Abstract
A highly sensitive assay of tRNA aminoacylation was developed that directly measures the fraction of aminoacylated tRNA by following amino acid attachment to the 3′-32P-labeled tRNA. When applied to Escherichia coli alanyl-tRNA synthetase, the assay allowed accurate measurement of aminoacylation of the most deleterious mutants of tRNAAla. The effect of tRNAAla identity mutations on both aminoacylation efficiency (kcat/KM) and steady-state level of aminoacyl-tRNA was evaluated in the absence and presence of inorganic pyrophosphatase and elongation factor Tu. Significant levels of aminoacylation were achieved for tRNA mutants even when the kcat/KM value is reduced by as much as several thousandfold. These results partially reconcile the discrepancy between in vivo and in vitro analysis of tRNAAla identity.
Two major approaches have been developed to identify nucleotides required for tRNA recognition by aminoacyl-tRNA (aa-tRNA) synthetases (aaRSs). The first involves introducing a suppressor anticodon into a tRNA of interest and then examining the suppressor activity of the point mutations in vivo (1). The second involves assaying aminoacylation activity of mutant tRNAs made by transcription in vitro (2). Both methods reveal a limited number of nucleotides critical for aminoacylation, the importance of which was confirmed by performing “swap” experiments in which the proposed nucleotides were transplanted into the body of another tRNA (2, 3). Subsequent analysis of aminoacylation rate in vitro or sequencing of the reporter proteins in vivo was used to confirm that change in tRNA identity had occurred. These two methods generally agree reasonably well with each other and correlate well with observed base-specific contacts in cocrystal structures of tRNA-aaRS complexes (4–6). However, there are several examples where the conclusions derived from in vitro and in vivo approaches disagree, the most notable of which is the recognition of tRNAAla by Escherichia coli alanyl-tRNA synthetase (AlaRS). Although no cocrystal structure is available, in vitro experiments with truncated RNA substrates (7, 8) and mutant tRNAAla (9) as well as identity-swap experiments (10, 11) strongly suggested that the G3⋅U70 pair is the major identity element of tRNAAla. The deleterious effect of mutations of this base pair significantly exceeds those of other tRNAAla recognition elements (8, 9, 12–15) and clearly was indicative of the predominant role of this base pair and in particular the exocyclic amino group of G3 in recognition. In contrast, in vivo experiments showed that many of same mutations in the G3⋅U70 base pair have only a moderate effect on the levels of aminoacylated tRNAAla in E. coli and consequently on the ability of the mutant tRNAs to function in protein synthesis (16, 17). A careful analysis of the in vivo experiments indicates that the observed discrepancy was not caused by the technical problems associated with the determining of tRNA aminoacylation levels in vivo, and therefore it was proposed that the discrepancy in some way was a consequence of differences in reaction conditions for aminoacylation in vitro and in vivo (17). These experiments lead to the idea that the recognition of tRNAAla depended more on particular properties of the structure of the tRNA acceptor stem than on base-specific interactions with the G3⋅U70 base pair (11, 16).
In an attempt to reconcile the different conclusions obtained by the two approaches, it is important to consider that the level of aminoacylation depends on many factors that contribute to both tRNA aminoacylation and tRNA deacylation rates (18). The value of the forward reaction rate will be determined by the activity of the aaRS under the reaction conditions chosen, the concentrations of amino acid, ATP, and tRNA substrates, and the presence of enzyme inhibitors. The value of the deacylation rate will be determined by the rate of the reverse reaction or hydrolysis catalyzed by the aaRS, the rate of base catalyzed hydrolysis of the activated aminoacyl linkage, and the rate of deacylation caused by incorporation into the translation machinery inside cells. When the overall forward rate greatly exceeds the reverse rate, the steady-state level of aa-tRNA is high. However, in the case of tRNA identity mutations that reduce the forward rate, the importance of the reverse rate becomes more prominent, because it is often less sensitive to the tRNA mutations. As the forward rate approaches the reverse rate, the steady-state level of aa-tRNA decreases. Although both the rate of aminoacylation and the level of aa-tRNA can be measured in vitro, only the level can be obtained from in vivo experiments. When one also considers that the factors that influence the forward and reverse rates are likely to be very different inside cells from those in a typical in vitro reaction, different identity mutations could aminoacylate to very different levels in the two systems.
A sensitive assay for the aminoacylation of tRNA is described that can be used to evaluate the activity of tRNA in vitro in a much broader range of reaction conditions. Similar to several other aaRSs (19–21), the PPi product generated in the reaction was found to be a potent inhibitor of AlaRS in vitro, and the presence of inorganic pyrophosphatase (PPase) dramatically increased the reaction rate under many reaction conditions. In addition, the initial rates and levels of aminoacylation of several tRNAAla identity mutations are differentially stimulated by the presence of PPase. When translational elongation factor Tu (EF-Tu) is also included in the reaction mixture to trap the aa-tRNA product, the steady-state level of aminoacylation of several of the tRNA mutants more closely reflects those observed in vivo. Thus, much of the discrepancy between the in vitro and in vivo data in the Ala system can be reconciled.
