Abstract
Duplexes of 21-nt RNAs, known as short-interfering RNAs (siRNAs), efficiently inhibit gene expression by RNA interference (RNAi) when introduced into mammalian cells. We show that siRNAs can be synthesized by in vitro transcription with T7 RNA polymerase, providing an economical alternative to chemical synthesis of siRNAs. By using this method, we show that short hairpin siRNAs can function like siRNA duplexes to inhibit gene expression in a sequence-specific manner. Further, we find that hairpin siRNAs or siRNAs expressed from an RNA polymerase III vector based on the mouse U6 RNA promoter can effectively inhibit gene expression in mammalian cells. U6-driven hairpin siRNAs dramatically reduced the expression of a neuron-specific β-tubulin protein during the neuronal differentiation of mouse P19 cells, demonstrating that this approach should be useful for studies of differentiation and neurogenesis. We also observe that mismatches within hairpin siRNAs can increase the strand selectivity of a hairpin siRNA, which may reduce self-targeting of vectors expressing siRNAs. Use of hairpin siRNA expression vectors for RNAi should provide a rapid and versatile method for assessing gene function in mammalian cells, and may have applications in gene therapy.
RNA interference (RNAi) has become a powerful and widely used tool for the analysis of gene function in invertebrates and plants (reviewed in ref. 1). Introduction of double-stranded RNA (dsRNA) into the cells of these organisms leads to the sequence-specific destruction of endogenous RNAs that match the dsRNA. During RNAi, long dsRNA molecules are processed into 19- to 23-nt RNAs known as short-interfering RNAs (siRNAs) that serve as guides for enzymatic cleavage of complementary RNAs (2–10). In addition, siRNAs can function as primers for an RNA-dependent RNA polymerase that synthesizes additional dsRNA, which in turn is processed into siRNAs, amplifying the effects of the original siRNAs (11, 12). Although the overall process of siRNA inhibition has been characterized, the specific enzymes that mediate siRNA function remain to be identified.
In mammalian cells, dsRNA is processed into siRNAs (13–16), but RNAi with dsRNA has not been successful in most cell types because of nonspecific responses elicited by dsRNA molecules longer than about 30 nt (17). However, Tuschl and coworkers (13, 18) recently made the remarkable observation that transfection of synthetic 21-nt siRNA duplexes into mammalian cells effectively inhibits endogenous genes in a sequence-specific manner. These siRNA duplexes are too short to trigger the nonspecific dsRNA responses, but they still cause destruction of complementary RNA sequences (19). It is not known whether siRNAs in mammalian cells also prime synthesis of dsRNA to form additional siRNAs. The recent discovery of large numbers of microRNA genes (reviewed in ref. 20) raises the prospect that the cellular machinery necessary for siRNA inhibition in mammalian cells may be linked to normal processes of gene regulation.
In the hope of applying siRNA inhibition to studies of neurogenesis and neuronal differentiation, we have explored the possibility of synthesizing siRNAs within mammalian cells by using an expression vector. An siRNA expression vector would facilitate transfection experiments in cell culture, as well as allow the use of transgenic or viral delivery systems. As a first step, we evaluated siRNA designs better suited to expression vectors, such as hairpin RNAs, in which both strands of an siRNA duplex would be included within a single RNA molecule. We used T7 in vitro transcription from oligonucleotide templates (21) as an inexpensive and rapid procedure for synthesizing conventional and hairpin siRNAs, as well as mutant versions of these molecules. We have observed inhibition by the in vitro transcribed siRNAs and hairpin siRNAs by using transfection into mouse P19 cells, a model system for neuronal differentiation.
For synthesis of siRNAs in cells, we wanted to express short RNAs with defined ends. Transcriptional termination by RNA polymerase III occurs at runs of four consecutive T residues in the DNA template (22, 23), providing a mechanism to end an siRNA transcript at a specific sequence. In addition, previous studies demonstrated that RNA polymerase III-based expression vectors can be used to synthesize short RNA molecules in mammalian cells (24, 25). Although most genes transcribed by RNA polymerase III require cis-acting regulatory elements within their transcribed regions, the regulatory elements for the U6 small nuclear RNA gene are contained in a discrete promoter located 5′ to the U6 transcript (26). In using an expression vector with a mouse U6 promoter, we have found that both hairpin siRNAs and pairs of single-stranded siRNAs expressed in cells can inhibit gene expression. Inhibition by hairpin siRNAs expressed from the U6 promoter was more effective than the other methods tested, including the transfection of in vitro-synthesized siRNA duplexes.
Materials and Methods
siRNA Synthesis.
