where the electron density of the inhibitor is clear and complete.
Comparison of 5PAH and 5PAA Bound to PGI.
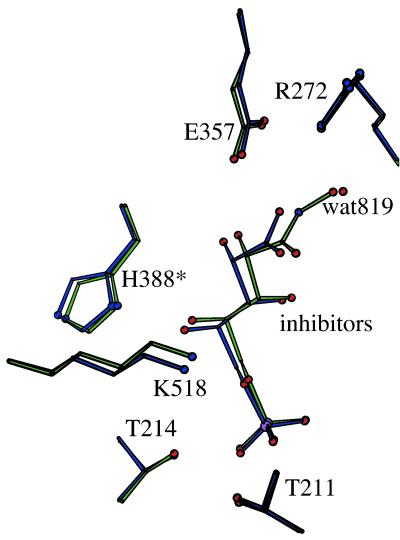
The conformations of the two inhibitors, 5PAA and 5PAH, bound to rabbit PGI are very similar. The rms deviation for all atoms of 5PAA superposed on the corresponding atoms of 5PAH is 0.036 Å. The main differences are in the region near O1, which is an atom not found in 5PAA. Instead, in the structure of the rabbit PGI/5PAA complex, water819 was proposed to be bound in approximately the position expected for the O1 of the cis-enediol(ate) intermediate (43). Indeed, in the PGI/5PAH complex, the N-OH oxygen atom of the inhibitor is located in place of water819 and makes hydrogen bonds to Arg-272.
Superposition of all atoms of 24 key active site amino acid residues of subunit B (residues 156–159, 209–217, 270–272, 357–358, 511–514, and 517–518) and His-388 from subunit A of the PGI/5PAH and PGI/5PAA structures results in an rms deviation of 0.15 Å. In the superposed structures, the active site amino acids are found in the same conformations, and the phosphorus atoms and the oxygen atoms of the inhibitor phosphoryl groups lie on top of each other and form similar hydrogen bonds to active site amino acid residues and ordered water molecules. However, the position of the C1 end of the inhibitors differs, because of the presence of the 5PAH terminal oxygen instead of water819 (Fig. 3). In the PGI/5PAA structure, the distance between water819 and the carboxylate oxygen Oɛ2 of 5PAA is 2.6 Å, which is close to a minimum length for a hydrogen bond. However, the length of the corresponding N-O covalent bond in the PGI/5PAH structure is 1.14 Å shorter. As a result, C1, C2, C3, and the corresponding hydroxyl groups of 5PAH are shifted away from Glu-357 and toward Arg-272 in comparison to how 5PAA is bound in the active site so that there is a 0.7 Å difference in the positions of the 5PAH nitrogen atom and the corresponding atom in 5PAA (Oɛ2).
Figure 3.
A ball and stick model of the active site of the PGI/5PAH complex (green) overlayed onto the active site of the rabbit PGI/5PAA complex (blue). The active site contains amino acid residues from subunit B and His-388 (marked by an asterisk) from subunit A. The inhibitor molecules are tilted with respect to each other.
The relative tilt of the two inhibitors results in differences in the lengths of the hydrogen bonds between some of the atoms of the inhibitors and active site amino acids that have been proposed to perform key steps in the catalytic mechanism (Table 2). Glu-357 has been proposed to transfer the proton between C1 and C2 during the isomerization step, and Arg-272 has been proposed to stabilize the cis-enediol(ate) intermediate. Despite a small increase in the distance between the inhibitor and Glu-357, the PGI/5PAH crystal structure is consistent with the mechanism of proton transfer we proposed (43). For comparison, in the crystal structure of yeast TIM complexed with the analogous inhibitor phosphoglycolohydroxamic acid [PGH; PDB ID code 7TIM (46), discussed below], the glutamate that transfers the proton during the isomerization step is positioned at about the same distance from the ligand. The nitrogen atom of PGH, corresponding to the C1 of the intermediate, is 3.2 Å away from Oɛ1 of Glu-165, whereas the analogous distance in PGI/5PAH is 3.3 Å. Atom C1 of PGH, corresponding to C2 of the intermediate, is 3.3 Å away from Oɛ2 of Glu-165, and the distance between the analogous atoms in the PGI/5PAH structure is also 3.3 Å. The increased proximity of Arg-272 to the hydroxamic acid group in the PGI/5PAH structure emphasizes the importance of this residue.
