Abstract
By combining the pairwise interactions between proteins, as predicted by the conserved co-occurrence of their genes in operons, we obtain protein interaction networks. Here we study the properties of such networks to identify functional modules: sets of proteins that together are involved in a biological process. The complete network contains 3,033 orthologous groups of proteins in 38 genomes. It consists of one giant component, containing 1,611 orthologous groups, and of 516 small disjointed clusters that, on average, contain only 2.7 orthologous groups. These small clusters have a homogeneous functional composition and thus represent functional modules in themselves. Analysis of the giant component reveals that it is a scale-free, small-world network with a high degree of local clustering (C = 0.6). It consists of locally highly connected subclusters that are connected to each other by linker proteins. The linker proteins tend to have multiple functions, or are involved in multiple processes and have an above average probability of being essential. By splitting up the giant component at these linker proteins, we identify 265 subclusters that tend to have a homogeneous functional composition. The rare functional inhomogeneities in our subclusters reflect the mixing of different types of (molecular) functions in a single cellular process, exemplified by subclusters containing both metabolic enzymes as well as the transcription factors that regulate them. Comparative genome analysis, thus, allows identification of a level of functional interaction between that of pairwise interactions, and of the complete genome.
Genomic associations between genes reflect functional associations between their proteins (1–8). Furthermore, the strength of the genomic associations correlates with the strength of the functional associations: genes that frequently co-occur in the same operon in a diverse set of species are more likely to physically interact than genes that occur together in an operon in only two species (7), and proteins linked by gene fusion or conservation of gene order are more likely to be subunits of a complex than are proteins that are merely encoded in the same genomes (3, 7). Other types of associations have been used for network studies, but these focus on certain specific types of functional interactions, like subsequent enzymatic steps in metabolic pathways (9), or physical interactions (10–13). In contrast, genomic associations cover a relatively wide range of functional associations between proteins (3, 7). They reflect what selection regards as functionally interacting proteins, and can therefore be regarded as an alternative measure of functional interaction. Different types of genomic association have been introduced: gene fusion (3, 4), conservation of gene order (2, 6, 14, 15), in silico recognition of shared regulatory elements (16, 17), and co-occurrence of genes (phylogenetic profiles) (5, 18, 19). Of these, we focus here on conserved gene order, which currently in prokaryotes is the most powerful type, having both a large coverage and a high selectivity (7, 14, 16). When we iteratively connect genes by this type of genomic association (14), a network of associations appears (Fig. 1). In this network the nodes are orthologous groups of genes, and the edges are the genomic associations between these groups. It has been suggested before, that by such iterative approaches, one would be able to obtain all of the proteins involved in a biological process (6, 14, 20). All of the proteins from a pathway such as the purine biosynthesis could thus be extracted with only one potential “false positive,” a hypothetical protein (6). However, with more and more genomes becoming available, such iterative linking tends to connect nearly all proteins either directly or indirectly to each other, and indeed, in our analysis the orthologous groups involved in purine biosynthesis become part of a “giant component” containing 1,611 orthologous groups. As manual expert curation to separate clusters from each other (6) may not be feasible in the long run, we seek here an automatic procedure to separate the giant component into subnetworks that would correspond to functional modules. Our analysis of the global and local properties of the giant component reveals that it consists of locally highly connected subnetworks that are connected to each other with linkers. By splitting up the network at these linkers, we identify a level of organization of proteins that lies between pairwise interactions and the complete network, and that can be regarded as a functional module: a set of proteins involved in the same biological process.
Figure 1.