For in vitro transcription, 40-nt DNA template oligonucleotides were designed to produce 21-nt siRNAs. siRNA sequences of the form GN17CN2 were selected for each target, because efficient T7 RNA polymerase initiation requires the first nt of each RNA to be a G (21). The last two nt form the 3′ overhang of the siRNA duplex and were changed to U for the sense strand (ref. 13; for sequences, see Fig. 1 A and B and Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). For hairpin siRNAs, only the first nt needs to be a G (Fig. 2A). Each template and a 20-nt T7 promoter oligonucleotide (Fig. 1B) were mixed in equimolar amounts, heated for 5 min at 95°C, then gradually cooled to room temperature in annealing buffer (10 mM Tris⋅HCl/100 mM NaCl). In vitro transcription was carried out by using the AmpliScribe T7 High Yield Transcription Kit (Epicentre Technologies, Madison, WI) with 50 ng of oligonucleotide template in a 20-μl reaction for 6 h or overnight. RNA products were purified by QIAquick Nucleotide Removal kit (Qiagen, Chatsworth, CA). For annealing of siRNA duplexes, siRNA strands (150–300 ng/μl in annealing buffer) were heated for 5 min at 95°C and then cooled slowly to room temperature. Short RNA products from the in vitro transcription reactions (21) sometimes reduced transfection efficiency (unpublished observations), so siRNA duplexes and hairpin siRNAs were gel purified by using 4% NuSieve GTG agarose (BMA Biomedicals). RNA duplexes were identified by comigration with a chemically synthesized RNA duplex of the same length and recovered from the gel by β-agarase digestion (New England Biolabs). The DhGFP1 siRNAs were chemically synthesized (Dharmacon Research, Lafayette, CO), deprotected as directed by the manufacturer, and annealed as described above. RNAs were quantified by using RiboGreen fluorescence (Molecular Probes).
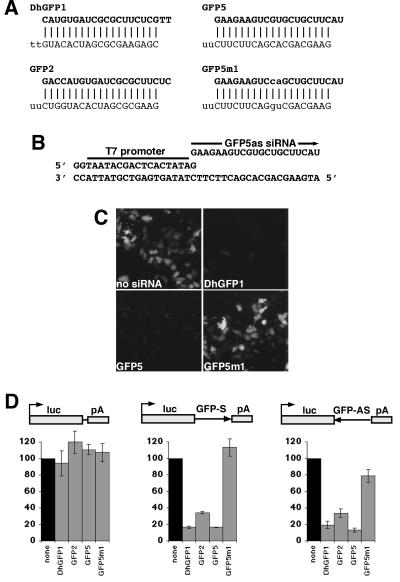
Figure 1.
RNA interference using 21-nt siRNAs synthesized by in vitro transcription. (A) Sequences and expected duplexes for siRNAs targeted to GFP. Both DhGFP1 strands were chemically synthesized, whereas other siRNA strands were synthesized by in vitro transcription with T7 RNA polymerase. GFP5m1 contains a two-base mismatch with the GFP target. Nucleotides corresponding to the antisense strand of GFP are in bold; nucleotides mismatched with the target are lowercase. (B) An example of a DNA oligonucleotide template for T7 transcription. (C) GFP fluorescence was effectively reduced by cotransfection of either the DhGFP1 or GFP5 siRNAs with a GFP expression vector but not by the GFP5m1 siRNA. (D) siRNA inhibition of luciferase activity from vectors with and without GFP sequences inserted into the 3′ untranslated region of luciferase (luc, luciferase; pA, SV40 polyadenylation site). siRNAs synthesized either chemically or by in vitro transcription were similarly effective at inhibiting luciferase when GFP sequences were present in the luciferase mRNA, whereas the mismatched GFP5m1 siRNA did not inhibit effectively. The no-siRNA control (none) is set to 100% for each set of transfections. Data are averaged from three experiments with SE indicated.
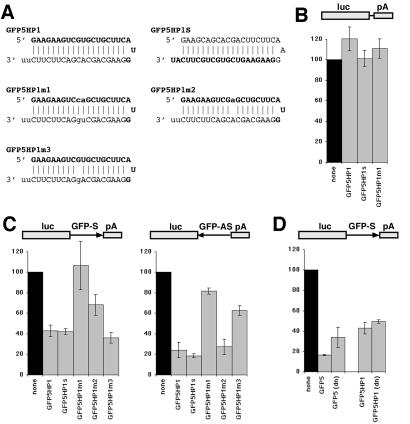
Figure 2.
RNA interference using hairpin siRNAs synthesized by in vitro transcription. (A) Sequences and expected structures for the hairpin siRNAs to GFP (notation as in Fig. 1). GFP5HP1m2 and GFP5HP1m3 contain single-base mismatches with the sense and antisense strands of GFP respectively, whereas GFP5HP1m1 contains a two-base mismatch identical to GFP5m1 (see Fig. 1A). (B–D) Hairpin siRNA inhibition of luciferase activity (see legend for Fig. 1D). (B) CS2+luc was not inhibited by hairpin siRNAs. (C) GFP5HP1 and GFP5HP1S inhibited luciferase from both sense and antisense targets. The GFP5HP1m1 hairpin did not effectively inhibit luciferase activity from vectors containing either strand of GFP in the luciferase mRNA, whereas GFP5HP1m2 and GFP5HP1m3 reduced inhibition only for the mismatched strand. (D) Denaturation (dn) of the GFP5 siRNA reduced inhibition of a luciferase-GFP target, whereas denaturation of GFP5HP1 did not significantly alter inhibition.