Table 2.
Distances between atoms in 5PAH or 5PAA and PGI (in Å)
| Atoms of the intermediate | Atoms of inhibitors
|
Glu-357 Oɛ1
|
Glu-357 Oɛ2
|
Arg-272 NH1
|
||||
|---|---|---|---|---|---|---|---|---|
| 5PAH | 5PAA | 5PAH | 5PAA | 5PAH | 5PAA | 5PAH | 5PAA | |
| C1 | N | Oɛ2 | 3.0 | 2.7 | 3.3 | 3.2 | 3.8 | 4.3 |
| C2 | C1 | C1 | 3.3 | 3.2 | 4.3 | 4.3 | 4.5 | 4.9 |
| O1 | ON | H2O | 3.6 | 3.7 | 3.6 | 3.9 | 2.9 | 2.8 |
| O2 | O1 | Oɛ1 | 3.9 | 3.6 | 5.1 | 5.0 | 4.4 | 4.7 |
Comparison of PGI/5PAH to Complexes of TIM and XYL with Analogous Inhibitors.
Derivatives of hydroxamic acids were successfully used in previous crystallographic and biochemical studies of other sugar isomerases. XYL, TIM, and PGI catalyze analogous reactions, even though biochemical and crystallographic studies of these three enzymes have shown that details of the isomerization reaction are different.
Hydroxamic acids were shown to be the most potent inhibitors of all three enzymes. For rabbit PGI, the Ki(5PAH) is 0.2 μM, whereas the KM(F6P) is 100 μM (45). For yeast TIM, the Ki(PGH) is 15 μM and the KM is 1500 μM (53). Chicken TIM has a Ki(PGH) of 7.1 μM and the KM is 470 or 970 μM, depending on the direction of reaction (54). Ki(PGH) for trypanosomal TIM is about 8 μM, whereas KM is 250 or 1200 μM, depending on the direction of reaction (55). XYL from Streptomyces olivochromogenes has a low affinity for its natural substrates: the KM equals 5 mM for d-xylose and 290 mM for d-glucose (56). For d-threonohydroxamic acid (THA), the Ki ≤ 100 nM (47).
TIM (EC 5.3.1.1) catalyzes the reversible interconversion of the three carbon sugars d-glyceraldehyde 3-phosphate and dihydroxyacetone phosphate. As in PGI, the mechanism of the isomerization reaction proceeds through formation of a cis-enediol(ate) intermediate that differs only in the length of the sugar backbone. Also, there is no ring-opening step in the reaction catalyzed by TIM.