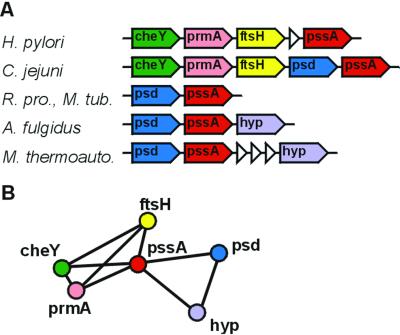
Going from conserved gene order to networks of genomic association. (A) The conserved gene order of six orthologous groups in six species. Genes with same color and name belong to the same orthologous group. The small empty triangles denote genes that do not have conserved gene order. The correspondence of the full species names to the ones used in the figure is as follows: H. pylori, Helicobacter pylori 2669; C. jejuni, Campylobacter jejuni NCTC11168; R. pro., Rickettsia prowazekii; M. tub., Mycobacterium tuberculosis Rv; A. fulgidus, Archaeoglobus fulgidus; M. thermoauto., Methanobacterium thermoautotrophicum. (B) The corresponding network. We consider two orthologous groups to have a connection if they co-occur in the same potential operon two or more times.
Methods
Orthologous Groups.
To define conserved gene order through comparative genomics, we must determine the equivalent genes across genomes (18): i.e., which genes are orthologous to each other (21). For 38 genomes (for which species, see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org) we constructed orthologous groups by iterative clustering of genes that (i) are significant (Smith–Waterman, E < 0.01) homologs (ii), are best bidirectional hits, and (iii) have conserved gene order (14). When genes in an orthologous group contain nonoverlapping hits to other genes in that group, the group is split in two to reflect the domain nature of its composition. Subsequently, any two orthologous groups A and B are merged into one group A-B if at least two independent best bidirectional hits exist between genes from group A and group B. Finally, genes that do not belong to any group are added to a group if, and only if, a strong triangular pairwise-orthology relation exists between the gene and the genes from that group. Due to the combined requirement of best bidirectional hits and conservation of gene order, the iterative usage of the pairwise-orthology relations is expected to give reliable results (14). Although we use the clusters of orthologous groups (COG) functional categories (see below), we did not use the COG orthologous groups themselves, allowing us to (i) use conserved neighborhood as an additional criterion for orthology prediction, and (ii) include orthologous groups that occur only in two species. As a result of this approach, the average size of our orthologous groups is smaller and hence probably functionally more uniform than that of the COGs.
Note that orthology is evolutionarily defined, meaning that one orthologous group can (and often does) contain different functions. The conflict of function versus orthology is one of the reasons that the network arises in the first place. We therefore try to tackle this conflict using linkers (see below). Other approaches explicitly try to assemble genes with one function into one group like the “role groups” as introduced by Overbeek et al. (6).
Quantifying the Functional Homogeneity of (Sub)clusters.
To asses whether our (sub)clusters have functional and predictive relevance, we examined their functional composition. Functional categories for our orthologous groups were obtained by comparing them to the COG database (19). When members of a group were annotated in a COG of a certain functional category, this category was assigned to our orthologous groups. Subsequently, we quantified the functional homogeneity of a (sub)cluster by the entropy of its frequency distribution of functional categories: the sum of the frequencies of the functional categories within a cluster times the logarithms of those frequencies. The stronger a cluster is dominated by a single or by a few functional categories, the lower the entropy, which becomes zero when a cluster contains only a single functional category. Entropy is dependent on the number of elements in a group—e.g., 10 orthologous groups that all fall in different functional categories will have a lower entropy than a set of 20 orthologous groups, and would thus be considered more homogeneous. To assess the statistical significance of the (sub)cluster functional homogeneities, we therefore created randomly drawn samples of all observed cluster and subcluster sizes, and computed their entropy to compare them with the observed entropies in the (sub)clusters.
Measuring the Local Connectivity, C, and Average Path Length, L.
To assess whether it is at all feasible to separate our network, consisting of orthologous groups (the nodes) and genomic associations (the edges), into subclusters, we examined two important parameters that describe its topology: C and L. C is the local connectivity, or degree of local clusteredness; it is computed by first counting all pairs of associations (cases where orthologous group A is linked to group B and to group C), subsequently counting how often these pairs are closed (B is linked to C), then, dividing the second count by the first count (22). L is the average shortest path length between orthologous groups. To obtain L we compute the shortest path between all pairs of orthologous groups, and subsequently compute the average (22).
