Abstract
RecJ, a 5′ to 3′ exonuclease specific for single-stranded DNA, functions in DNA repair and recombination systems. We determined the crystal structure of RecJ bound to Mn2+ ion essential for its activity. RecJ has a novel fold in which two domains are interconnected by a long helix, forming a central groove. Mn2+ is located on the wall of the groove and is coordinated by conserved residues characteristic of a family of phosphoesterases that includes RecJ proteins. The groove is composed of residues conserved among RecJ proteins and is positively charged. These findings and the narrow width of the groove indicate that the groove binds single- instead of double-stranded DNA.
All living cells have DNA repair and recombination systems for the maintenance of genomic integrity (1). Some DNA exonucleases have key roles in such systems. Of these, RecJ, a 5′ to 3′ exonuclease specific for single-stranded DNA (ssDNA), acts in homologous recombination, base excision repair (BER), and methyl directed mismatch repair (MMR) (1–6). RecJ functions in the RecF pathway in homologous recombination and is essential in recombination-dependent replication (3, 7). RecJ initiates homologous recombination by degrading the DNA unwound by RecQ helicase at double-stranded DNA breaks (3). When the replication fork undergoes DNA damage, RecJ and RecQ also degrade lagging strands on the replication fork, interrupting DNA replication (7). After the damage is repaired, DNA replication is resumed by RecF protein. No other exonucleases can replace RecJ as an interrupter of replication (7). In addition to the assistance of recombinational repair, RecJ can remove a 5′-terminal 2′-deoxyribose-5-phosphate residue that is produced in the BER pathway (4). RecJ also functions in MMR by removing ssDNA containing mismatch lesion (5, 6).
RecJ proteins and its homologues contain five conserved motifs (8). Genomic database screening showed that these motifs are ubiquitous in bacterial, archaeal, and eukaryotic genomes and belong to a family of predicted phosphoesterases, the DHH family (8). The biochemical activities of only two proteins in this family have been characterized, those of RecJ exonuclease and the exopolyphosphatase of Saccharomyces cerevisiae (scPPX1) (8, 9). Of the other members of the DHH family, these worth noting are the PRUNE gene product of Drosophila melanogaster, the mutation of which leads to a lethal association with a single mutant copy of Killer-of-prune, and mycoplasmal MgPA, which encodes an cytadherence-associated protein (8). Because RecJ requires Mg2+ ion for its activity, some Asp and His residues in these motifs are believed to be involved in metal binding (8, 10). Mutation analysis of Escherichia coli RecJ (ecRecJ) showed that all of the motifs are necessary for exonuclease activity (10). Several homologues, including scPPX1, lack motif V, and this may differentiate the functions of individual homologues (8). As the tertiary structure of neither RecJ nor any of its homologues is available, the function of these motifs and the mode of interaction with DNA are unknown.
We describe the crystal structure of the RecJ from Thermus thermophilus HB8 bound to Mn2+ ion. Its structure suggests both particular roles for the characteristic motifs in RecJ homologues and the molecular mechanism of its exonuclease activity.
Methods
Crystallization and Data Collection.
Cd-ttRecJ (the truncated ttRecJ corresponding to the core domain composed of 40–463 residues) and selenomethionyl (Se-Met) cd-ttRecJ were purified as described (11). A 10-mg⋅ml−1 sample of protein was dissolved in a solution containing 20 mM Tris⋅HCl, 100 mM NaCl, 0.1 mM EDTA, 10% glycerol, at pH 7.5. Crystals of cd-ttRecJ were grown by hanging drop vapor diffusion at 20°C. A 0.4-μl portion of a 100 mM MnCl2 solution was added to 2 μl of the protein solution, and the mixture was combined with 2 μl of the reservoir solution (1.0 M ammonium sulfate/0.2 M sodium/potassium tartrate/50 mM sodium citrate/10% glycerol, pH 5.8). The solution obtained was equilibrated against the reservoir solution, producing polyhedral crystals. The Se-Met cd-ttRecJ was crystallized by the same method. The native and Se-Met cd-ttRecJ crystals were cubic, with the space group P213, and the unit cell dimensions a = b = c = 141.6 Å. This crystal form has a very high solvent content (76%), i.e., VM is 5.12 Å3/Da, when one cd-ttRecJ molecule is in an asymmetric unit. This was confirmed by the following analysis.