Materials and Methods
Full-length histidine-tagged E. coli AlaRS was overexpressed from plasmid pQE-alaS-6H (22) and purified as described (23) with several modifications. Chromatography on a MonoQ 5/5 column was used instead of DEAE-Sepharose and an additional gel-filtration step on a Superdex-75 column, equilibrated in 50 mM potassium phosphate, pH 7.0, containing 50 mM NaCl, 1 mM DTT, and 10% glycerol, was included. The resulting enzyme preparation, free of trace RNase and PPase contamination, was stored at −20°C in 50% glycerol. The presence of PPase was tested by performing aminoacylation in the presence of 30 μM [γ-32P]ATP and separation of labeled reaction products on polyethylenimine-cellulose in 0.8 M LiCl. Only labeled PPi was observed in the enzyme preparations devoid of PPase contamination. AlaRS concentration was measured by active site titration (24). Yeast phenylalanyl-tRNA synthetase (PheRS) was purified from baker's yeast as described (25). Terminal tRNA nucleotidyl transferase was purified from E. coli DH5a cells containing pTC9 plasmid (provided by C. McHenry, University of Colorado Health Science Center, Boulder, CO). Thermus thermophilus EF-Tu was purified from an overproducing strain bearing a modified pEFTu10 plasmid (provided by A. Benerjee and M. Makinen, University of Chicago, Chicago).
YFA2 (double-specific Phe/Ala-tRNA) and CA0 (tRNAAla) were obtained by in vitro transcription of the corresponding plasmids (26). Transcription templates for mutants of YFA2 tRNA were obtained by PCR of the YFA2 plasmid using mutated oligonucleotides. Transcription was performed as described (26), and tRNAs were purified on 10% denaturing polyacrylamide gel to a single-nucleotide resolution. Before aminoacylation, tRNAs were folded by heating for 3 min at 65°C in water followed by the addition of MgCl2 to 15 mM and slow cooling to room temperature. The microhelix of tRNAAla (7) was obtained from Dharmacon (Lafayette, CO).
tRNAs 32P-labeled at the 3′-terminal internucleotide linkage was prepared by an ATP-PPi exchange reaction catalyzed by E. coli tRNA-terminal nucleotidyl transferase. Reactions (100 μl) contained 50 mM glycine-HCl buffer, pH 9.0, 10 mM MgCl2, 1 μM tRNA, 1.5 μM [α-32P]ATP [3,000 Ci/mmol (1 Ci = 37 GBq), NEN], 50 μM sodium PPi, and 30 μg/ml tRNA-terminal nucleotidyl transferase. After incubation at 37°C for 5 min, yeast PPase was added to a final concentration of 10 units/ml, and the reaction mixture was incubated for an additional 30 sec before quenching by phenol extraction. After ethanol precipitation, 3′-32P-labeled tRNAs were purified by denaturing polyacrylamide gel electrophoresis in 10% gels. A typical reaction recovered 100 μCi of 3′-[32P]tRNA with a specific activity of 1,000 Ci/mmol.
Incorporation of 3H-labeled amino acids was measured by either spotting aliquots of a reaction on paper filters or filtration through nitrocellulose filters using a dot-blot device, and incorporated radioactivity was measured by either liquid scintillation counting or exposure of the filter to the tritium screen of the PhosphorImager, respectively (26). Aminoacylation levels by PheRS were determined in reactions containing 50 mM Hepes, pH 7.5, 30 mM KCl, 15 mM MgCl2, 2 mM ATP, 0.5 mM DTT, and 10 μM [3H]Phe (65 Ci/mmol) by using 100 nM PheRS and 1 μM tRNA. The individual kinetic constants for aminoacylation by AlaRS were determined by measuring activity at 37°C in reactions containing 50 mM Hepes, pH 7.0, 30 mM KCl, 15 mM MgCl2, 2 mM ATP, 0.5 mM DTT, and 20 μM [3H]Ala (50 Ci/mmol). Depending on the substrate, 0.2 - 5 μM of tRNA, and 0.01–1 μM AlaRS were used. When indicated, reaction mixtures contained 10 units/ml yeast PPase and 2 μM EF-Tu. The concentration of EF-Tu was determined by an active site titration (27).
Incorporation of nonlabeled Ala onto 3′-32P-labeled tRNA was measured by determining the ratio between Ala-AMP and AMP by TLC after digestion of aminoacylated tRNA samples with nuclease P1. Typical reactions (10 μl) contained at least 0.01 μCi of 3′-[32P]tRNA and the desired concentration of nonlabeled tRNAs. Aminoacylation conditions were identical to those described above except that 1 mM of nonradioactive Ala was used. All components except the tRNA were mixed in 5 μl and preincubated for 1 min at 37°C, and the reaction was initiated by the addition of 5 μl of tRNA solution. One-microliter aliquots were withdrawn at the desired times and quenched into 4 μl of 200 mM sodium acetate, pH 5.0, containing 1 unit/μl of nuclease P1 (Sigma) at 0°C. The quenched aliquots were kept on ice until the aminoacylation time course was completed and then incubated together for 10 min at room temperature to digest the tRNA. Quench efficiency was established in separate experiments; no further aminoacylation was observed after the aminoacylation mixture was injected in acetate buffer without P1 nuclease. Approximately 1 μl of each P1 digestion reaction was spotted on 10-cm polyethylenimine-cellulose plates (Sigma) that had been prewashed with water. 32P-labeled AMP and aminoacyl-AMP were separated by TLC in glacial acetic acid/1M NH4Cl/H2O (5:10:85). The radioactivity was analyzed by using a PhosphorImager (Molecular Dynamics).
Results
An Improved Assay for tRNA Aminoacylation.