Defining Linkers and Delineating Subclusters by Using Linkers.
To split our giant component into subclusters, we exploit the existence of linkers. Linkers are here defined as orthologous groups with mutually exclusive associations. First, we mark them by clustering for each orthologous group (A) all of the orthologous groups (N) it is connected to the group by the conservation of gene order. If, in the absence of A, these orthologous groups, N, fall into two or more subsets, then A is considered a linker. Subsequently, we perform single linkage for all of the orthologous groups, except that now the orthologous groups marked as linkers are not allowed to bring in new members: the single-linkage clustering is not allowed to run through linkers. As a final step, we connect orthologous groups that are not allocated into a group to all of the subclusters they hit, but without subsequently linking those subclusters to each other. By this procedure most linkers end up in multiple clusters. The exceptions arise when (i) linkers link to other linkers, in which case the clusters are split between the linkers instead of “at the linkers,” and (ii) two sets of orthologous groups can locally be linked only by the linker, but at a larger distance (via a detour) can also be linked in a dense grid by other orthologous group. In the latter case the cluster would not be split up and the linker would be member of only one cluster.
Signifiance of the Overrepresentation of Multiple Enzyme Commission (EC) Numbers in Linkers by Using a Binned χ2 Test.
Genes are assigned EC numbers based on their annotation in the SWISS-PROT proteomes (23). To estimate the significance of the fact that orthologous groups classified as linkers contain more genes but also contain more EC numbers we perform a binned χ2 test (24) instead of a normal χ2 test. This means that instead of testing the significance of the overrepresentation of multiple EC numbers for the total data set, we perform it for bins containing restricted sets of orthologous groups with a similar number of members. The summed χ2 test value is then compared to the expected value with a number of degrees of freedom (v) equal to the number of bins.
Results
Global Properties.
The primary object of our study, the nodes in our network, is orthologous groups of genes, which are stringently defined by using both relative levels of sequence similarity as well as conservation of genomic context (see Methods). When defining as a significant link (edge) between two orthologous groups that they co-occur with each other in the same potential operon (run, see Fig. 1) in two or more species that are not closely related (6, 14, 15), we find 3,033 orthologous groups with 8,178 pairwise significant associations in 38 species. These 3,033 orthologous groups of genes contain 29,211 genes of the 53,926 genes that have orthologs in at least two genera and of a total of 82,360 genes in these 38 species. The functional composition of the genes for which we find genomic associations appears to be unbiased relative to the complete set of genes. In terms of functional categories it is the same as the complete COG database (19)—e.g., 10.6% of the COGs and 10.3% of our orthologous groups with significant associations belong to the “energy production and conversion” category. When we iteratively connect all orthologous groups to each other by means of their genomic associations, we find one large cluster consisting of 1,611 orthologous groups (Fig. 6). All of the other clusters are much smaller: the second largest consists of 32 orthologous groups, followed by 34 clusters of sizes 6–15, and 481 clusters of 5 or less (see www.bork.embl-heidelberg.de/Docu/Modules/smalldisjoint.html for these clusters). The large cluster contains 23,430 genes, implying that 80% of the genes that have significant links belong to the large network. This cluster is a so-called “giant component” as is often observed in random networks (12). The graph layout suggests that more abundant proteins predominantly occur in the center of this large cluster (Fig. 6). The giant component contains many different orthologous groups and thus, unsurprisingly, also a mix of functions. The smaller disjoint clusters, on the other hand, seem to be functionally meaningful: 88% of the disjoint smaller clusters have a more homogeneous functional composition in terms of COG functional category (19) than that of a random cluster of the same size (P ≪ 0.001, sign test, see Methods). Thus, the small clusters reflect functional clusters, and we consider them to be functional modules.