The crystals were soaked in a cryoprotectant solution (1.0 M ammonium sulfate, 0.2 M sodium/potassium tartrate/50 mM sodium citrate/10 mM MnCl2/20% glycerol/20 mM Tris⋅HCl/100 mM NaCl, pH 5.8), then flash cooled by a Rigaku (Tokyo) cryostream cooler. All x-ray data were collected at 100 K with a Mar charge-coupled device detector and synchrotron radiation at BL41XU (SPring-8, Hyogo, Japan). Diffraction data were collected for one crystal at two wavelengths that sandwich the absorption edge of manganese to identify the Mn2+ ion. Processing of the diffraction images and scaling of the integrated intensities were done with the program HKL2000 (12). Data collection statistics are given in Table 1.
Table 1.
Crystallographic statistics
| Native | Se-Met peak | Native
|
||
|---|---|---|---|---|
| Mn2+ high remote | Mn2+ low remote | |||
| Data collection | ||||
| Wavelength, Å | 0.9795 | 0.9793 | 1.8808 | 1.9051 |
| Camera length, mm | 200 | 220 | 120 | 120 |
| Oscillation angle, deg | 0.5 | 0.5 | 0.5 | 0.5 |
| Exposure time, sec | 8 | 2 | 20 | 20 |
| Number of images | 180 | 180 | 114 | 114 |
| Resolution range, Å | 60.0–2.9 | 50.0–3.2 | 50.0–3.5 | 50.0–3.5 |
| Measured reflections | 584147 | 361587 | 214259 | 208042 |
| Unique reflections | 21198 | 29761 | 23281 | 23108 |
| Completeness, %* | 99.8 (100) | 99.6 (99.9) | 99.9 (99.5) | 99.9 (100) |
| Rsym, %*† | 4.9 (27.9) | 7.2 (29.1) | 8.0 (26.6) | 8.3 (19.3) |
| Phase statistics | ||||
| Phasing power | 2.17 | |||
| Figure of merit | 0.384 | |||
| Model statistics | ||||
| Number of amino acids | 385 | |||
| R factor, %‡ | 22.8 | |||
| Rfree, %‡ | 25.9 | |||
| r.m.s. deviation | ||||
| Bond lengths, Å | 0.008 | |||
| Bond angles, deg | 1.51 | |||
Values in parentheses are for the outermost resolution shell.
Rsym = ∑hkl∑i|Ii(hkl)–〈I(hkl)〉|/∑hkl∑iIi(hkl), where 〈Ihkl〉 is the mean intensity of multiple Ii(hkl) observations for symmetry-related reflections.
R factor = ∑∥Fobs| − |Fcalc∥/∑|Fobs|. Rfree was calculated with 5% of the data.
X-Ray Structure Determination.
The structure of RecJ was solved by single-wavelength anomalous dispersion technique by using Se atoms. The positions of four Se atoms were determined from a Bijvoet difference Patterson map. Atomic parameters of these atoms were refined at 3.2-Å resolution with the program cns 1.0 (13) to calculate the initial map. Subsequent phase extension to 2.9 Å against the native data and density modification facilitated tracing of the polypeptide chain. The model was built with the aid of the known sequence by use of the program o (14) and refined by the program cns 1.0 (13). The four Se atom sites deduced from the difference Patterson map were coincided with the four sulfur sites of the methionine residues in the model. The fifth largest peak surrounded by the conserved residues in the Bijvoet difference Fourier map for the Se-Met cd-ttRecJ crystal was subsequently identified as that of Mn2+ ion by an anomalous scattering experiment. X-ray absorption fine structure measurement determined the Mn2+ absorption edge to be 1.8920 Å. A map based on Bijvoet differences measured at λ = 1.8808 Å (f" ≈ 3.4) showed a significant electron density at the site (Fig. 1c), whereas that at λ = 1.9051 Å (f" ≈ 0.46) gave no significant density (data not shown). Mn2+ was added to the model in the subsequent refinement. The respective final crystallographic R factor and Rfree values are 22.8 and 25.9% at 2.9-Å resolution. No water molecule was included in the model because of the limited resolution of the diffraction data. Residues were 83.4% in the most favored regions of the Ramachandran plot and 0.3% in the disallowed regions, as determined by the program procheck (15).