The aminoacylation of tRNA traditionally is assayed by measuring the esterification of 3H- or 14C-labeled amino acids onto tRNA by acid precipitation of aa-tRNA product on paper or nitrocellulose filters followed by liquid-scintillation counting. Although reproducible and reliable, this assay has several shortcomings. First, the fraction of aminoacylated tRNA is determined indirectly by calculating the amount of labeled amino acid on the filter from the specific activity and counting efficiency and then dividing by the amount of tRNA present in the aliquot. The calculation requires accurate determination of both the counting efficiency and the aliquot volume. Secondly, the high KM for amino acids for many synthetases makes it very difficult to perform reactions at a saturating concentration of amino acids without the use of prohibitive amounts of radioactivity. Because both the amino acid and tRNA often participate in the organization of the active sites of aaRS (3, 28–32), a saturating concentration of one of the substrates is required for the proper determination of the kinetic constants for the other substrate. Finally, the sensitivity of the traditional assay is limited, especially with poor tRNA substrates that show low rates of aminoacylation. The low sensitivity also prevents accurate analysis of aminoacylation reactions with low concentrations of tRNA and amino acids.
Because the aim of this study is to measure aminoacylation of severely deficient tRNAAla mutants, the above shortcomings of the traditional aminoacylation assay become crucial. We previously designed an assay using an acid polyacrylamide gel to follow the aminoacylation of 3′-32P-labeled tRNA with nonradioactive amino acid (23). Although this assay had high sensitivity and directly measured the fraction of aa-tRNA, it was labor-intensive, and the gel-separation step generally required the use of bimolecular tRNAs substrates with a nick in T-stem, limiting its universality. We report here an aminoacylation assay that combines the advantages of a gel assay with the high throughput and universality of the traditional assay.
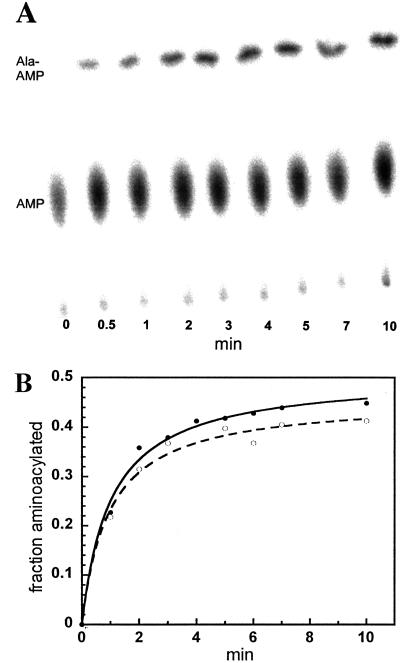
The assay employs a 3′-32P-labeled full-length tRNA prepared by an exchange reaction with [α-32P]ATP and PPi catalyzed by E. coli tRNA-terminal nucleotidyl transferase. The resulting tRNA contains a unique 32P at position 76, the 3′-terminal residue. This labeling reaction proceeds efficiently with modified or unmodified tRNAs, tRNA mutants, and many tRNA subfragments containing a 3′-terminal CCA sequence. Aminoacylation of the 3′-[32P]tRNA is performed as usual by using any desired concentration of nonradioactive amino acid, and time points are quenched in a P1 nuclease solution in acetate buffer, pH 5.0, that stops the reaction and stabilizes the aminoacyl bond. Nuclease P1 digestion in this buffer generates the mixture of 32P-labeled AMP and aminoacyl-AMP, which reflects the relative amounts of aminoacylated and nonacylated tRNA at the particular time point of reaction. These two radioactive products are separated by TLC, and their ratio is determined by using a PhosphorImager. Fig. 1A shows a TLC plate analyzing the time course of aminoacylation of 3′-[32P]YFA2 (26) by E. coli AlaRS using 1 mM nonradioactive Ala. Because the assay measures the fraction of Ala-AMP (and thus of Ala-YFA2) at each time point directly, the error-prone determination of specific activity and aliquot size is avoided.
Figure 1.
Time course of YFA2 aminoacylation by E. coli AlaRS measured by our TLC assay or the traditional incorporation of [3H]Ala. Aminoacylation was performed at 1 μM tRNA, 20 μM Ala, and 5 (A) or 10 nM AlaRS (B). (A) Time course of 3′-32P-labeled YFA2 aminoacylation analyzed by TLC. (B) Aminoacylation measured in a double-label experiment either by TLC (●) or 3H incorporation (○).
To confirm that the TLC assay gives results comparable with the traditional assay, the aminoacylation of 3′-32P-labeled YFA2 was performed with 20 μM [3H]Ala. Duplicate time points were withdrawn and analyzed by both assays. The time course gives identical apparent rates (Fig. 1B). A similar agreement of the two assays was obtained when YFA2 was aminoacylated with Phe by yeast PheRS (data not shown).
Another immediate advantage of the assay is the possibility to directly measure the KM for amino acid in aminoacylation reaction. AlaRS was reported to have a low affinity for amino acid with a KM for Ala at 240 μM as measured by ATP-PPi exchange (33). When measured in the tRNA aminoacylation reaction, the KM for Ala is 47 μM in the absence and 8 μM in the presence of PPase. These data agree well with a previous observation that an increase in Ala concentration from 10 μM to 1 mM translated in only a 3-fold increase in aminoacylation rate (23). A similar KM for Ala (13 μM) in the presence of PPase was obtained by using the traditional assay.
Because both assays give identical results and the KM for Ala is low, both assays were used interchangeably in the remainder of this work.
Activation of AlaRS by PPase.