Cell Culture and Transfections.
Mouse P19 cells (27) were cultured as described (28). For transfection, cells were plated on dishes coated with murine laminin (Invitrogen) at 70–90% confluency without antibiotics. Transfections were performed with Lipofectamine 2000 (Invitrogen) as directed by the manufacturer. For inhibition of green fluorescent protein (GFP), 1.6 μg CS2+eGFP (28) was cotransfected with 200 ng siRNAs per 35-mm dish. Cells were fixed 19–20 h after transfection. For inhibition of neuronal β-tubulin, 1.0 μg biCS2+MASH1/GFP was cotransfected with either 200 ng siRNAs or 0.8 μg of each U6 siRNA vector per 35-mm dish. Media was replaced with OPTI-MEM1 (Invitrogen) supplemented with 1% (vol/vol) FBS 8–14 h after transfection and changed 3 days after transfection. Cells were fixed 3.5–4 days after transfection.
Expression Plasmids.
Plasmids were constructed by using standard techniques. The U6 promoter (26) was isolated by PCR from mouse genomic DNA with the oligonucleotides CCCAAGCTTATCCGACGCCGCCATCTCTA and GGGATCCGAAGACCACAAACAAGGCTTTTCTCCAA. An introduced Bbs1 site (underlined) allowed insertion of siRNA sequences at the first nucleotide of the U6 transcript. The U6 promoter was cloned into the vector RARE3E (29). siRNA and hairpin siRNA sequences were synthesized as two complementary DNA oligonucleotides, annealed, and ligated between the Bbs1 and XbaI sites (for sequences, see Fig. 4A and supporting information). The biCS2+MASH1/GFP vector is a variant of CS2 (30, 31) that contains both rat MASH1 (32) and enhanced GFP (EGFP; CLONTECH) coding sequences, expressed in divergent orientations from a shared simian CMV IE94 enhancer. CS2+luc contains the luciferase gene from pGL3 (Promega) inserted into the CS2+ vector (30, 31).
Reporter Assays.
Approximately 500 nt from the 3′ end of the EGFP-coding region was inserted into CS2+luc plasmid after the luciferase stop codon in sense (CS2+luc-GFP-S) and antisense (CS2+luc-GFP-AS) orientation. In 12-well plates, 500 ng CS2+luc, CS2+luc-GFP-S, or CS2+luc-GFP-AS were cotransfected with 150 ng siRNAs and 500 ng CS2+cβgal (31) per well; 150–200 ng of siRNA gave near maximal inhibition (data not shown). Reporter activity was assayed 19–20 h after transfection by using the Dual-Light system (Applied Biosystems). Luciferase activity was normalized to β-galactosidase activity to control for transfection efficiency variation. To test the effect of denaturation on siRNA function, siRNAs were diluted to 3 ng/μl, heated to 95° for 5 min, cooled on ice, and diluted for transfection.
Immunohistochemistry.
Cells were fixed as described (28). Antibody dilutions: mouse monoclonal TuJ1 antibody (CRP, Cumberland, VA) against neuronal class III β-tubulin 1:2,000, mouse monoclonal 16A11 (Molecular Probes) against HuC/D 1:500, and Alexa Fluor 546 goat anti-mouse IgG secondary antibody (Molecular Probes) 1:4,000. Cells were photographed with a video camera on an inverted microscope and the images were digitized. Cell counts for GFP and HuC/D were performed with NIH IMAGE software. TuJ1-labeled cells were counted manually. The number of antibody-labeled cells was normalized to the number of GFP-expressing cells for each field of view.
Results
Synthesis of siRNAs by in Vitro Transcription.
To test the ability of RNAs generated by in vitro transcription to function as siRNAs, we synthesized complementary pairs of 21-nt RNAs with T7 RNA polymerase and partially single-stranded DNA oligonucleotide templates (Fig. 1 A and B; ref. 21). Each pair of 21-nt siRNA strands was synthesized separately and annealed to create a 19-nt siRNA duplex, with two nt 3′ overhangs at each end, as described (see Materials and Methods for details of synthesis, purification, and quantitation). As a rapid assay for siRNA function, we tested the ability of either T7 or chemically synthesized siRNA duplexes to inhibit the expression of GFP in a transient transfection. siRNAs and a GFP-expression vector were cotransfected into mouse P19 cells, and GFP expression was assessed by epifluorescence. DhGFP1, a duplex of chemically synthesized siRNAs, and GFP5, a T7-synthesized siRNA duplex, both efficiently reduced GFP expression. To confirm that inhibition was sequence specific, we tested GFP5m1, a T7-synthesized siRNA duplex with a two-base mismatch in each strand located at the presumptive cleavage site in the GFP target (2, 5). The GFP5m1 siRNA duplex did not reduce GFP fluorescence (Fig. 1C). To quantify siRNA-mediated inhibition, we inserted part of the GFP gene into the 3′ untranslated region of the luciferase reporter in the CS2+luc expression vector, in sense (CS2+luc-GFP-S) and antisense (CS2+luc-GFP-AS) orientations (Fig. 1D). Based on studies in Drosophila extracts, we expected that siRNA duplexes would inhibit a mammalian mRNA containing either sense or antisense target sequences. Although cotransfection of the DhGFP1 or GFP5 siRNA duplexes did not inhibit luciferase activity from the CS2+luc vector (which does not contain matching sequences), both siRNA duplexes reduced luciferase expression by 5- to 7-fold from the CS2+luc-GFP-S and CS2+luc-GFP-AS vectors (Fig. 1D). This result indicates that a T7-synthesized siRNA can inhibit gene expression in mammalian cells as effectively as a chemically synthesized siRNA. GFP2, another T7-synthesized siRNA duplex directed against a different sequence in GFP (partially overlapping the DhGFP1 target), also reduced luciferase activity, although slightly less effectively. Cotransfection of the mismatched GFP5m1 siRNA duplex did not inhibit luciferase activity from CS2+luc-GFP-S at all, consistent with its lack of effect on GFP fluorescence, whereas it inhibited luciferase activity from CS2+luc-GFP-AS only slightly.