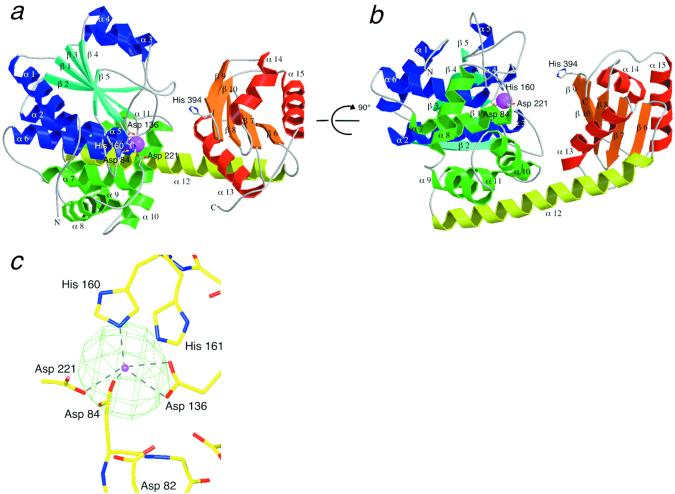
Figure 1.
The RecJ structure. (a, b) Ribbon diagrams of RecJ. b is viewed at an orientation rotated 90° from a. β-strands in domain I are shown in cyan; α-helices in the α/β structure in domain I, blue; α-helices in the helix-rich region in domain I, green; β-strands in domain II, orange; α-helices in domain II, red; and α12, yellow. The Mn2+ ion is shown by a magenta ball. The side chains of the residues bound to Mn2+ are indicated by a stick model. (c) The Mn2+-binding site. The electron density (green) of the Mn2+ ion derived from the Bijvoet difference (λ = 1.8808 Å) is superimposed on the model. The position of the Mn2+ ion is shown by a magenta ball. The distances to Mn2+ are Asp-82, 4.55 and 4.68 Å; Asp-84, 2.39 Å; Asp-136, 2.64, and 2.63 Å; His-160, 2.24 Å; His-161, 3.22 Å; and Asp-221, 2.50 Å. a and b were drawn with MOLSCRIPT (31) and RASTER 3D (32), c with O (14) and MOLRAY (33).
Results and Discussion
Overall Structure.
Because the full-length RecJ protein from T. thermophilus (ttRecJ) was expressed as an inclusion body, we expressed the catalytic core domain of ttRecJ corresponding to the region from 40 to 463 (cd-ttRecJ) as a soluble form (11). Cd-ttRecJ had all five motifs and 5′ to 3′ exonuclease activity in the presence of a metal ion (11). The crystals of cd-ttRecJ were grown in the presence of Mn2+ ion, and its structure was determined at 2.9-Å resolution (Table 1). The central region of cd-ttRecJ was well ordered as regards electron density, but the nine N- and 30 C-terminal residues were disordered.
Cd-ttRecJ has two domains (I and II) connected by a long α-helix (α 12) (Fig. 1 a and b). This is a novel folding; a search of the structural database, the dali server, for the whole protein found no similar folding (16). The N-terminal domain (domain I) is further divided into two regions. One folds into the α/β structure, in which five parallel β-strands (β 1–5) are flanked by α-helices (α 1–6), although the helix between β 4 and β 5 is lacking. The other region is composed of five helices (α 7–11). The C-terminal domain (domain II), which is smaller than domain I, consists of five mixed β-strands (β 6–10) flanked by three α-helices (α 13–15). A large groove exists between the two domains. A few interactions maintain this structure; three hydrogen bonds are present between domains I and II, six between α 12 and domain I, and two between α 12 and domain II.
Metal Coordination.