To evaluate the effect of PPase on the AlaRS reaction, it is necessary to remove all traces of PPase activity contaminating the AlaRS preparation. The presence of PPase is detected easily by including [γ-32P]ATP in the aminoacylation reaction and analyzing the reaction products for the presence of [32P]phosphate by TLC. Although His-6-tagged AlaRS (26) purified from E. coli lysates by nickel-affinity chromatography was practically homogeneous (>95%) when analyzed on SDS gels, it contained some PPase activity that required two additional column-purification steps to remove. The resulting PPase–free AlaRS only produces 32P-labeled PPi during aminoacylation reactions.
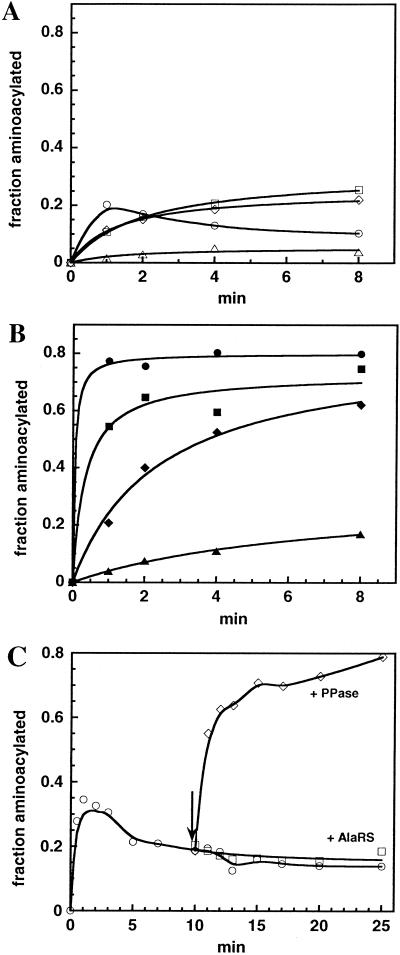
The kinetic properties of the tRNA aminoacylation reaction by the PPase–free AlaRS suggest that enzyme inhibition occurs. In reactions containing 1 μM tRNA, which approximately equals its KM, and saturating ATP and Ala, the initial rate of formation of Ala-tRNA is only proportional to an AlaRS concentration below 25 nM (Fig. 2A). At the same time, the extent of tRNA aminoacylation reaches the plateau level significantly lower than in the presence of PPase in reaction mixture (Fig. 2B), which is an indication of strong product inhibition. When higher concentrations of AlaRS are added, the measured initial rate reaches a constant value, probably because the reaction reaches the plateau level very fast, resulting in a steady-state level of Ala-tRNA of ≈30%, which might reflect the equilibrium state between deacylation rate and slow aminoacylation rate catalyzed by enzyme. This behavior is not caused by enzyme denaturation, because the addition of fresh enzyme does not increase these levels (Fig. 2C) significantly. In contrast, the addition of PPase after the reaction reaches plateau results in stimulation of aminoacylation such that complete aminoacylation is achieved (Fig. 2C). When PPase is added to the reactions, reaction rates are stimulated dramatically such that initial rates are proportional to AlaRS concentration and steady-state levels of Ala-tRNA approaching 90% are achieved (Fig. 2B). Although the experiments done in Fig. 2 were with YFA2, a tRNAPhe derivative that is fully active with AlaRS (26), similar results were obtained with modified or unmodified E. coli tRNAAla as well as a microhelix substrate.
Figure 2.
Effect of PPase on tRNA aminoacylation by E. coli AlaRS. (A and B) Aminoacylation was performed at 1 μM YFA2 and 0.25 (▵), 2.5 (⋄), 25 (□), or 250 (○) nM AlaRS. Open symbols, no PPase added; closed symbols, in the presence of PPase. (C) Time course of YFA2 aminoacylation in the absence of PPase (○). Aminoacylation was performed at 100 nM AlaRS and 1 μM tRNA. After 10 min, 10 units/ml PPase (♦) or 100 nM AlaRS (□) were added.
The activation of AlaRS by PPase presumably is because the PPi generated in the first adenylation step of the reaction is an effective inhibitor of the overall reaction. Indeed, when PPi is added to aminoacylation reactions under conditions similar to those described for Fig. 2, strong inhibition is observed. The presence of 4–8 μM PPi caused a 50% reduction in the aminoacylation rate of YFA2 and CA0 tRNAs and microhelix (data not shown). Whether pyrophosphate inhibits the overall reaction at the adenylation step via product inhibition, at the transfer step, or by a more complicated mechanism awaits a thorough kinetic analysis. Nevertheless, it is clear that if the goal is to evaluate the relative efficiency of different tRNA substrates by AlaRS, PPi inhibition of the enzyme should be prevented by the addition of PPase.
Aminoacylation of tRNA Mutants.
The effect of identity mutations on the rate of aminoacylation by E. coli AlaRS was studied in the background of the dual-specific YFA2 tRNA (which is a derivative of yeast tRNAPhe, which contains all the major AlaRS identity elements present in the acceptor stem). The kinetic constants for the aminoacylation of YFA2 tRNA by AlaRS are essentially identical to those of tRNAAla (26). The major reason for using YFA2 in these studies is that it also is an excellent substrate for yeast PheRS, an enzyme that is almost insensitive to acceptor stem mutations but very sensitive to the overall folded state of the tRNA. It permits the aminoacylation by PheRS to be used as a control to confirm that the low rate and level of aminoacylation of a given mutant by AlaRS is a direct result of change in the identity nucleotide and not the result of improper synthesis or folding of tRNA. All but one of the mutant tRNAs used in this study were at least 85% active according to the levels of aminoacylation by PheRS. The only exception was the C3⋅A70 mutant, where phenylalanylation never exceeded 60%, suggesting the presence of an inactive fraction.