With more genomes becoming available, we expect that smaller clusters will merge with each other, and with the giant component. Thus, there is an ever-increasing need to identify subclusters within the giant component. A first step is to probe the idea of whether the giant component contains a substructure. We do this by measuring the standard connectivity parameter C (22), which is the observed fraction of cases where, if node (i.e., orthologous group) a is connected to node b as well as node c, then, nodes b and c are also connected to each other. We find its C to be 0.60. This finding suggests that the large disjoint set is locally highly clustered, as a simulated, random network with the same number of nodes and the same number of connections has a C of only 0.005 (see Methods). Moreover, this C suggests that there are (sub)modules in the large cluster, which might be retrievable (see below).
The local connectivity (C) is actually close to that of a regular network —for example, a regular ring lattice— which is 0.75 (22). However, unlike in such a regular network, we here find that L, the shortest path in terms of the number of links between all pairs of two orthologous groups, is 5.15: by following on average 5.15 genomic associations, one can move from an orthologous group to any other. This is just slightly higher than the 3.75 steps that we, on average, find in randomly created networks with the same number of nodes and the same number of connections. This combination of L being somewhat higher than Lrandom, and C ≫ Crandom, indicates that our network of genomic associations is a “small-world network” (22). This type of network is characterized as between random and completely regular, as it contains properties of both: it is random to the extent that the L is low, while at the same time it is regular because of a relatively high C.
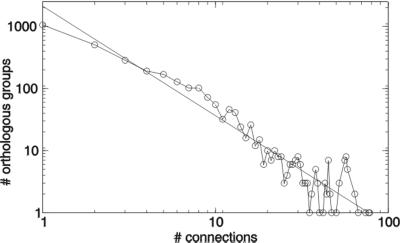
The distribution of the number associations of each orthologous group follows a power law: many orthologous groups have only one or two connections, and only a very few have many connections (Fig. 2). Aside from being a small-world network, this is therefore also a scale-free network, there is no characteristic number of connections per node (25).
Figure 2.
Distribution of the number of associations per orthologous group. The drawn line is a power law fit to the data.
Linkers.
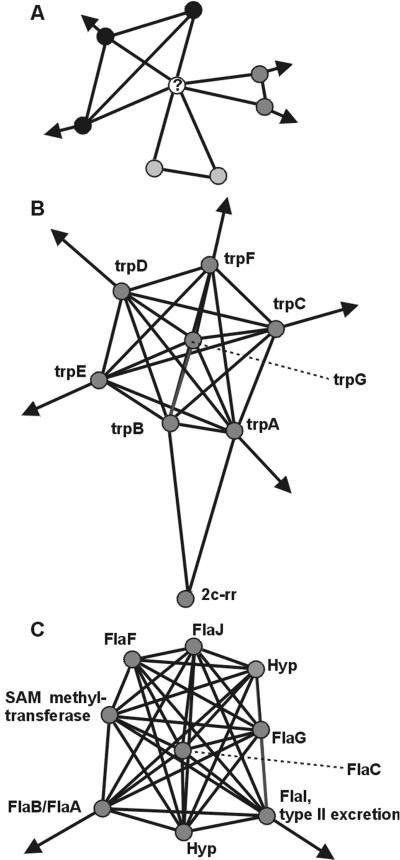
The high local connectivity parameter C indicates that there, potentially, are subclusters in the network. To separate these subclusters from each other, we identified orthologous groups with a specific type of local network topology: linkers. A linker is an orthologous groups with local mutually exclusive associations (see Methods). In other words, a linker connects two (or more) sets of orthologous groups that, at least locally in the network around the linker, are connected only that linker (Fig. 3A). All told (i.e., in the large cluster and the disjoint clusters), we found 425 linkers that locally connect at least two different sets. Linkers are expected to have multiple functions and/or to play a role in different processes. To test if they indeed have multiple functions, we determined which orthologous groups were annotated in the SWISS-PROT proteomes (23) as having multiple EC numbers. This analysis revealed that linkers contained a significant overrepresentation of orthologous groups with multiple EC numbers, even when correcting for greater average size of the groups (2.3 times as many, P < 0.05, see Methods). Thus, the local network topology of linkers also indeed reflects their (multi)functionality. It should be noted that a linker does represent a group of orthologous proteins. The multifunctionality of a linker does, therefore, not necessarily reside in the individual members of the group. The concept of orthology and its operational implementations have relevance to the evolutionary history of a group of genes, and do not necessarily imply that the proteins within an orthologous group have identical functions. The different functions in a linker can, therefore, also be distributed over the different members. Without huge experimental efforts it is impossible to derive the precise molecular function of every protein, and therefore, to answer the question as to what extent the individual proteins in a linker node are all multifunctional. We have therefore developed an operational approach that overcomes the complications that could arise from the multifunctionality of orthologous groups in predicting functional modules from genome data. The proteins in linkers can be shown to be more essential than those in nonlinkers in an individual organism: Mutations in Saccharomyces cerevisiae genes that reside in linkers have a significantly higher potential to be lethal (P < 0.05; ref. 26) than mutations in genes that do not reside in linkers.