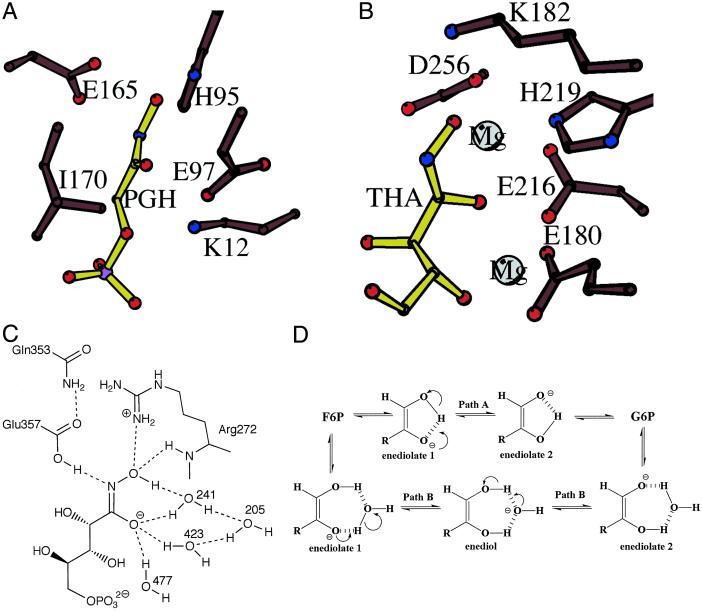
Crystal structures of TIM from chicken, yeast, and trypanosome, 1.8, 1.9, and 2.5 Å resolution, respectively, were solved in complex with PGH (46, 57, 58). The active sites of the three structures are identical; therefore, below we refer to only the yeast enzyme (Fig. 4A). From biochemical and crystallographic studies of TIM it was proposed that, like PGI, TIM uses a proton transfer mechanism for isomerization and a glutamate residue (Glu-165) to abstract a proton from the C1 position and transfer it to C2.
Figure 4.
Ball and stick models of (A) yeast TIM with PGH (yellow) bound in the active site (from PDB ID code 7TIM) and (B) yeast XYL with THA (yellow) bound in the active site (from PDB ID code 2GYI). (C) Schematic diagram of the PGI active site with hydrogen bonds between the inhibitor and active site residues indicated by dotted lines. The inhibitor is proposed to bind as its hydroximate form. (D) Schematic diagram illustrating two possible mechanisms for the transfer of a proton between O1 and O2 atoms of the enediolate intermediates. Path A shows intramolecular proton transfer, and path B shows intermolecular proton transfer by a nearby water molecule.
Although the majority of the amino acid residues comprising the active sites of PGI and TIM differ, the side chains of Glu-357 of PGI and Glu-165 of TIM are on the same side of the inhibitor molecule, pointing toward the planar hydroxamic acid group. In the PGI/5PAH crystal structure, Glu-357 is slightly farther from the ligand and its carboxylate group is rotated 90o with respect to Glu-165 of TIM/PGH. However, proton transfer is likely to occur in the same way because in both structures the inhibitor is oriented so that the pro-R hydrogen on C1 of the substrate would be transferred to the syn sp2 orbital of the carboxylate. In TIM, His-95 was proposed to act as an electrophile to polarize the carbonyl group in the catalysis of the enolization reaction of the substrate dihydroxyacetone phosphate. In addition, His-95 was postulated to stabilize the cis-enediol(ate) intermediate(s) by mediating proton transfer between the oxygen atoms on C1 and C2 (59), through a possible imidazole/imidazolate pair acting as an acid/base catalyst (60). Intramolecular proton transfer between O1 and O2 of the two enediolate intermediates has also been proposed as a possible alternative (61), however reported as less energetically favorable according to some other theoretical studies (62, 63).
PGI also has one histidine residue (His-388) in the active site, but, unlike in TIM, it is too far from the hydroxamic acid moiety of 5PAH to play a role in proton transfer. Instead, based on analysis of a PGI/F6P complex structure, His-388 was proposed to participate in the ring-opening step of the cyclic substrates (44). In the PGI/5PAH crystal structure, no active site residue is situated close to O1 of the inhibitor. However, it is likely stabilized by interactions with a network of water molecules: water241, water423, and water477 (Figs. 2B and 4C). Hence, our structure suggests that, in the case of PGI, there is no need for a nearby residue for proton transfer to occur between the substrate oxygen atoms on C1 and C2. This observation leads us to propose two possible mechanisms: (i) intramolecular proton transfer between the O1 and O2 atoms of the enediolate intermediates (Fig. 4D, path A), and (ii) acid/base catalysis by a water molecule as a H2O/OH− pair (which may occur in a concerted or nonconcerted manner), in analogy with the imidazole/imidazolate pair of TIM (Fig. 4D, path B).