The aminoacylation properties of a selection of eight variants of YFA2 containing mutations in identity nucleotides are shown in Table 1. In the absence of PPase, their kcat/KM values are reduced severely with respect to the parent tRNA and are difficult to measure accurately. When PPase is included in the reaction, the kcat/KM value for all tRNAs is increased. This “activation” by PPase ranges from 4- to 6-fold for the native tRNAs and the moderate A73U mutation to at least 20-fold for the more deleterious mutations. The values obtained in the absence of PPase generally agree quite well with kcat/KM determinations for many of the same mutations in E. coli tRNAAla (9). In any case, it is clear that Although their loss in catalytic efficiency is less in the presence of PPase, the initial rate of aminoacylation of many of the tRNAAla identity mutation is reduced severely.
Table 1.
Effect of PPase on catalytic efficiency of tRNA aminoacylation
| tRNA |
kcat/KM
|
Catalytic efficiency loss (no PPase), fold | Activation in the presence of PPase, fold | |
|---|---|---|---|---|
| +PPase | −PPase | |||
| YFA2 | 1 | 0.23 | 1 | 4.4 |
| CA0 | 1.9 | 0.63 | 0.37 | 3.0 |
| Microhelix | 0.048 | 0.009 | 25 | 5.3 |
| U73 | 0.036 | 0.006 | 38 | 6.0 |
| G73 | 0.017 | 0.00077 | 299 | 22 |
| C2⋅G71 | 0.013 | 0.00062 | 371 | 21 |
| A70 | 0.0030 | 0.00013 | 1,769 | 23 |
| C70 | 0.00047 | nd | >10,000 | >10 |
| A3 | 0.00051 | nd | >10,000 | >10 |
| C3⋅A70* | 0.0023 | 0.00012 | 1,916 | 19 |
| C3⋅C70 | 0.012 | 0.00080 | 2,875 | 15 |
Catalytic efficiency loss is calculated as the ratio of (kcat/KM)YFA2/(kcat/KM)varian. Activation in the presence of PPase is calculated as the ratio of (kcat/KM)+PPase/(kcat/KM)−PPase for each substrate. nd, not determined.
The level of aminoacylation of this mutant with PheRS is constantly ≈65% compared to more than 85% for all other mutants.
EF-Tu complexed with GTP binds aa-tRNAs with high affinity and acts as a product trap by greatly reducing the rate of deacylation of the activated aminoacyl linkage (34). The half-life of Ala-tRNAAla under our reaction conditions increases from 4.5 to >100 min when EF-Tu is added (data not shown). Although the presence of EF-Tu does not affect the forward rate of the aminoacylation reaction, much higher levels of aa-tRNA are observed at steady state. This effect is dramatic especially for tRNA identity mutations where the forward rate is slow. In Table 2, the steady-state levels of aa-tRNA achieved for the set of identity mutations are compared at 1 μM tRNA, a high (0.5 μM) concentration of AlaRS with and without PPase, and an excess of EF-Tu⋅GTP. As would be expected from the data in Table 1, the presence of PPase increased the steady-state level of aa-tRNA for the wild type and all the mutant tRNAs by increasing the forward rate of the reaction. For several of the identity mutations, the level can be increased further by the addition of EF-Tu. Quite high levels of aminoacylation can be achieved with comparatively poor tRNA substrates. For example, the C3⋅A70 mutation can be aminoacylated to nearly 80% despite a greater than 400-fold loss in the initial rate of the reaction. However, for several tRNA identity mutations (A3 or C70) only very low levels of aminoacylation can be achieved even in the presence of PPase and EF-Tu.
Table 2.
Aminoacylation levels of tRNAAla mutants in vitro and in vivo
| tRNA mutant | Catalytic efficiency loss | Level of aminoacylation, %*
|
||
|---|---|---|---|---|
| In vitro | In vitro (+EF-Tu and PPase) | In vivo† | ||
| YFA2 (wild type)‡ | 1 | 25 | 100 | 100 |
| G73 | 58 | 14 | 93 | nd |
| C3⋅C70 | 83 | 4.1 | 52 | 72 |
| A70 | 338 | 8 | 51 | 81 |
| C3⋅A70§ | 436 | 2.0 | 25 | 74 |
| A3 | 1,952 | <0.1 | 3.8 | 3.8 |
| C70 | 2,145 | <0.1 | 4.2 | 2.1 |
The aminoacylation level was measured at 1 μM tRNA and 250 nM AlaRS; EF-Tu was added to 2 μM.
In vivo data are taken from ref. 17. Numbers are normalized to the aminoacylation level of wild-type tRNA. nd, not determined.
Aminoacylation in vivo is measured in the background of wild-type tRNAAla.
The aminoacylation level of this mutant with Phe was 65% compared to more than 85% for all other tRNAs. Number in the table corresponds to the corrected value.