Inhibition of Gene Expression by Hairpin siRNAs.
We wanted to determine whether a short hairpin RNA could function like an siRNA duplex composed of two siRNA strands. We used T7 in vitro transcription to synthesize variants of the GFP5 siRNAs in which the two siRNA strands were contained within a single hairpin RNA, with the sequence for each strand connected by a loop of three nucleotides (Fig. 2A). In GFP5HP1, the GFP5 antisense siRNA (corresponding to the antisense strand of GFP) is located at the 5′ end of the hairpin RNA, whereas in GFP5HP1S, the GFP5 sense siRNA is at the 5′ end of the hairpin RNA. The loop sequence for each vector is a continuation of the 5′ end siRNA in the hairpin. Each hairpin RNA ended with two unpaired U residues that did not match the target strand. As a control for sequence specificity, we synthesized the GFP5HP1m1 hairpin RNA, which has a two-base mismatch with GFP (analogous to the GFP5m1 siRNA duplex). All hairpin RNAs migrated on a nondenaturing gel with the same mobility as the annealed DhGFP1 or GFP5 siRNA duplexes, consistent with synthesis of full-length RNA (data not shown). When cotransfected into cells with luciferase vectors, both the GFP5HP1 and GFP5HP1S hairpin RNAs inhibited luciferase activity from the CS2+luc-GFP-S and CS2+luc-GFP-AS vectors but not from the CS2+luc vector (Fig. 2 B and C). The order of the sense and antisense strands within the hairpin RNA did not alter inhibition, although neither hairpin RNA was as effective as the GFP5 siRNA duplex. As expected, the GFP5HP1m1 hairpin RNA was completely ineffective in the inhibition of luciferase expression from CS2+luc-GFP-S, and it inhibited luciferase expression from CS2+luc-GFP-AS only slightly. These effects are identical to the effects of the GFP5m1 siRNA on luciferase activity from these vectors (Fig. 1D). These observations, as well as additional observations described below, suggest that a hairpin siRNA molecule functions similarly to an siRNA duplex. We considered the possibility that two hairpin siRNA molecules might function as a longer RNA duplex rather than as a single-molecule hairpin siRNA. If the hairpin RNA functioned primarily as a single RNA molecule, it should be resistant to denaturation, because both “strands” of the siRNA are covalently linked, whereas denaturation of the GFP5 siRNA should reduce inhibition. We compared the inhibition of luciferase activity from CS2+luc-GFP-S by the GFP5 siRNA duplex and the GFP5HP1 hairpin siRNA after denaturation immediately before transfection (Fig. 2D). Although inhibition by the GFP5 duplex decreased, GFP5HP1 inhibition remained unchanged, consistent with the hypothesis that GFP5HP1 functions primarily as a single RNA molecule. The failure of denaturation to completely prevent GFP5 siRNA duplex inhibition may reflect reannealing of the two strands during transfection or inside cells.
Like siRNA duplexes, hairpin siRNAs can effectively inhibit RNAs complementary to either the sense or antisense siRNA sequences (Fig. 2C). It may be useful to inhibit a target RNA, but not its complement (e.g., to prevent self-targeting of a retroviral vector expressing an siRNA hairpin). We tested the effect of single-base changes in either the antisense (GFP5HP1m2) or sense (GFP5HP1m3) sequences of the GFP5HP1 hairpin (Fig. 2A) on the inhibition of luciferase activity from CS2+luc-GFP-S and CS2+luc-GFP-AS. In each case, the ability of the hairpin to inhibit the GFP strand complementary to the mismatched sequence was reduced, although inhibition of the perfectly matched GFP strand was unaffected (Fig. 2C). Thus, a hairpin siRNA can target an RNA without targeting its complement equally, and basepairing within a hairpin siRNA duplex need not be perfect to trigger inhibition. Although a single base mismatch in the hairpin siRNA provided only partial strand specificity, it may be possible to increase specificity with additional mismatched bases.