The water molecule or molecules directly involved in the catalysis of the proton transfer between O1 and O2 are not clear. The best candidate is water241, which is ideally situated between O1 and O2 of 5PAH (3.1 and 3.2 Å, respectively). Protonation of enediolate 1 would yield the enediol intermediate and water241 in the form of a hydroxide anion (Fig. 4D). Deprotonation of O1H would lead to enediolate 2. Superposition of the hydroxamic acid atoms of both PGI/5PAH and TIM/PGH structures places the oxygen atom of water241 at a position close to the nitrogen atom Nɛ of His-95 (≈0.7 Å), which supports the hypothesis of a direct involvement of water241 in proton transfer. Another variation of path B would be to consider water423, the closest water molecule to O2 (2.6 Å). After protonation of enediolate 1 to give the enediol intermediate, the resulting hydroxide anion could be indirectly involved, through water205 (2.7 Å from water423), in the O1H deprotonation of the enediol by water241 (3.2 Å from water205, and 3.1 Å from O1) to give enediolate 2 (Fig. 4D). Finally, water477, which is 2.9 Å from O2, seems to stabilize the enediol(ate) intermediate.
In addition to His-95, Lys-12 of TIM is proposed to stabilize the intermediate by interacting with O2. The importance of this lysine was confirmed by the inability of the enzyme to bind substrate if Lys-12 was substituted by methionine (53). In PGI, Lys-518 is in the active site but it interacts with O4 and O5 of the ligand and, together with His-388, participates in the ring-opening step. Instead, Arg-272 in PGI is thought to stabilize the enedio(ate) intermediate through interaction with O1. However, Arg-272 is located too far from O2 (4.4 Å).
Natural substrates of XYL (EC 5.3.1.5) are the five carbon sugars d-xylose and d-xylulose, but it is also able to catalyze the interconversion of d-glucose and d-fructose. Other than the presence of the phosphoryl group in the PGI substrates, the two enzymes catalyze analogous transformations, and the intermediate of the isomerization reaction catalyzed by PGI is proposed to resemble the transition-state intermediate of the XYL isomerization reaction. The mechanisms used by XYL and PGI differ significantly: XYL uses two coordinated ions of Mg2+, Mn2+, or Co2+ to transfer intramolecularly a hydrogen atom between the C1 and C2 positions by a hydride-shift mechanism through a transition state in which O1, C1, C2, O2, and the transferring hydride are coplanar.
The structure of a XYL/THA complex was solved at 1.6 Å resolution (Fig. 4B) (47). Despite the presence of five aspartate and three glutamate residues in the active site, none of them participates in catalysis directly. Instead, these amino acids provide an extensive web of hydrogen-bonding interactions for binding the metal ions. Still, XYL and PGI share one feature: a positively charged residue compensates for the possible negative charge of the C1 end of the inhibitor or transition state. In XYL, Lys-182 makes a hydrogen bond to O1 of the ligand. Another interesting similarity between XYL and PGI is that both enzymes have been proposed to use a histidine residue to catalyze ring-opening of the bound cyclic substrates (64) and thereafter this histidine interacts with the hydroxyl on C5 of the linear substrate. However, it should be noted that later studies of XYL challenged the involvement of a histidine in the ring-opening step (65, 66). It was also reported that the XYL active site base that transfers the proton between O1 and O2 is assumed to be a hydroxide ion ligated to one of the metal ions (64, 67). This situation is analogous to what we propose to occur in PGI, for which lowering of the pKa of water423 would not be achieved by a metal ion, but by the surrounding network of water molecules.
PGI, TIM, and XYL share some common mechanistic features: PGI and XYL may both use a histidine for catalysis of the ring-opening of their respective substrates, and TIM and PGI catalyze the enolization step with a glutamate. However, it is still remarkable to observe three globally different mechanisms displayed by three enzymes of the same class for the catalysis of similar and apparently simple transformations. These differences could be related to the nature of the substrate itself, i.e., acyclic or cyclic and phosphorylated or nonphosphorylated. However, the origin of the differences may be more complicated than that. For instance, phosphomannose isomerase is thought to work somewhat like PGI but has a catalytic zinc ion in its active site (68), and l-fucose isomerase, which also needs a metal cofactor (Mn2+) for the isomerization of its nonphosphorylated substrates, is thought to belong to the enediol-type aldose ketose isomerases (69). Further studies will have to be carried out before forming conclusions about the existence of different mechanistic subclasses of aldose ketose isomerases.