Discussion
A new assay protocol for measuring the formation of aa-tRNA catalyzed by aaRSs is presented. tRNAs containing a single 3′-terminal 32P are aminoacylated with nonradioactive amino acid and the reaction products subjected to nuclease P1 digestion to generate labeled aminoacyl-AMP and AMP. Separation of these labeled nucleotides by TLC and subsequent counting give the fraction of tRNA that was aminoacylated. There are several reasons why this assay is a major improvement over the traditional assay, which used radiolabeled amino acid and precipitation of the aa-tRNA product. First, by using 32P-labeled tRNA instead of 3H- or 14C-labeled amino acid, our assay is much more sensitive and uses the more economical [α-32P]ATP as a source of radiolabel for all aaRSs. In addition, the assay allows directly measuring aminoacylation when radioactive amino acid is unavailable, such as in the case of most unnatural amino acids. The high sensitivity of the assay permits detection of low levels of aminoacylation typical of mutant tRNAs and tRNA fragments. The broad substrate specificities of tRNA-terminal nucleotidyl transferase and T4 RNA ligase also permit virtually any RNA fragment to be 3′-32P-labeled. Second, by using labeled tRNA, the fraction of aa-tRNA is measured directly instead of being calculated from the specific activity of the amino acid and the concentration of tRNA in the aliquot. These changes greatly improve the accuracy of the assay, because the error-prone determination of counting efficiency and aliquot volume is avoided. Third, the assay permits a much broader range of reaction conditions to be accessed. For example, unlike the conventional assay, aminoacylation rates can be measured easily at saturating amino acid concentration. Furthermore, because 3′-[32P]tRNA of very high specific activity can be prepared, aminoacylation can be performed under presteady-state conditions by using enzyme excess provided that the rapid reaction rate can be measured. These advantages make this assay a valuable tool for the study of the mechanism of aaRSs. Besides aminoacylation, the assay might be used to measure tRNA deacylation rates, catalyzed by aaRS in a posttransfer editing reaction (35). In addition, the assay can be adapted easily for the assay of other enzymes that use aa-tRNA as a substrate including tRNAfMet formylase (36), tRNA-transamidase (37), and possibly selenocysteine syntase (38). It should be possible also to adapt the assay to measure the rate of peptide-bond formation catalyzed by the ribosome.
PPi generated in the adenylation step of AlaRS substantially inhibits the initial rate of aminoacylation of tRNA. Although the precise mechanism of this inhibition remains to be established, the phenomenon complicates the interpretation of all aminoacylation experiments. Similar to most synthetases, AlaRS can perform the first step of the reaction, but the Ala-AMP product remains bound tightly to the enzyme, preventing multiple turnovers. When tRNA is present in the reaction, Ala-AMP is turned over, and the amount of PPi increases, ultimately slowing the overall rate. Thus, the degree to which PPi inhibits a given reaction depends on the reaction conditions used, the efficacy of the tRNA substrate, and even the extent that the reaction has progressed. Here it is shown that under conditions typically used for tRNA identity experiments, PPase inhibition of AlaRS is pronounced particularly when tRNA substrates containing deleterious identity mutations are used. Because the goal of in vitro tRNA identity experiments are to measure the rate of aminoacylation of mutant tRNAs accurately, it is appropriate to perform experiments in the presence of an excess of added PPase.
In the case of AlaRS, the kcat/KM values of tRNA identity mutations were increased from 6- to 20-fold when PPase was added to the reaction. Although the activation was not uniform among the mutations, our understanding of tRNA recognition by AlaRS was not changed significantly. The G3⋅U70 base pair remains a critical identity element. In contrast, in the case of yeast PheRS, the addition of PPase selectively increased the kcat/KM of mutations of G20 to a degree that this residue no longer could be considered a recognition nucleotide (19). Because it is likely that all tRNA synthetases to some degree are inhibited by PPi, in vitro experiments defining tRNA recognition nucleotides should be reexamined for PPase effects. In the case of E. coli seryl-tRNA synthetase, PPase was found to uniformly increase kcat/KM of all substrates tested (20). Because tRNA synthetase preparations are contaminated often by PPase, it is possible that most of the previously identified tRNA recognition nucleotides will not change.
When aminoacylation reactions by AlaRS are supplemented with PPase to increase the forward rate and EF-Tu⋅GTP to reduce the deacylation rate, high levels of aminoacyl tRNA can be observed for all but the most deleterious tRNaAla mutations, which agrees with the previous observation that high levels of aa-tRNA are observed in vivo for the same mutations (16). This result in turn explains why Ala is inserted efficiently into proteins by tRNAAla suppressors containing most identity mutations (17). Thus, tRNAs do not necessarily need to be good substrates in vitro to achieve high levels of aminoacylation in vivo.
Although PPase and EF-Tu are present in E. coli, it is unclear whether they are actually important in maintaining the high levels of mutant aa-tRNA inside cells. Intracellular EF-Tu concentration approximately equals total tRNA concentration (39, 40) and therefore potentially could protect aa-tRNA from deacylation. However, because tRNA cycles through the translation apparatus in ≈1 sec (41, 42), it seems unlikely that EF-Tu plays an important role in retarding tRNA deacylation except perhaps with tRNAs that are not consumed effectively in translation such as certain suppressor tRNAs. The role of PPi and PPase in the regulation of aminoacylation efficiency in vivo also is ambiguous. PPase is an essential protein, and experiments regulating its intracellular concentration reveal that cellular growth is inhibited when PPi concentrations exceed 2–3 mM (43). Surprisingly, typical PPi concentrations in log-phase E. coli cells are between 0.5 and 1 mM (43, 44), well above the concentration at which it fully inhibits tRNA synthetases in vitro (21, 45). Perhaps the majority of the PPi is sequestered in an intracellular compartment such that it is unavailable to inhibit aaRSs. Finally, it is possible that other cellular factors play a role in defining aminoacylation levels in vivo. Nonspecific tRNA-binding proteins analogous to yeast Arc1p (46) or eubacterial TRBP111 (47, 48) might be candidates for this role.