Inhibition of Neuronal β-Tubulin Expression by T7 siRNAs and Hairpin siRNAs.
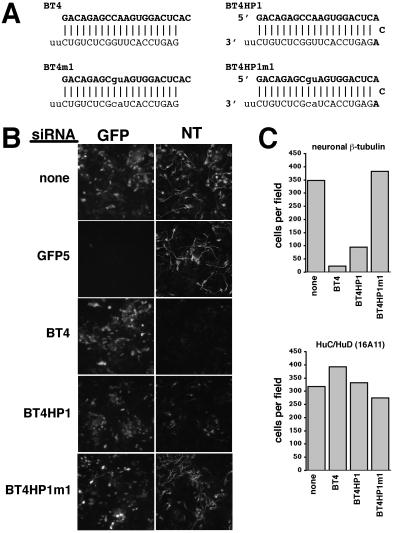
The ability of T7-synthesized siRNAs and hairpin siRNAs to inhibit endogenous gene expression was tested with a cell culture model of neuronal differentiation. We have previously shown that uncommitted mouse P19 cells can be converted into differentiated neurons by the transient expression of neural basic helix-loop-helix (bHLH) transcription factors (28). An abundant and readily detectable protein marker of neuronal differentiation expressed in these neurons is the neuron-specific β-tubulin type III recognized by the monoclonal antibody TuJ1 (33), referred to here as neuronal β-tubulin. We synthesized both an siRNA duplex and a hairpin siRNA directed against the same target sequence in the 3′ untranslated region of the mRNA for neuronal β-tubulin (GenBank accession no. AF312873; Fig. 3A). Mouse P19 cells were cotransfected with the siRNAs and biCS2+MASH1/GFP, a vector that expresses the neural bHLH protein MASH1 and GFP. Cotransfection of the siRNA duplex against neuronal β-tubulin substantially reduced the number of neuronal β-tubulin-expressing cells detected by indirect immunofluorescence (≈17-fold), but it did not alter GFP expression (Fig. 3 B and C). In contrast, cotransfection of the GFP5 siRNA duplex reduced GFP expression, but it did not alter neuronal β-tubulin expression. Cotransfection of the hairpin siRNA against neuronal β-tubulin also reduced the number of neuronal β-tubulin-expressing cells (≈4-fold), although not as effectively as the double-stranded siRNA. The decrease in the number of neuronal β-tubulin-expressing cells did not reflect either cell death or a failure of the transfected cells to differentiate, because the number of transfected cells expressing the HuC/HuD RNA-binding proteins (markers of neuronal differentiation recognized by the monoclonal antibody 16A11) did not decrease (Fig. 3C). Cotransfection of either an siRNA duplex or a hairpin siRNA that contained a two-base mismatch with the neuronal β-tubulin mRNA did not reduce the number of neuronal β-tubulin-expressing cells (Fig. 3 and data not shown).
Figure 3.
RNA interference with neuronal β-tubulin using in vitro-synthesized siRNAs and hairpin siRNAs. (A) Sequences and expected structures for the siRNAs and hairpin siRNAs against neuronal β-tubulin (notation as in Fig. 1). (B) GFP fluorescence and neuronal β-tubulin expression detected by indirect immunofluorescence in mouse P19 cells 4 days after cotransfection with biCS2+MASH1/GFP and various siRNAs. GFP5 reduced GFP expression to undetectable in most cells without altering neuronal β-tubulin (NT), whereas BT4 and BT4HP1 reduced the number of neuronal β-tubulin-expressing cells without altering GFP expression. The mismatched siRNA BT4HP1m1 had no effect on GFP or neuronal β-tubulin. (C) Number of cells per field of view expressing detectable neuronal β-tubulin or the HuC/HuD neuronal RNA-binding proteins detected by indirect immunofluorescence after cotransfection of biCS2+MASH1/GFP and BT4, BT4HP1, or BT4HP1m1 siRNAs. An average from three fields is shown for each transfection. Neuronal β-tubulin and HuC/HuD were scored in parallel transfections, and cell numbers were normalized to the number of GFP-expressing cells in each field to control for transfection efficiency. Data are from a representative experiment.
Inhibition of Neuronal β-Tubulin Expression with U6 siRNA Expression Vectors.