The structures and mechanisms of PGI, TIM, and XYL illustrate that similarities or differences in overall protein fold do not necessarily correspond to similarities or differences, respectively, in active site architecture or catalytic mechanism even if the substrates are very similar. TIM and XYL have the same structural fold, an α/β barrel, but substantially different mechanisms, whereas TIM and PGI have different structural folds but are closer in terms of catalytic mechanism. The similarities between the mechanisms for TIM and PGI bring to mind the convergent evolution of the catalytic triad used by proteases for peptide bond hydrolysis. The two different methods of stabilizing the enediol(ate) intermediate in the sugar isomerases, however, show that the two folds do not necessarily need to provide all of the same functional groups to catalyze the same reaction.
Conclusions
As was seen in previous crystallographic studies of TIM and XYL with PGH and THA, respectively, the new crystal structure of the PGI/5PAH complex shows that 5PAH, the most potent PGI inhibitor reported to date, is a good structural mimic of the cis-enediol(ate) catalytic intermediate for PGI. The new crystal structure provides better information about how the reaction intermediate is likely to bind in the active site than was seen with crystal structures of PGI/5PAA complexes (41, 43) and provides an insight into the mechanism of proton transfer between the O1 and O2 atoms of the intermediate. The most significant differences are in the positioning of the C1 and C2 region of the inhibitors, the region that corresponds to the double bond in the proposed cis-enediol(ate) catalytic intermediate. Therefore, the new lengths of the hydrogen bonds determined from the PGI/5PAH structure are likely to reflect, to date, the most accurate hydrogen bond pattern between PGI and the reaction intermediate.
Acknowledgments
We thank the BioCARS staff for support and assistance in data collection (Advanced Photon Source Sector 14, Argonne National Labs). This work was partially supported by a Univ. of Illinois (Chicago) Campus Research Board award (to C.J.J.) and by a Univ. of Illinois Cancer Center/American Cancer Society Institutional Research grant (to C.J.J.).
Abbreviations
- PGI
phosphoglucose isomerase
- 5PAA
5-phospho-d-arabinonate
- G6P
d-glucose 6-phosphate
- F6P
d-fructose 6-phosphate
- 5PAH
5-phospho-d-arabinonohydroxamic acid
- TIM
triosephosphate isomerase
- XYL
xylose isomerase
- PGH
phosphoglycolohydroxamic acid
- THA
d-threonohydroxamic acid
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1KOJ).