Our results demonstrate that tRNAAla mutations whose cause is a significant decrease in aminoacylation rate do not always have a dramatic effect on the steady-state level of aa-tRNA. Experiments in vivo (16) have shown that an aminoacylation level as low as 50% is sufficient for mutant tRNAAla to maintain cell growth. These data include several tRNAAla mutations that show a several hundred-fold decrease in catalytic efficiency in vitro. In other words, tRNAAla does not need to be a really efficient substrate to function successfully in vivo. This situation is not unique to the Ala system. The lack of 2-thiouracil modification at position 34 reduces the kcat/KM value of aminoacylation of tRNAGlu by 520-fold (49), whereas the same unmodified tRNA is aminoacylated up to 60% in vivo (50). Similarly, tRNAThr mutants with a 100-fold decrease in aminoacylation efficiency were able to substitute wild-type E. coli tRNAThr (51). The effect of most known tRNA “identity” mutations on the kcat/KM value of aminoacylation measured in vitro is mostly within 10–1,000-fold range (4), and thus many of those tRNA mutants might be aminoacylated in vivo to sufficient levels to maintain their function. Apparently, the limitations imposed on tRNAs by their cognate aaRS are not as strict as it was thought as long as significant variations in aminoacylation efficiency of tRNA are tolerated. Of course, it is possible that the effect of even a small decrease in aminoacylation efficiency on the cellular phenotype might become apparent when cells are placed under extreme conditions or in competition experiments. Alternatively, a decrease in aminoacylation efficiency by cognate synthetases might result in misacylation by noncognate aaRS, increasing the error rate. Finally, in its life cycle tRNA interacts with numerous other proteins (and RNAs) involved in its primary biochemical function (aaRS, EF-Tu, and ribosome), maturation, modification, and degradation. As a result, tRNA should be adapted to confer to the structural requirements imposed by all these interactions in a way to comply with the general requirements of the organism to ensure the certain level of translation efficiency. Thus, tRNA mutations, which do not affect the aminoacylation level directly, might not be neutral for other tRNA functions.
Acknowledgments
We thank Haruichi Asahara and Kevin Polach for reading the manuscript. This work was supported by National Institutes of Health Grant GM 37552 (to O.C.U.).
Abbreviations
- aa-tRNA
aminoacyl-tRNA
- aaRS
aminoacyl-tRNA synthetase
- AlaRS
alanyl-tRNA synthetase
- PPase
inorganic pyrophosphatase
- EF-Tu
translational elongation factor Tu
- PheRS
phenylalanyl-tRNA synthetase
References
- 1.Normanly J, Kleina L G, Masson J M, Abelson J, Miller J H. J Mol Biol. 1990;213:719–726. doi: 10.1016/S0022-2836(05)80258-X. [DOI] [PubMed] [Google Scholar]
- 2.Sampson J R, DiRenzo A B, Behlen L S, Uhlenbeck O C. Science. 1989;243:1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- 3.Schulman L H, Pelka H. Science. 1988;242:765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- 4.Giege R, Sissler M, Florentz C. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rould M A, Perona J J, Soll D, Steitz T A. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 6.Ruff M, Krishnaswamy S, Boeglin M, Poterszman A, Mitschler A, Podjarny A, Rees B, Thierry J C, Moras D. Science. 1991;252:1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- 7.Francklyn C, Schimmel P. Nature (London) 1989;337:478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 8.Musier-Forsyth K, Usman N, Scaringe S, Doudna J, Green R, Schimmel P. Science. 1991;253:784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- 9.Beuning P J, Yang F, Schimmel P, Musier-Forsyth K. Proc Natl Acad Sci USA. 1997;94:10150–10154. doi: 10.1073/pnas.94.19.10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou Y M, Schimmel P. Nature (London) 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 11.McClain W H, Chen Y M, Foss K, Schneider J. Science. 1988;242:1681–1684. doi: 10.1126/science.2462282. [DOI] [PubMed] [Google Scholar]
- 12.Fischer A E, Beuning P J, Musier-Forsyth K. J Biol Chem. 1999;274:37093–37096. doi: 10.1074/jbc.274.52.37093. [DOI] [PubMed] [Google Scholar]
- 13.Beuning P J, Gulotta M, Musier-Forsyth K. J Am Chem Soc. 1997;119:8397–8402. [Google Scholar]
- 14.Shi J P, Schimmel P. J Biol Chem. 1991;266:2705–2708. [PubMed] [Google Scholar]
- 15.Liu H, Kessler J, Peterson R, Musier-Forsyth K. Biochemistry. 1995;34:9795–9800. doi: 10.1021/bi00030a017. [DOI] [PubMed] [Google Scholar]
- 16.Gabriel K, Schneider J, McClain W H. Science. 1996;271:195–197. doi: 10.1126/science.271.5246.195. [DOI] [PubMed] [Google Scholar]
- 17.McClain W H, Jou Y Y, Bhattacharya S, Gabriel K, Schneider J. J Mol Biol. 1999;290:391–409. doi: 10.1006/jmbi.1999.2884. [DOI] [PubMed] [Google Scholar]