Figure 3.
Parts of the network. Each filled circle is an orthologous group of genes, each thick line is a significant association. The dotted line is used to connect a circle to its gene name. The arrows in A mean that these orthologous groups have connections outside the focus of the panel, while the arrows in B and C denote that an orthologous group has an association to another orthologous group that is not part of the subcluster as delineated by our method. (A) Schematic example of the local network topology around a linker. The orthologous group with the “?” is the linker. The three other sets of circles of the same color are the mutually exclusive associated sets of orthologous groups. (B) The tryptophan subcluster as retrieved by our approach. The node labeled “2c-rr” is a predicted two-component response regulator. (C) Archaeal flagellum subcluster. We predict the two orthologous groups without clear predicted function to also have a role in the archaeal flagellum. The genes in the hypothetical orthologous group are: PF_353433, PAB1376, PH0544, and MJ0905. The genes in the S-adenosylmethionine (SAM)-dependent methyltransferase orthologous group are PF_352470, PAB1377, PH0545, and MJ0906.
Delineating Functional Modules by Using Linkers.
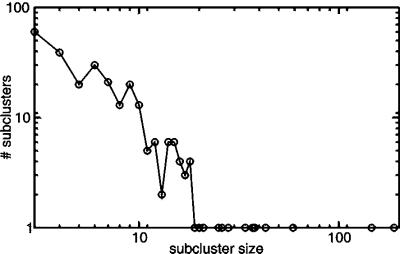
The presence of substructure suggests it should be possible to delineate subclusters in the large cluster. Because linkers reflect their affiliation to multiple processes in their local network topology, they provide a straightforward way to split this giant component. We thus split the large cluster by performing single linkage for all orthologous groups, except that linkers are not allowed to bring in new members (see Methods). With this approach the large cluster is split into 265 smaller subclusters (see www.bork.embl-heidelberg.de/Docu/Modules/subclus.html for a listing of these subclusters). The size distribution of the clusters (Fig. 4) reveals that the sizes are distributed better, albeit the two largest subclusters of size 146 and 189 seem to be outliers. These might reflect imperfect delineation. Still, 27.4% and 18.3% of the 189 orthologous groups belong, respectively, to the “cell motility and secretion” and “cell envelope biogenesis, outer membrane” category, indicating some recurring theme in this largest subcluster. In general, of the derived subclusters, 70% have a more homogeneous functional composition in terms of COG functional category than that of a random cluster of the same size (P ≪ 0.001, sign test). Moreover, nearly all are more homogeneous than the large cluster from which they stem. Because 271 orthologous groups in the giant component have an EC number, we explicitly looked at another measure of cellular process: metabolic pathway. Checking how often pairs of enzymes in the same subcluster are also in the same pathway as defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG) (27), as compared with pairs of enzymes that are in different subclusters, we found 50% of the within-subcluster enzyme pairs to be in the same pathway versus 9% of the between-subclusters pairs. Among the subclusters are well known cases such as the tryptophan biosynthesis genes. Our approach successfully delineated this subcluster despite multiple tryptophan biosynthesis genes being linked to other genes and thereby to the large cluster (Fig. 3B).