For expression of siRNAs and hairpin siRNAs in mammalian cells, we were concerned that sequence extensions at either end of an siRNA would prevent inhibition. Therefore, we constructed an expression vector based on the mouse U6 promoter in which we could insert a sequence after the first nt of the U6 transcript (a G). By selecting siRNA sequences that begin with G, it is possible to express siRNAs in this vector that precisely match the target gene, except for the four 3′ end U residues from RNA polymerase III termination (Fig. 4 A and B). The terminal U residues were used as 3′ overhanging ends for both siRNAs and hairpin siRNAs, because the overhanging ends of an siRNA need not match its target sequence, and their length can be varied from at least 2 to 4 nt (2, 5, 12). All of our T7-synthesized siRNAs began with G (Fig. 3A), so we used the same sequences to target neuronal β-tubulin in the U6 expression system. The U6-BT4s and U6-BT4as vectors are expected to express 21-nt complementary single-stranded RNAs with 19 nt corresponding to the sense or antisense strands of the BT4 siRNA duplex (each U6 vector expresses one siRNA strand), whereas the U6-BT4HP1, U6-BT4HP2, and U6-BT4HP2m1 vectors are expected to express 45-nt hairpin siRNAs (Fig. 4B). The U6-BT4HP2 contains a one-base mismatch in the sense strand of the hairpin siRNA, analogous to the GFP5HP1m3 siRNA (Fig. 2A), whereas the antisense strand of U6-BT4HP2m1 contains a two-base mismatch with neuronal β-tubulin.
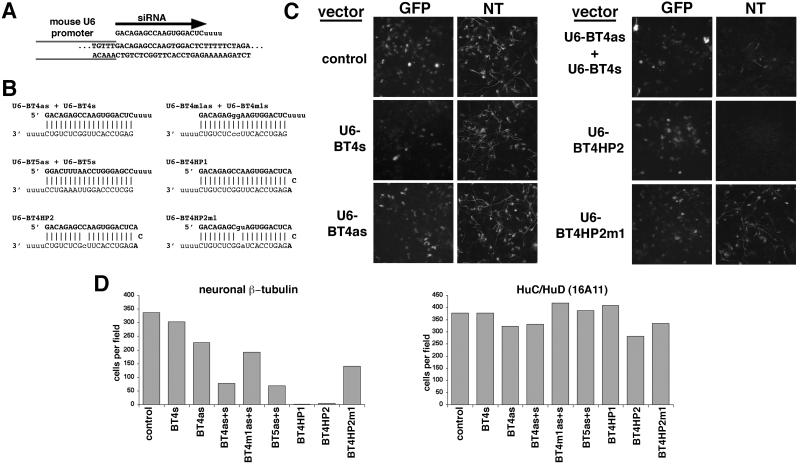
Figure 4.
RNA interference using siRNAs and hairpin siRNAs expressed in mouse P19 cells from a U6 RNA polymerase III promoter. (A) An example of the transcribed region of a mouse U6 promoter siRNA vector (U6-BT4as). The first nucleotide of the U6 transcript corresponds to the first nucleotide of the siRNA, whereas the siRNA terminates at a stretch of 5 T residues in the vector. (B) Sequences for the siRNAs and hairpin siRNAs to neuronal β-tubulin synthesized from the U6 vector. Expected RNA duplexes are shown for the hairpin siRNAs and for pairs of single-strand siRNAs (notation as in Fig. 1). (C) GFP fluorescence and indirect immunofluorescence for neuronal β-tubulin (NT) 4 days after cotransfection of the indicated U6 vectors and biCS2+MASH1/GFP. (D) Number of cells with detectable neuronal β-tubulin and HuC/HuD after cotransfection of biCS2+MASH1/GFP and various U6 vectors (using parallel transfections; details as in Fig. 3). The expression of either siRNA hairpin reduces the number of neuronal β-tubulin positive cells about 100-fold, whereas cotransfection of two vectors expressing individual siRNA strands reduces the number of neuronal β-tubulin cells about 5-fold. HuC/D expression is not altered by the hairpin siRNAs.
Cotransfection of the U6-BT4as and U6-BT4s vectors reduced the number of neuronal β-tubulin-expressing cells generated by biCS2+MASH1/GFP about four-fold (Fig. 4 C and D). In addition, the intensity of fluorescence was reduced for most cells with detectable neuronal β-tubulin, suggesting decreased levels of expression (Fig. 4C). The U6-BT4as and U6-BT4s vectors had little or no effect on the number of neuronal β-tubulin-expressing cells when cotransfected individually with biCS2+MASH1/GFP, indicating that both U6-driven siRNA strands are required for effective inhibition (Fig. 4 C and D). We also tested a vector in which the two siRNA strands were expressed from tandem U6 promoters on a single plasmid. This vector inhibited neuronal β-tubulin with approximately the same efficiency as cotransfection of the U-BT4as and U6-BT4s vectors, suggesting that cotransfection efficiency is not a limiting factor for inhibition (data not shown). Cotransfection of the U6-BT5as and U6-BT5s vectors (Fig. 4B), which express two complementary siRNA strands targeted against a different sequence in neuronal β-tubulin, reduced the number of expressing cells with similar efficiency to U6-BT4as and U6-BT4s (Fig. 4D).