- 18.Dietrich A, Kern D, Bonnet J, Giege R, Ebel J P. Eur J Biochem. 1976;70:147–158. doi: 10.1111/j.1432-1033.1976.tb10965.x. [DOI] [PubMed] [Google Scholar]
- 19.Khvorova A, Motorin Y, Wolfson A D. Nucleic Acids Res. 1999;27:4451–4456. doi: 10.1093/nar/27.22.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sampson J R, Saks M E. Nucleic Acids Res. 1993;21:4467–4475. doi: 10.1093/nar/21.19.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dignam J D, Deutscher M P. Biochemistry. 1979;18:3165–3170. doi: 10.1021/bi00581a039. [DOI] [PubMed] [Google Scholar]
- 22.Ribas de Pouplana L, Schimmel P. Biochemistry. 1997;36:15041–15048. doi: 10.1021/bi971788+. [DOI] [PubMed] [Google Scholar]
- 23.Wolfson A D, Pleiss J A, Uhlenbeck O C. RNA. 1998;4:1019–1023. doi: 10.1017/s1355838298980700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fersht A R, Ashford J S, Bruton C J, Jakes R, Koch G L, Hartley B S. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 25.Khvorova A M, Motorin Yu A, Wolfson A D. FEBS Lett. 1992;311:139–142. doi: 10.1016/0014-5793(92)81385-y. [DOI] [PubMed] [Google Scholar]
- 26.Pleiss J A, Wolfson A D, Uhlenbeck O C. Biochemistry. 2000;39:8250–8258. doi: 10.1021/bi0001022. [DOI] [PubMed] [Google Scholar]
- 27.Louie A, Jurnak F. Biochemistry. 1985;24:6433–6439. doi: 10.1021/bi00344a019. [DOI] [PubMed] [Google Scholar]
- 28.Perona J J, Rould M A, Steitz T A. Biochemistry. 1993;32:8758–8771. doi: 10.1021/bi00085a006. [DOI] [PubMed] [Google Scholar]
- 29.Rath V L, Silvian L F, Beijer B, Sproat B S, Steitz T A. Structure (London) 1998;6:439–449. doi: 10.1016/s0969-2126(98)00046-x. [DOI] [PubMed] [Google Scholar]
- 30.Ibba M, Hong K W, Sherman J M, Sever S, Soll D. Proc Natl Acad Sci USA. 1996;93:6953–6958. doi: 10.1073/pnas.93.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eiler S, Dock-Bregeon A, Moulinier L, Thierry J C, Moras D. Eur J Biochem. 1999;18:6532–6541. doi: 10.1093/emboj/18.22.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delagoutte B, Moras D, Cavarelli J. EMBO J. 2000;19:5599–5610. doi: 10.1093/emboj/19.21.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park S J, Schimmel P. J Biol Chem. 1988;263:16527–16530. [PubMed] [Google Scholar]
- 34.Pingoud A, Urbanke C, Krauss G, Peters F, Maass G. Eur J Biochem. 1977;78:403–409. doi: 10.1111/j.1432-1033.1977.tb11752.x. [DOI] [PubMed] [Google Scholar]
- 35.Eldred E W, Schimmel P R. J Biol Chem. 1972;247:2961–2964. [PubMed] [Google Scholar]
- 36.Schmitt E, Panvert M, Blanquet S, Mechulam Y. EMBO J. 1998;17:6819–6826. doi: 10.1093/emboj/17.23.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tumbula D L, Becker H D, Chang W Z, Soll D. Nature (London) 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 38.Leinfelder W, Forchhammer K, Veprek B, Zehelein E, Bock A. Proc Natl Acad Sci USA. 1990;87:543–547. doi: 10.1073/pnas.87.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furano A V. Proc Natl Acad Sci USA. 1975;72:4780–4784. doi: 10.1073/pnas.72.12.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jakubowski H. J Theor Biol. 1988;133:363–370. doi: 10.1016/s0022-5193(88)80327-8. [DOI] [PubMed] [Google Scholar]
- 41.Jakubowski H, Goldman E. J Bacteriol. 1984;158:769–776. doi: 10.1128/jb.158.3.769-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gouy M, Grantham R. FEBS Lett. 1980;115:151–155. doi: 10.1016/0014-5793(80)81155-0. [DOI] [PubMed] [Google Scholar]
- 43.Chen J, Brevet A, Fromant M, Leveque F, Schmitter J M, Blanquet S, Plateau P. J Bacteriol. 1990;172:5686–5689. doi: 10.1128/jb.172.10.5686-5689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kukko-Kalske E, Lintunen M, Inen M K, Lahti R, Heinonen J. J Bacteriol. 1989;171:4498–4500. doi: 10.1128/jb.171.8.4498-4500.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Airas R K, Cramer F. Eur J Biochem. 1986;160:291–296. doi: 10.1111/j.1432-1033.1986.tb09970.x. [DOI] [PubMed] [Google Scholar]
- 46.Simos G, Sauer A, Fasiolo F, Hurt E C. Mol Cell. 1998;1:235–242. doi: 10.1016/s1097-2765(00)80024-6. [DOI] [PubMed] [Google Scholar]
- 47.Morales A J, Swairjo M A, Schimmel P. EMBO J. 1999;18:3475–3483. doi: 10.1093/emboj/18.12.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomanbhoy T, Morales A J, Abraham A T, Vortler C S, Giege R, Schimmel P. Nat Struct Biol. 2001;8:344–348. doi: 10.1038/86228. [DOI] [PubMed] [Google Scholar]
- 49.Madore E, Florentz C, Giege R, Sekine S, Yokoyama S, Lapointe J. Eur J Biochem. 1999;266:1128–1135. doi: 10.1046/j.1432-1327.1999.00965.x. [DOI] [PubMed] [Google Scholar]
- 50.Kruger M K, Sorensen M A. J Mol Biol. 1998;284:609–620. doi: 10.1006/jmbi.1998.2197. [DOI] [PubMed] [Google Scholar]
- 51.Saks M E, Sampson J R, Abelson J. Science. 1998;279:1665–1670. doi: 10.1126/science.279.5357.1665. [DOI] [PubMed] [Google Scholar]