Figure 4.
Occurrence distribution of the number of subcluster sizes derived from the giant component. Most subclusters are of size 3. The biggest subclusters seem to be outliers and, thus, might indicate a failure of our method to correctly split them.
Not only can we retrieve known pathways and processes such as tryptophan biosynthesis, but we can also use the subclusters for function prediction. For example, one orthologous group of unknown function and a group for which only its general molecular function is known, SAM-dependent methyltransferase, falls into a subcluster exclusively consisting of archaeal flagellum (28) genes (Fig. 3C). These two orthologous groups and the archaeal genes with which they cluster, occur only in archaea. They can thus be predicted to have a role in the assembly, regulation, or motility of the archaeal flagellum. In general, moving from a gene-based to a comprehensive view of genomic associations, by delineating subclusters, allows us to make better predictions for the process to which a gene belongs. This is because, by introducing a cut-off in the list of genes indirectly associated to a gene, we define a set of genes from which we can take the common functional denominator.
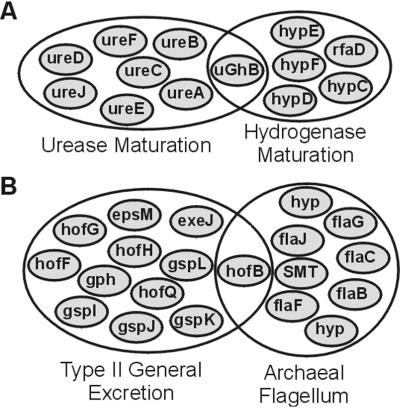
In contrast to conventional hierarchical clustering, in our approach orthologous groups (the linkers) can belong to multiple subclusters. Due to associations beyond their immediate local topology, not all linkers are necessarily assigned to different subclusters (see Methods). We find that 210 linkers of the set of 425 are part of multiple subclusters. As mentioned above, the expected underlying cellular reason for linkers to be in multiple subclusters is multifunctionality on a molecular or a cellular process level. For example, in the maturation of the nickel-containing enzymes urease and hydrogenase, one orthologous group performs two related, but different molecular functions (29). It turns out that this group achieves this specialization by duplication, leading to different functional associations and assignment to two different subclusters (Fig. 5A). Even when the molecular function among the proteins in one orthologous group is the same, it can perform this function within multiple cellular processes, like the integral membrane protein transport orthologous group involved in type II protein excretion pathway, as well as the archaeal flagellum (Fig. 5B). This constellation reflects our expectations for linkers in general; not only do they prevent the random linkage of two subclusters, they provide a handle for dissecting the complex functional and evolutionary relations between cellular processes.
Figure 5.
Venn diagrams of linkers in multiple subclusters. Each small ellipse is an orthologous group. The big ellipses circumscribe the subclusters as our approach delineates them. Orthologous groups are named by a gene name of a prominent member. A shows the two subclusters of which the hypF/ureG orthologous group is a member. This orthologous group is named uGhB in this figure. B shows the two subclusters, of which the integral membrane protein transport orthologous group (hofB) is a member. Note that one of the two subclusters is the archaeal flagellum subcluster from Fig. 3C.