Cotransfection of either of the hairpin siRNA expression vectors (U6-BT4HP1 or U6-BT4HP2) with biCS2+MASH1/GFP resulted in a 100-fold reduction in cells with detectable neuronal β-tubulin staining (Fig. 4 C and D). This inhibition was more effective than either cotransfection of the U6-BT4as and U6-BT4s vectors together, or cotransfection of in vitro-synthesized siRNAs (compare with Fig. 3 B and C). Similar results also were obtained with a variant of U6-BT4HP2 in which the loop sequence was extended to 4 nucleotides (data not shown). In contrast, neuronal β-tubulin expression was only slightly reduced by cotransfection of the mismatched hairpin-expression vector U6-BT4HP2m1 (Fig. 4 C and D). Expression of the HuC/HuD neuronal RNA-binding proteins and GFP were not altered by the U6 siRNA or hairpin siRNA-expression vectors (Fig. 4 C and D), indicating that the inhibition of neuronal β-tubulin by the U6-BT4HP1 and U6-BT4HP2 vectors is specific.
Discussion
The use of siRNAs to inhibit gene expression in mammalian cells is a promising new approach for the analysis of gene function (13, 18, 19). Here, we have demonstrated that siRNAs can be synthesized by in vitro transcription or in cells from a U6-expression vector, providing economical alternatives to chemical synthesis of siRNAs. We also found that hairpin siRNAs can inhibit gene expression in mammalian cells. Inhibition by hairpin siRNAs was sequence-specific, as a two-base mismatch between an in vitro-synthesized hairpin siRNA and its target abolished inhibition, and even a single-base mismatch in one hairpin strand allowed differential inhibition of sense and antisense target strands. These observations are consistent with prior reports of siRNA duplex specificity (5, 13).
Production of a hairpin siRNA from a transfected U6-expression vector was the most effective method tested for inhibition of neuronal β-tubulin protein expression in differentiating mouse P19 cells, resulting in a 100-fold decrease in the number of cells with detectable protein. The cells without detectable neuronal β-tubulin were viable and expressed other markers of neuronal differentiation, although we have not analyzed additional aspects of differentiation (e.g., neurite outgrowth rate) in detail. It should be noted that neuronal β-tubulin expression is not detected until 2 days after transfection of bHLH-expression vectors in most cells (28). This delay probably allowed time for the expression of the hairpin siRNA from the U6 vector before target gene expression and may have facilitated the detection of neuronal β-tubulin inhibition, because turnover of preexisting protein was not required.
Inhibition of neuronal β-tubulin by a hairpin siRNA expressed from the U6 promoter in transfected cells was more effective than inhibition by two siRNA strands expressed from separate U6 vectors. It is thought that two siRNAs must form a duplex for inhibition of a target gene by RNAi (2, 4, 5, 10, 13). Although we have not assessed duplex formation in cells, cotransfection of both sense and antisense U6 siRNA vectors was required for effective inhibition, consistent with a requirement for duplex formation. Formation of a duplex by the folding back of a hairpin siRNA should be rapid and efficient, whereas formation of a duplex between two siRNA strands synthesized separately within a cell is likely to be less efficient. We speculate that duplex formation is the limiting event for inhibition by siRNAs synthesized within cells, thus permitting the hairpin design to function more effectively. In contrast, inhibition by a transfected siRNA duplex comprised of two in vitro-synthesized siRNA strands was somewhat more effective than transfection of an in vitro-synthesized hairpin siRNA against the same target sequence. Possibly recognition of the siRNA duplex by the cellular machinery that mediates RNAi and/or other events subsequent to duplex formation are more efficient with a duplex composed of two separate siRNAs. Recognition of a target sequence by an siRNA strand includes unwinding of the siRNA duplex and formation of a new duplex with the target RNA (4). For hairpin siRNA molecules, this process may be less efficient. Alternately, hairpin siRNAs may need processing, such as cleavage within the loop, before functioning. Such processing could be similar to the maturation of hairpin RNA precursors for microRNAs (20). Also, it is possible that synthesis of siRNAs in the nucleus directs these molecules to cellular compartments distinct from those accessible to siRNAs introduced by lipid-mediated transfection, thus altering their effectiveness (34).
The ability to inhibit gene expression with hairpin siRNAs synthesized in mammalian cells is likely to have broad application. In particular, this approach should facilitate studies of gene function in transfectable cell lines. We also expect that this approach will be adaptable to situations for which delivery of in vitro-synthesized siRNAs may not be practical, such as primary cell cultures, studies in intact animals, and gene therapy. The U6 siRNA-expression cassette we have used is small (<400 nt), and should be suitable for delivery into cells by DNA-based viral vectors (22, 35). The ability to design hairpin siRNAs with strand specificity also may permit the inclusion of hairpin siRNAs in retroviral vectors containing a U6 promoter (36) without self-targeting of the viral genomic RNA. The combination of a marker gene and one (or more) U6 hairpin-expression cassettes in a viral vector would facilitate single-cell or mosaic analysis of gene function. This approach should be particularly valuable for tissue or stage-specific analysis of genes with broad roles in development. Finally, our observations demonstrate that it is possible to inhibit a neuron-specific gene in a model system for neuronal differentiation, suggesting that it will be possible to apply RNAi to studies of neurogenesis and differentiation in mammals.
Supplementary Material
Acknowledgments
We thank Anne Vojtek, Robert Davis, and Dan Goldman for helpful discussions and comments on the manuscript. We thank Chris Hart for assistance with cloning of the mouse U6 promoter. This work was supported by NIH Grant NS38698 and the University of Michigan Frontiers in Neuroscience.