The electron density map of RecJ showed a significant density surrounded by several residues. Using a Bijvoet difference Fourier map, we identified the density as that of the Mn2+ ion. This Mn2+ is coordinated by Asp-84, Asp-136, His-160, and Asp-221 (Fig. 1c). Notably, these amino acids are all in motifs I–IV (Fig. 2) and are perfectly conserved in the DHH family (8). They are situated in the loops or terminals of the α-helices and β-strands. Mutation analysis for ecRecJ showed that all four residues are essential for the exonuclease activity of this protein (10). Exonuclease activity of RecJ depends on Mg2+, Mn2+, or Co2+, not on Ca2+ or Zn2+ (11). This metal requirement is the same as that of scPPX1 (9). Although Mg2+ is considered to be a natural cofactor of RecJ, the coordination by the His residue in the RecJ structure is more suited to Mn2+ than Mg2+, because Mg2+ prefers the harder ligands of carboxyl oxygens (17, 18).
Figure 2.
Secondary structure and sequence alignment. Seven RecJ proteins (T. thermophilus, E. coli, Aquifex aeolicus, Helicobacter pylori, Haemophilus influenzae Rd, Thermotoga maritima, and Synechocystis sp) were aligned by the program CLUSTAL W (34). For clarification, the sequences of T. thermophilus RecJ (ttRecJ), E. coli RecJ (ecRecJ), and H. influenzae RecJ (hiRecJ) are shown within the 40–463 region in ttRecJ. Residues conserved in the seven RecJ proteins are colored blue; conserved substitutions, light blue; semiconserved substitutions, cyan. The five motifs proposed by Aravind and Koonin (8) are designated by red boxes. Perfectly conserved Asp and His residues in the five motifs are highlighted in red. Residues binding to the Mn2+ ion are indicated by red asterisks. Two additional motifs, proposed by Sutera et al. (10), are represented by green boxes. Residues that may be involved in ssDNA binding are indicated by purple asterisks. The secondary structures of cd-ttRecJ are shown above the sequences; horizontal bars indicate α-helices, arrows β-strands. Broken lines indicate disordered regions. The color scheme for each segment is the same as that in Fig. 1a.
Many nucleases bind two or more metal ions at their active sites (18–23). Anomalous difference maps of native cd-ttRecJ at three wavelengths (0.9795, 1.8808, and 1.9051 Å) showed no significant density other than one Mn2+, indicating that RecJ has only one metal ion, like DNase I and ExoIII (24, 25). E. coli DNA polymerase I binds to a second metal ion when deoxynucleoside monophosphate is present (19). Because Asp-82 and -80 are near the Mn2+ site and perfectly conserved in seven RecJ proteins, there is the possibility that they bind to a second metal ion in the presence of nucleotide or ssDNA.
The DNA-Binding Site.
DNA is thought to be bound in the groove at the center of RecJ because the motifs and metal site are clustered on the wall of the groove. The groove is approximately 11 Å wide, 15 Å deep, and 23 Å long, too narrow for the groove to bind to double-stranded DNA (dsDNA), the diameter of which is ≈20 Å (26). The crystal structure of exonuclease I (ExoI), which degrades ssDNA in the 3′ to 5′ direction, has been reported (27). This enzyme is monomeric, like RecJ, and has a narrow groove (<14 Å in width) in the center of its molecule. The narrow grooves common to both RecJ and Exo I may be characteristic of the exonucleases specific for ssDNA.