Just as gene function prediction by genomic context methods is complementary to that by homology determination (6), a functional classification based on genomic context is complementary to one that is based on molecular function. Hence come differences that we observe between classification systems that are (largely) based on homology relations [e.g., domain databases such as a simple modular architecture research tool (SMART) (30), or an orthology/domain database such as COGs (19)], and a system that is based on genomic context. Such conflicting classifications should be interpreted not as errors in either one of the systems, but rather in terms of the difference in conceptual approach. For example, we find one subcluster that contains three enzymes from amino sugar metabolism catalyzing sequential steps, together with a transcriptional regulator of hitherto unknown specificity. Based on this finding, we expect this regulatory orthologous group (consisting of PA3757, yvoA, XF1461, and DRA0211), to regulate the enzymes. In the COG category scheme this is an inhomogeneity, as the regulator belongs to the “transcription” category, whereas the enzymes are “carbohydrate transport and metabolism.” More generally we see that, whereas in the COG classification scheme, transcription falls into one functional class, in our classification it is spread out over 78 subclusters. In only 4 (1.6%) of the subclusters are transcription genes the largest group within that cluster. This observation illustrates the complementarity of a genomic context based classification scheme as well as the potential of this approach to assign proteins to cellular processes for which the molecular functions are known.
Discussion
General Network Properties.
The network of pairwise genomic associations derived from conserved gene order exhibits interesting network features that can be interpreted in terms of the functional relations between the genes. There is a large dominant cluster that spans most of the genes. The values of C and L in the network have important implications respectively for the identification of functional modules and for the connectedness of the processes in a cell. Although the low L, i.e., the low number of associations to get from one orthologous group to any other group, suggests that the functions of all proteins are intimately connected, the high local connectivity, C, indicates that one can still identify functional modules, and thus draw boundaries between the various processes. The power law in the number of connections indicates that it is also a scale-free network (25). Such a network is thought to emerge when a network has grown by preferentially attaching new genes/nodes to already existing highly connected, genes/nodes (25). This evolutionary scenario is also supported by the predominance of widespread, and thus presumably older, orthologous groups in the “center” of the large cluster (see Fig. 6). The global network properties that we find have recently also been described for other complex large-scale biological interaction networks (9–13), and protein domain evolutionary networks (31). We thus conclude that the small-world and scale-free properties are general for biological networks.
Local Network.
We analyze orthologous groups in terms of the specific network topology that surrounds them. Orthologous groups with locally mutually exclusive network associations, so-called linkers, reflect their different genomic associations by having significant overrepresentation of genes with multiple EC numbers. In addition they contain more lethal mutations, probably because they link various processes and/or have roles in multiple processes. They are crucial points both in the functional as well as the genomic association network, making them promising targets for antimicrobial drugs. In general, the local association network around orthologous groups reflects their functional embedding. It should be noted that our linkers are not comparable to the “hubs” introduced in ref. 9. The discrepancy lies not only in the fact that hubs are substrates (including ATP, NAD, H2O, etc.) as opposed to our linkers, which are orthologous groups of genes, but also, more importantly, in that linkers link different processes (i.e., different sets of orthologous groups), whereas hubs merely link a large number of entities.
Subclusters and Functional Classification.
That one could obtain all of the proteins involved in a biological process by an iterative search for conserved gene order has been suggested before (6, 14, 20). Actually, it is not so straightforward, as such an iterative search tends to connect “everything with everything.” This trend is likely to only get worse with more genomes becoming available. However, the topology of these genomic association networks suggests a natural way of splitting genes into meaningful subclusters, in a manner that also allows certain genes to belong to different modules. The thereby retrieved subclusters reflect known processes. More importantly, these subclusters improve function predictions for hypothetical genes and assign genes with a known molecular function to a biological process. The clusters and subclusters can serve as the basis for a new concept for functional classification that is defined by comparative genome analysis and that is complementary to one that is based on molecular function. Ultimately, this work should contribute to an integration of the different levels of functional description (32), with the aim of obtaining a natural classification scheme for proteins and cellular processes (33).