Abbreviations
- RNAi
RNA interference
- dsRNA
double-stranded RNA
- siRNA
short-interfering RNA
- GFP
green fluorescent protein
References
- 1.Sharp P A. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir S M, Lendeckel W, Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parrish S, Fleenor J, Xu S, Mello C, Fire A. Mol Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- 4.Nykanen A, Haley B, Zamore P D. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir S M, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond S M, Bernstein E, Beach D, Hannon G J. Nature (London) 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 7.Zamore P D, Tuschl T, Sharp P A, Bartel D P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 8.Bass B L. Nature (London) 2001;411:428–429. doi: 10.1038/35078175. [DOI] [PubMed] [Google Scholar]
- 9.Yang D, Lu H, Erickson J W. Curr Biol. 2000;10:1191–1200. doi: 10.1016/s0960-9822(00)00732-6. [DOI] [PubMed] [Google Scholar]
- 10.Boutla A, Delidakis C, Livadaras I, Tsagris M, Tabler M. Curr Biol. 2001;11:1776–1780. doi: 10.1016/s0960-9822(01)00541-3. [DOI] [PubMed] [Google Scholar]
- 11.Sijen T, Fleenor J, Simmer F, Thijssen K L, Parrish S, Timmons L, Plasterk R H, Fire A. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 12.Lipardi C, Wei Q, Paterson B M. Cell. 2001;107:297–307. doi: 10.1016/s0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- 13.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 14.Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Proc Natl Acad Sci USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S, Tutton S, Pierce E, Yoon K. Mol Cell Biol. 2001;21:7807–7816. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paddison P J, Caudy A A, Hannon G J. Proc Natl Acad Sci USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson H D, Mathews M B. Biochimie. 1996;78:909–914. doi: 10.1016/s0300-9084(97)86712-0. [DOI] [PubMed] [Google Scholar]
- 18.Harborth J, Elbashir S M, Bechert K, Tuschl T, Weber K. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 19.Hutvagner G, McLachlan J, Pasquinelli A E, Balint E, Tuschl T, Zamore P D. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 20.Grosshans H, Slack F J. J Cell Biol. 2002;156:17–22. doi: 10.1083/jcb.200111033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tazi J, Forne T, Jeanteur P, Cathala G, Brunel C. Mol Cell Biol. 1993;13:1641–1650. doi: 10.1128/mcb.13.3.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Booth B L, Jr, Pugh B F. J Biol Chem. 1997;272:984–991. doi: 10.1074/jbc.272.2.984. [DOI] [PubMed] [Google Scholar]
- 24.Noonberg S B, Scott G K, Garovoy M R, Benz C C, Hunt C A. Nucleic Acids Res. 1994;22:2830–2836. doi: 10.1093/nar/22.14.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Good P D, Krikos A J, Li S X, Bertrand E, Lee N S, Giver L, Ellington A, Zaia J A, Rossi J J, Engelke D R. Gene Ther. 1997;4:45–54. doi: 10.1038/sj.gt.3300354. [DOI] [PubMed] [Google Scholar]
- 26.Reddy R. J Biol Chem. 1988;263:15980–15984. [PubMed] [Google Scholar]
- 27.McBurney M W. Int J Dev Biol. 1993;37:135–140. [PubMed] [Google Scholar]
- 28.Farah M H, Olson J M, Sucic H B, Hume R I, Tapscott S J, Turner D L. Development (Cambridge, UK) 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- 29.Davis R L, Turner D L, Evans L M, Kirschner M W. Dev Cell. 2001;1:553–565. doi: 10.1016/s1534-5807(01)00054-5. [DOI] [PubMed] [Google Scholar]
- 30.Rupp R A, Snider L, Weintraub H. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 31.Turner D L, Weintraub H. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 32.Johnson J E, Birren S J, Anderson D J. Nature (London) 1990;346:858–861. doi: 10.1038/346858a0. [DOI] [PubMed] [Google Scholar]
- 33.Lee M K, Tuttle J B, Rebhun L I, Cleveland D W, Frankfurter A. Cell Motil Cytoskeleton. 1990;17:118–132. doi: 10.1002/cm.970170207. [DOI] [PubMed] [Google Scholar]
- 34.Bertrand E, Castanotto D, Zhou C, Carbonnelle C, Lee N S, Good P, Chatterjee S, Grange T, Pictet R, Kohn D, et al. RNA. 1997;3:75–88. [PMC free article] [PubMed] [Google Scholar]
- 35.Potter P M, McKenzie P P, Hussain N, Noonberg S, Morton C L, Harris L C. Mol Biotechnol. 2000;15:105–114. doi: 10.1385/MB:15:2:105. [DOI] [PubMed] [Google Scholar]
- 36.Ilves H, Barske C, Junker U, Bohnlein E, Veres G. Gene. 1996;171:203–208. doi: 10.1016/0378-1119(96)00075-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.