References
- 1.Salas M, Vinuela E, Sols A. J Biol Chem. 1965;240:561–568. [PubMed] [Google Scholar]
- 2.Schray K J, Benkovic S J, Benkovic P A, Rose I A. J Biol Chem. 1973;248:2219–2224. [PubMed] [Google Scholar]
- 3.Rose I A, O'Connell E L, Schray K J. J Biol Chem. 1973;248:2232–2234. [PubMed] [Google Scholar]
- 4.Seeholzer S H. Proc Natl Acad Sci USA. 1993;90:1237–1241. doi: 10.1073/pnas.90.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan M J. FEBS Lett. 1981;130:124–126. doi: 10.1016/0014-5793(81)80679-5. [DOI] [PubMed] [Google Scholar]
- 6.Kugler W, Lakomek M. Balliere's Clin Haematol. 2000;13:89–101. doi: 10.1053/beha.1999.0059. [DOI] [PubMed] [Google Scholar]
- 7.Read J, Pearce J, Li X, Muirhead H, Chirgwin J, Davies C. J Mol Biol. 2001;309:447–463. doi: 10.1006/jmbi.2001.4680. [DOI] [PubMed] [Google Scholar]
- 8.Hardré R, Salmon L, Opperdoes F R. J Enzyme Inhib. 2000;15:509–515. doi: 10.3109/14756360009040706. [DOI] [PubMed] [Google Scholar]
- 9.Chaput M, Claes V, Portetelle D, Cludts I, Cravador A, Burny A, Gras H, Tartar A. Nature (London) 1988;332:454–455. doi: 10.1038/332454a0. [DOI] [PubMed] [Google Scholar]
- 10.Faik P, Walker J I, Redmill A A, Morgan M J. Nature (London) 1988;332:455–457. doi: 10.1038/332455a0. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe H, Takehana K, Date M, Shinozaki T, Raz A. Cancer Res. 1996;56:2960–2963. [PubMed] [Google Scholar]
- 12.Xu W, Seiter K, Feldman E, Ahmed T, Chiao J W. Blood. 1996;87:4502–4506. [PubMed] [Google Scholar]
- 13.Matsumoto I, Staub A, Benoist C, Mathis D. Science. 1999;286:1732–1735. doi: 10.1126/science.286.5445.1732. [DOI] [PubMed] [Google Scholar]
- 14.Cao M-J, Osatomi K, Matsuda R, Ohkubo M, Hara K, Ishihara T. Biochem Biophys Res Commun. 2000;272:485–489. doi: 10.1006/bbrc.2000.2803. [DOI] [PubMed] [Google Scholar]
- 15.Yakirevich E, Naot Y. Biol Reprod. 2000;62:1016–1023. doi: 10.1095/biolreprod62.4.1016. [DOI] [PubMed] [Google Scholar]
- 16.Jeffery C J. Trends Biochem Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- 17.Parr C W. Nature (London) 1956;177:1401. doi: 10.1038/1781401a0. [DOI] [PubMed] [Google Scholar]
- 18.Grazi E, De Flora A, Pontremoli S. Biochem Biophys Res Commun. 1960;2:121–125. [Google Scholar]
- 19.Chirgwin J M, Noltmann E A. J Biol Chem. 1975;250:7272–7276. [PubMed] [Google Scholar]
- 20.Chirgwin J M, Parsons T F, Noltmann E A. J Biol Chem. 1975;250:7277–7279. [PubMed] [Google Scholar]
- 21.Rose I A. Adv Enzymol Relat Areas Mol Biol. 1975;43:491–517. doi: 10.1002/9780470122884.ch6. [DOI] [PubMed] [Google Scholar]
- 22.Howell E E, Schray K J. Mol Cell Biochem. 1981;37:101–107. doi: 10.1007/BF02354933. [DOI] [PubMed] [Google Scholar]
- 23.Shapiro T A, Talalay P. Exp Parasitol. 1982;54:196–201. doi: 10.1016/0014-4894(82)90127-8. [DOI] [PubMed] [Google Scholar]
- 24.Marchand M, Kooystra U, Wierenga R K, Lambeir A-M, van Beeumen J, Opperdoes F R, Michels P A M. Eur J Biochem. 1989;184:455–464. doi: 10.1111/j.1432-1033.1989.tb15038.x. [DOI] [PubMed] [Google Scholar]