The sequence nonspecificity of RecJ suggests that it mainly interacts with DNA through the DNA backbone. Of the DNA metabolizing enzymes, the NH group of the Arg residues, the NH
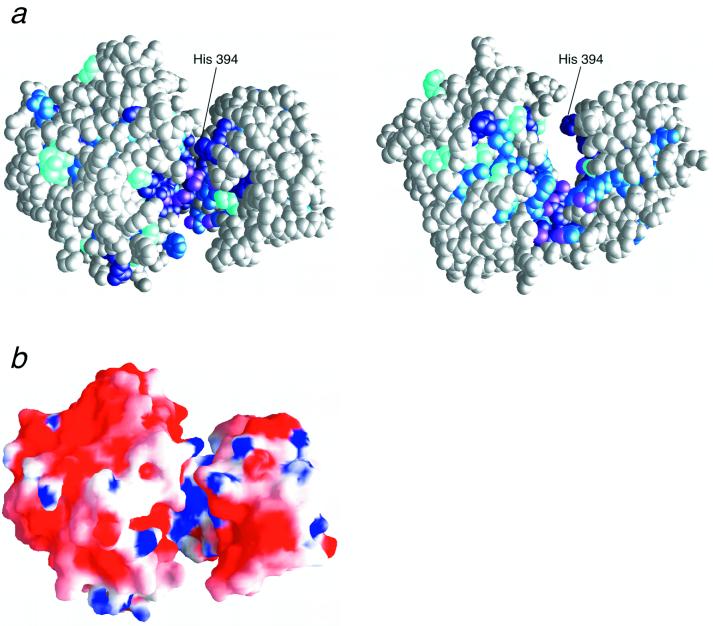
group of the Arg residues, the NH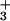 group of the Lys residues, or the NH2 group of the Asn and Gln residues interact with the DNA backbone (26). As shown in Fig. 3a, the groove is surrounded by the highly conserved residues: Arg-110, Arg-277, Arg-310, Arg-350, Arg-370, Asn-273, Asn-307, and Gln-311. These residues extend downward from the Mn2+ site (Fig. 3a). The surface of the groove is positively charged because of the conserved Arg residues, although cd-ttRecJ is acidic (pI = 5.5) and surrounded mainly by negatively charged residues (Fig. 3b).
group of the Lys residues, or the NH2 group of the Asn and Gln residues interact with the DNA backbone (26). As shown in Fig. 3a, the groove is surrounded by the highly conserved residues: Arg-110, Arg-277, Arg-310, Arg-350, Arg-370, Asn-273, Asn-307, and Gln-311. These residues extend downward from the Mn2+ site (Fig. 3a). The surface of the groove is positively charged because of the conserved Arg residues, although cd-ttRecJ is acidic (pI = 5.5) and surrounded mainly by negatively charged residues (Fig. 3b).
Figure 3.
Conserved residues and the electrostatic surface of RecJ. (a) Orthogonal views of the CPK model of RecJ. The left and right views are in the same respective orientations as in Fig. 1 a and b. The extent of conservation is indicated by differences in the shades of blue, as described in Fig. 2. The NH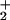 group of the Arg residue and NH2 group of the Asn and Gln residues, which are available for ssDNA binding, are shown in purple. (b) The electrostatic surface potential of RecJ viewed from the same orientation as in Fig. 1a. These figures were drawn with GRASP (35).
group of the Arg residue and NH2 group of the Asn and Gln residues, which are available for ssDNA binding, are shown in purple. (b) The electrostatic surface potential of RecJ viewed from the same orientation as in Fig. 1a. These figures were drawn with GRASP (35).
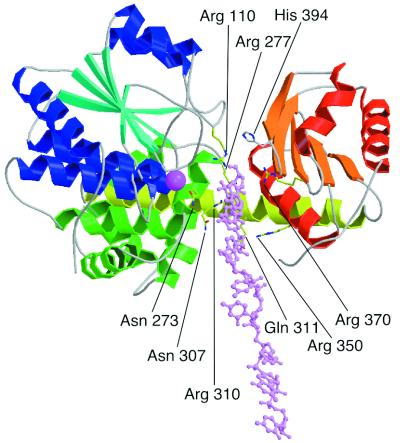
A DNA-binding model based on the characteristic features of the groove is shown in Fig. 4. RecJ degrades ssDNA from the 5′ terminus, and cleavage of DNA should occur around the Mn2+ site. This proposal is consistent with the present model, in which the Mn2+ site is close to the site at which the phosphodiester bond in the bound ssDNA is cleaved. Sequence alignment identified two additional motifs in the RecJ-related superfamily. One has Asn-273 and Arg-277, the other Arg-370 (Fig. 2) (10). These motifs face the groove (Fig. 4). In the three RecJ paralogs of Methanococcus janasschii, the two with the motif that includes Asn-273 and Arg-277 can complement an E. coli recJ mutation, whereas the other that does not have a motif does not have such activity (28). Moreover, mutation of the Arg residue of ecRecJ in the other motif (Arg-370 in ttRecJ) lost its exonuclease activity (10). These results offer strong support for the proposal that the two additional motifs are responsible for the binding to ssDNA.
Figure 4.