Supplementary Material
Abbreviations
- COG
clusters of orthologous groups
- EC
Enzyme Commission
- SAM
S-adenosylmethionine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dandekar T, Snel B, Huynen M, Bork P. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 2.Overbeek R, Fonstein M, D'Souza M, Pusch G D, Maltsev N. In Silico Biol. 1998;1:0009. http://www.bioinfo.de/isb/1998/01/0009 , http://www.bioinfo.de/isb/1998/01/0009. . [PubMed] [Google Scholar]
- 3.Enright A J, Iliopoulos I, Kyrpides N C, Ouzounis C A. Nature (London) 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 4.Marcotte E M, Pellegrini M, Ng H L, Rice D W, Yeates T O, Eisenberg D. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 5.Pellegrini M, Marcotte E M, Thompson M J, Eisenberg D, Yeates T O. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overbeek R, Fonstein M, D'Souza M, Pusch G D, Maltsev N. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huynen M, Snel B, Lathe W, III, Bork P. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanai I, Derti A, DeLisi C. Proc Natl Acad Sci USA. 2001;98:7940–7945. doi: 10.1073/pnas.141236298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong H, Tombor B, Albert R, Oltvai Z N, Barabasi A L. Nature (London) 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwikowski B, Uetz P, Fields S. Nat Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 12.Wagner A. Mol Biol Evol. 2001;18:1283–1292. doi: 10.1093/oxfordjournals.molbev.a003913. [DOI] [PubMed] [Google Scholar]
- 13.Lappe M, Park J, Niggemann O, Holm L. Bioinformatics. 2001;17:S149–S156. doi: 10.1093/bioinformatics/17.suppl_1.s149. [DOI] [PubMed] [Google Scholar]
- 14.Snel B, Lehmann G, Bork P, Huynen M A. Nucleic Acids Res. 2000;28:3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolf Y I, Rogozin I B, Kondrashov A S, Koonin E V. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 16.McGuire A M, Church G M. Nucleic Acids Res. 2000;28:4523–4530. doi: 10.1093/nar/28.22.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Terai, G., Takagi, T. & Nakai, K. (2001) Genome Biol.2, research0048. [DOI] [PMC free article] [PubMed]
- 18.Huynen M A, Bork P. Proc Natl Acad Sci USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tatusov R L, Natale D A, Garkavtsev I V, Tatusova T A, Shankavaram U T, Rao B S, Kiryutin B, Galperin M Y, Fedorova N D, Koonin E V. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lathe W C, III, Snel B, Bork P. Trends Biochem Sci. 2000;25:474–479. doi: 10.1016/s0968-0004(00)01663-7. [DOI] [PubMed] [Google Scholar]
- 21.Fitch W M. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 22.Watts D J, Strogatz S H. Nature (London) 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 23.Apweiler R, Biswas M, Fleischmann W, Kanapin A, Karavidopoulou Y, Kersey P, Kriventseva E V, Mittard V, Mulder N, Phan I, Zdobnov E. Nucleic Acids Res. 2001;29:44–48. doi: 10.1093/nar/29.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kendall M, Stuart A. Distribution Theory, The Advanced Theory of Statistics. Vol. 1. London: Griffin; 1977. pp. 243–247. [Google Scholar]
- 25.Barabasi A L, Albert R. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 26.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas N A, Bardy S L, Jarrell K F. FEMS Microbiol Rev. 2001;25:147–174. doi: 10.1111/j.1574-6976.2001.tb00575.x. [DOI] [PubMed] [Google Scholar]
- 29.Olson J W, Mehta N S, Maier R J. Mol Microbiol. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 30.Schultz J, Milpetz F, Bork P, Ponting C P. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wuchty S. Mol Biol Evol. 2001;18:1694–1702. doi: 10.1093/oxfordjournals.molbev.a003957. [DOI] [PubMed] [Google Scholar]
- 32.Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan Y. J Mol Biol. 1998;283:707–725. doi: 10.1006/jmbi.1998.2144. [DOI] [PubMed] [Google Scholar]
- 33.Benner S A, Gaucher E A. Trends Genet. 2001;17:414–418. doi: 10.1016/s0168-9525(01)02320-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.