- 25.Hardré R, Salmon L. Carbohydr Res. 1999;318:110–115. doi: 10.1016/s0008-6215(99)00100-7. [DOI] [PubMed] [Google Scholar]
- 26.Schnackerz K D, Noltmann E A. J Biol Chem. 1970;245:6417–6423. [PubMed] [Google Scholar]
- 27.O'Connell E L, Rose I A. J Biol Chem. 1973;248:2225–2231. [PubMed] [Google Scholar]
- 28.Riordan J F, McElvany K D, Borders C L., Jr Science. 1977;195:884–886. doi: 10.1126/science.190679. [DOI] [PubMed] [Google Scholar]
- 29.Gibson D R, Gracy R W, Hartman F C. J Biol Chem. 1980;255:9369–9374. [PubMed] [Google Scholar]
- 30.Lu H S, Talent J M, Gracy R W. J Biol Chem. 1981;256:785–792. [PubMed] [Google Scholar]
- 31.Palmieri R H, Gee D M, Noltmann E A. J Biol Chem. 1982;257:7965–7968. [PubMed] [Google Scholar]
- 32.Meng M, Chane T L, Sun Y J, Hsiao C D. Protein Sci. 1999;8:2438–2443. doi: 10.1110/ps.8.11.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Topper Y J. J Biol Chem. 1957;225:419–425. [PubMed] [Google Scholar]
- 34.Liemans V, Malaisse-Lagae F, Willem R, Malaisse W J. Biochim Biophys Acta. 1989;998:111–117. doi: 10.1016/0167-4838(89)90261-6. [DOI] [PubMed] [Google Scholar]
- 35.Meng M, Chen Y T, Hsiao Y Y, Itoh Y, Bagdasarian M. Eur J Biochem. 1998;257:500–505. doi: 10.1046/j.1432-1327.1998.2570500.x. [DOI] [PubMed] [Google Scholar]
- 36.Meng M, Lin H Y, Hsieh C J, Chen Y T. FEBS Lett. 2001;499:11–14. doi: 10.1016/s0014-5793(01)02507-8. [DOI] [PubMed] [Google Scholar]
- 37.Dyson J E D, Noltmann E A. J Biol Chem. 1968;243:1401–1414. [PubMed] [Google Scholar]
- 38.Rose I A, O'Connell E L. J Biol Chem. 1961;236:3086–3092. [PubMed] [Google Scholar]
- 39.Shaw P J, Muirhead H. J Mol Biol. 1977;109:475–485. doi: 10.1016/s0022-2836(77)80025-9. [DOI] [PubMed] [Google Scholar]
- 40.Sun Y J, Chou C C, Chen W S, Wu R T, Meng M, Hsiao C D. Proc Natl Acad Sci USA. 1999;96:5412–5417. doi: 10.1073/pnas.96.10.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou C C, Sun Y J, Meng M, Hsiao C D. J Biol Chem. 2000;275:23154–23160. doi: 10.1074/jbc.M002017200. [DOI] [PubMed] [Google Scholar]
- 42.Jeffery C J, Bahnson B J, Chien W, Ringe D, Petsko G A. Biochemistry. 2000;39:955–964. doi: 10.1021/bi991604m. [DOI] [PubMed] [Google Scholar]
- 43.Jeffery C J, Hardré R, Salmon L. Biochemistry. 2001;40:1560–1566. doi: 10.1021/bi0018483. [DOI] [PubMed] [Google Scholar]
- 44.Lee J H, Chang K Z, Patel V, Jeffery C J. Biochemistry. 2001;40:7799–7805. doi: 10.1021/bi002916o. [DOI] [PubMed] [Google Scholar]
- 45.Hardré R, Bonnette C, Salmon L, Gaudemer A. Bioorg Med Chem Lett. 1998;8:3435–3438. doi: 10.1016/s0960-894x(98)00621-0. [DOI] [PubMed] [Google Scholar]
- 46.Davenport R C, Bash P A, Seaton B A, Karplus M, Petsko G A, Ringe D. Biochemistry. 1991;30:5821–5826. doi: 10.1021/bi00238a002. [DOI] [PubMed] [Google Scholar]
- 47.Allen K N, Lavie A, Petsko G A, Ringe D. Biochemistry. 1995;34:3742–3749. doi: 10.1021/bi00011a032. [DOI] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. In: Macromolecular Crystallography, part A. Carter C W Jr, Sweet R M, editors. San Diego: Academic; 1997. pp. 307–326. [Google Scholar]