Model of the 8-mer ssDNA bound to RecJ. DNA is shown in pink stick model. RecJ is viewed from the same orientation as in Fig. 1a. Side chains of the possible residues for ssDNA binding are shown by stick models. This figure was drawn with MOLSCRIPT (31) and RASTER 3D (32).
The 5′ exonucleases of known structure: T5 5′ exonuclease, FEN-I, and the N-terminal domain of Taq polymerase I, have a common core structure and structure-specific endonuclease activity for the flap structure of DNA (20–22, 29). Their structures form a wide groove that can bind to double-stranded (ds)DNA, and helical arch or loop, into which ssDNA is threaded. These structural features, specific to both ssDNA and dsDNA, may show structure-specific endonuclease activity for the flap structure. In contrast, RecJ has only a narrow groove that may prefer ssDNA, indicative that RecJ has no structure-specific endonuclease activity for the flap structure. RecJ is believed to degrade the pseudo Y-structure of DNA (2). The putative DNA-binding region in RecJ is short, as long as 4 nucleotides, whereas that in ExoI is 12 nucleotides (27) (Fig. 4). ExoI cannot degrade ssDNA to a size smaller than 4 mer (30). The shortness of the DNA-binding region in RecJ may make it possible to degrade ssDNA completely in the pseudo Y-structure of DNA. This may be important for the bacterium not having flap endonuclease.
Implications for the Catalytic Properties of the DHH Family.
The coordination number of the Mg2+, Mn2+, and Co2+ ions is usually six (17). Because Mn2+ is coordinated by four amino acids, two of the sites at Mn2+ are vacant (not occupied by amino acid residues). In the nuclease reaction, the divalent metal ion may interact directly with the scissile phosphate of DNA or with water to stabilize the pentavalent phosphorous transition state (23). The two vacant sites are directed to the groove and may be spatially suitable for the nuclease reaction (Fig. 1).
The cleavage reaction of the phosphodiester bond requires nucleophilic water (23). Two mechanisms for the activation of water molecule have been proposed for nucleases (19, 20, 23–25). One is that the water molecule is activated by the bound metal ion, as exemplified by the 3′ to 5′ exonuclease of E. coli DNA polymerase I (19). Glu-357 in DNA polymerase I functions to position the water molecule in the proper direction for attack on the scissile phosphate (19). Asp-82 is within 5 Å of Mn2+ and therefore able to interact with an available water molecule at the vacant Mn2+ site. Mutation of aspartic acid at 81 in ecRecJ (Asp-82 in ttRecJ) to alanine results in the loss of exonuclease activity (10), which suggests that Asp-82 functions beneficially to position a water molecule for nucleophilic attack on the phosphodiester bond. The other proposed mechanism is the activation of water by amino acids, in which Mn2+ can bind to the oxygen of the scissile phosphate (24, 25). Lys, Arg, His, Glu, and Asp residues are candidates for activators of the water molecule. Asp-82 and His-161 are near Mn2+ and possibly are accessible to the phosphate of the DNA bound to Mn2+. In any case, Asp-82 and His-161 are candidates for activators of the water molecule.
RecJ Motifs.
The RecJ structure reveals that motifs I–IV are clustered and bind to the metal ion in domain I and that motif V is present in domain II (Fig. 1). Multiple sequence alignment showed that motifs I to IV are conserved in all of the DHH family members, whereas motif V is not present in several homologues, including scPPX1 (8). The metal requirements of RecJ and scPPX1 are identical, metal ion being essential for their activities (9, 11). These findings suggest that motifs I–IV are ubiquitous for phosphoesterase activity and that motif V is important for substrate specificity (8). Mutation of the conserved His residue in motif V (His-394 in ttRecJ) markedly decreased exonuclease activity (10). His-394 appears to be too far from the Mn2+ site (≈15 Å) to interact with the scissile DNA (Fig. 4). The side chain of His-394 protrudes into the groove from the opposite side of the Mn2+ site. This protrusion is most likely involved in the proper binding of DNA along the groove.
Acknowledgments
We thank Dr. Masahide Kawamoto of SPring-8 for valuable help with data collection by using synchrotron radiation at BL41XU. Part of this research was done with the approval of the SPring-8 Proposal Review Committee (No. 2001A0531-NL-np and 2001B0037-CL-np). This work was supported in part by Grants-in-Aid for Scientific Research, nos. 11169223 and 12680655 (to K.F.) and 1278042 (to R.M.), from the Ministry of Education, Culture, Sports, Science and Technology of Japan. A.Y. is the recipient of a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists (no. 809).
Abbreviations
- ssDNA
single-stranded DNA
- cd-ttRecJ
the truncated ttRecJ corresponding to the core domain composed of 40–463 residues
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates of cd-ttRecJ have been deposited in the Protein Data Bank (PDB ID code 1IR6).
References
- 1.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 2.Lovett S T, Kolodner R D. Proc Natl Acad Sci USA. 1989;86:2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kowalczykowski S C. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 4.Dianov G, Sedgwick B, Daly G, Olsson M, Lovett S, Lindahl T. Nucleic Acids Res. 1994;22:993–998. doi: 10.1093/nar/22.6.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper D L, Lahue R S, Modrich P. J Biol Chem. 1993;268:11823–11829. [PubMed] [Google Scholar]
- 6.Burdett V, Baitinger C, Viswanathan M, Lovett S T, Modrich P. Proc Natl Acad Sci USA. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courcelle J, Hanawalt P C. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 8.Aravind L, Koonin E V. Trends Biochem Sci. 1998;23:17–19. doi: 10.1016/s0968-0004(97)01162-6. [DOI] [PubMed] [Google Scholar]
- 9.Wurst H, Kornberg A. J Biol Chem. 1994;269:10996–11001. [PubMed] [Google Scholar]
- 10.Sutera V A, Jr, Han E S, Rajman L A, Lovett S T. J Bacteriol. 1999;181:6098–6102. doi: 10.1128/jb.181.19.6098-6102.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamagata A, Masui M, Kakuta Y, Kuramitsu S, Fukuyama K. Nucleic Acids Res. 2001;29:4617–4624. doi: 10.1093/nar/29.22.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 13.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 14.Jones T A, Thirup S. EMBO J. 1986;5:819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–293. [Google Scholar]
- 16.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 17.Glusker J P. Adv Protein Chem. 1991;42:1–76. doi: 10.1016/s0065-3233(08)60534-3. [DOI] [PubMed] [Google Scholar]
- 18.Hopfner K P, Karcher A, Craig L, Woo T T, Carney J P, Tainer J A. Cell. 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 19.Beese L S, Steitz T A. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosfield D J, Mol C D, Shen B, Tainer J A. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 21.Ceska T A, Sayers J R, Stier D, Suck D. Nature (London) 1996;382:90–93. doi: 10.1038/382090a0. [DOI] [PubMed] [Google Scholar]
- 22.Kim Y, Eom S H, Wang J, Lee D S, Suh W S, Steitz T A. Nature (London) 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 23.Kowvall R A, Matthews B W. Curr Opin Chem Biol. 1999;3:578–583. doi: 10.1016/s1367-5931(99)00012-5. [DOI] [PubMed] [Google Scholar]
- 24.Suck D, Oefner C. Nature (London) 1986;321:620–625. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- 25.Mol C D, Kuo C F, Thayer M M, Cunningham R P, Tainer J A. Nature (London) 1995;374:381–387. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 26.Sinden R R. DNA Structure and Function. San Diego: Academic; 1994. [Google Scholar]
- 27.Breyer W A, Matthews B W. Nat Struct Biol. 2000;7:1125–1128. doi: 10.1038/81978. [DOI] [PubMed] [Google Scholar]
- 28.Rajman L A, Lovett S T. J Bacteriol. 2000;182:607–612. doi: 10.1128/jb.182.3.607-612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Artymiuk P J, Ceska T A, Suck D, Sayers J R. Nucleic Acids Res. 1997;25:4224–4229. doi: 10.1093/nar/25.21.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brody R S. Biochemistry. 1991;30:7072–7080. doi: 10.1021/bi00243a006. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 32.Merrit E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 33.Harris M, Jones T A. Acta Crystallogr D. 2001;57:1201–1203. doi: 10.1107/s0907444901007697. [DOI] [PubMed] [Google Scholar]
- 34.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholls A, Sharp K, Honig B. Protein. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]