- 49.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 50.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 51.Brunger A T. Nature (London) 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 52.Collaborative Computational Project, No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 53.Lodi P J, Chang L C, Knowles J R, Komives E A. Biochemistry. 1994;33:2809–2814. doi: 10.1021/bi00176a009. [DOI] [PubMed] [Google Scholar]
- 54.Pompliano D L, Peyman A, Knowles J R. Biochemistry. 1990;29:3186–3194. doi: 10.1021/bi00465a005. [DOI] [PubMed] [Google Scholar]
- 55.Schliebs W, Thanki N, Eritja R, Wierenga R K. Protein Sci. 1996;5:229–239. doi: 10.1002/pro.5560050206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jenkins J, Janin J, Rey F, Chiadmi M, van Tilbeurgh H, Lasters I, De Maeyer M, Van Belle D, Wodak S J, Lauwereys M, et al. Biochemistry. 1992;31:5449–5458. doi: 10.1021/bi00139a005. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Z, Sugio S, Komives E A, Liu K D, Knowles J R, Petsko G A, Ringe D. Biochemistry. 1994;33:2830–2837. doi: 10.1021/bi00176a012. [DOI] [PubMed] [Google Scholar]
- 58.Noble M E, Zeelen J P, Wierenga R K. Proteins. 1993;16:311–326. doi: 10.1002/prot.340160402. [DOI] [PubMed] [Google Scholar]
- 59.Komives E A, Chang L C, Lolis E, Tilton R F, Petsko G A, Knowles J R. Biochemistry. 1991;30:3011–3019. doi: 10.1021/bi00226a005. [DOI] [PubMed] [Google Scholar]
- 60.Bash P A, Field M J, Davenport R C, Petsko G A, Ringe D, Karplus M. Biochemistry. 1991;30:5826–5832. doi: 10.1021/bi00238a003. [DOI] [PubMed] [Google Scholar]
- 61.Alagona G, Ghio C, Kollman P A. J Am Chem Soc. 1995;117:9855–9862. [Google Scholar]
- 62.Åqvist J, Fothergill M. J Biol Chem. 1996;271:10010–10016. doi: 10.1074/jbc.271.17.10010. [DOI] [PubMed] [Google Scholar]
- 63.Cui Q, Karplus M. J Am Chem Soc. 2001;123:2284–2290. doi: 10.1021/ja002886c. [DOI] [PubMed] [Google Scholar]
- 64.Whitlow M, Howard A J, Finzel B C, Poulos T L, Winborne E, Gilliland G L. Proteins Struct Funct Genet. 1991;9:153–173. doi: 10.1002/prot.340090302. [DOI] [PubMed] [Google Scholar]
- 65.Allen K N, Lavie A, Farber G K, Glasfeld A, Petsko G A, Ringe D. Biochemistry. 1994;33:1481–1487. doi: 10.1021/bi00172a026. [DOI] [PubMed] [Google Scholar]
- 66.Lambeir A M, Lauwereys M, Stanssens P, Mrabet N T, Snauwaert J, van Tilbeurgh H, Matthyssens G, Lasters I, De Maeyer M, Wodak S J, et al. Biochemistry. 1992;31:5459–5466. doi: 10.1021/bi00139a006. [DOI] [PubMed] [Google Scholar]
- 67.Lavie A, Allen K N, Petsko G A, Ringe D. Biochemistry. 1994;33:5469–5480. doi: 10.1021/bi00184a016. [DOI] [PubMed] [Google Scholar]
- 68.Cleasby A, Wonacott A, Skarzynski T, Hubbard R E, Davies G J, Proudfoot A E, Bernard A R, Payton M A, Wells T N C. Nat Struct Biol. 1996;3:470–479. doi: 10.1038/nsb0596-470. [DOI] [PubMed] [Google Scholar]
- 69.Seemann J E, Schulz G E. J Mol Biol. 1997;273:256–268. doi: 10.1006/jmbi.1997.1280. [DOI] [PubMed] [Google Scholar]
- 70.Esnouf R. J Mol Graph Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]