Abstract
Drosophila ovo/svb (dovo) is required for epidermal cuticle/denticle differentiation and is genetically downstream of the wg signaling pathway. Similarly, a mouse homolog of dovo, movo1, is required for the proper formation of hair, a mammalian epidermal appendage. Here, we provide biochemical evidence that movo1 encodes a nuclear DNA binding protein (mOvo1a) that binds to DNA sequences similar to those that dOvo binds to, further supporting the notion that mOvo1a and dOvo are genetically and biochemically homologous proteins. Additionally, we show that the movo1 promoter is activated by the lymphoid enhancer factor 1 (LEF1)/β-catenin complex, a transducer of wnt signaling. Collectively, our findings suggest that movo1 is a developmental target of wnt signaling during hair morphogenesis in mice, and that the wg/wnt-ovo link in epidermal appendage regulatory pathways has been conserved between mice and flies.
Keywords: hair follicle‖wnt‖wg‖mouse ovo1
Mouse and fly epidermis differ in their morphology and complexity, as do their respective epidermal appendages—hair in mice and denticles/bristles in flies. Whereas several signaling pathways (e.g., wnt/wg and shh/hh) appear to have conserved roles in patterning both hair follicles and denticles (for review, see ref. 1), less is known about the molecular events that directly participate in the morphogenesis of these epidermal appendages.
The Drosophila ovo/svb (dovo) gene represents a clear-cut link between signaling cues and downstream morphogenetic events in epidermis. dovo is normally expressed in denticle-producing epidermal cells, and its ectopic expression is sufficient to induce denticle formation (2). Loss-of-function dovo mutants develop denticles that are dramatically reduced in number and size (3), a phenotype distinct from that of the segment polarity mutants (e.g., wg, hh, and smo), suggesting that dovo functions in denticle morphogenesis rather than patterning. The zinc finger-containing dOvo proteins possess DNA-binding and transcription regulatory activities (4, 5), and likely regulate the expression of downstream genes involved in denticle formation. dovo transcription is regulated by the wg signaling pathway (2), central to which is the stabilization and nuclear translocation of cytoplasmic armadillo (Drosophila β−catenin) and the formation of a bipartite transcription activator complex between armadillo and a lymphoid enhancer factor/T cell factor (LEF/TCF) family member, dTCF/pangolin (for review, see refs. 6 and 7). Epistasis studies have positioned dovo downstream of armadillo and dTCF/pangolin (2). However, it is not clear whether dovo is a direct target of the Armadillo/dTCF complex, or whether intermediate factors are involved.
Genes that are related to dovo exist in other species, including mammals (8–10). A mouse ovo gene, movo1, is required for proper hair morphogenesis (10), raising the possibility that certain aspects of epidermal appendage differentiation are conserved between mice and flies. Whereas the genetic function of movo1 in epidermis is reminiscent of that of dovo, the biochemical function of mOvo1 protein(s) remains to be elucidated.
Components of the wnt signal transduction pathway have been functionally implicated in multiple events during hair follicle morphogenesis and differentiation. Ectopic expression of wnt3a in mice leads to hair defects (11). LEF1 knockout mice display a reduced number of hair follicles, and residual follicles fail to produce normal hair shafts (12). Conversely, ectopic expression of LEF1 leads to hair formation in ectopic locations (13). Expression of a stable form of β-catenin in skin leads to overt phenotypes such as de novo hair follicle morphogenesis and abnormal angling of protruding hairs (14), whereas conditional ablation of β-catenin in the epidermis or expression of an N-terminally truncated LEF1 that cannot associate with β-catenin results in defective hair morphogenesis (15, 16). These and other studies suggest that wnt signaling, LEF1, and β-catenin are required for formation of hair follicles during embryogenesis as well as for postnatal hair production. So far, little is known about the downstream targets of LEF1/β-catenin, and likewise of wnt signaling, in hair morphogenesis and in other developmental processes in mice. The genetic link between wnt/wg signaling and ovo raises the question as to whether movo1 might be a direct target for LEF1 and β-catenin.
Here, we show that movo1 encodes a sequence-specific DNA binding protein that accumulates in the nuclei of differentiating epidermal and hair follicle cells, and binds DNA sequences similar to the dOvo recognition sequence. We examine the regulation of movo1 expression, and demonstrate that the LEF1/β-catenin complex activates movo1 promoter in vitro, implicating movo1 as a direct wnt/LEF1/β-catenin target during hair morphogenesis.
Materials and Methods
Cloning and Sequence Analyses.
Cloning of movo1 cDNAs and genomic fragments has been reported previously (10). Two movo1 cDNA sequences have been assigned the following accession numbers: 2.3 kb cDNA (AF13804) and 1.9 kb cDNA (AF13805). Sequence of the movo1 promoter fragment has been submitted to GenBank (accession no. AF487891).
Chromosomal Localization.
Metaphase chromosome spreads were prepared from mouse embryonic stem cells. Colcemid was added at 0.01 μg/ml to the cells for 30 min, and cells were harvested by using a 50:50 mixture of 0.075 M KCl:1% citrate. Probe labeling, DNA hybridization, and antibody detection were carried out by using previously described methods (17). An 11.5-kb movo1 genomic clone was labeled with digoxigenin-16-dUTP (Roche Boehringer Mannheim), and hybridized together with a biotin-labeled mouse chromosome 19-specific probe (Oncor).
Transient Transfection Assays.
UG1 mouse keratinocytes (14) at passages 19–28 were seeded in 12-well plates and transfected at 40–50% confluence with Fugene 6 Transfection Reagent (Roche Boehringer Mannheim). A total of 0.45 μg of plasmid DNA containing various combinations (as indicated in figure legends) of plasmids was used to transfect each well. Construction of expression vectors producing human LEF1 or Xenopus β-catenin has been described previously (18, 19). A representative experiment uses 0.1 μg of pGL3-movo1 (movo1 promoter-luciferase construct), 0.2 μg of LEF1 expression vector, 0.1 μg of β-catenin expression vector, 0.05 μg of β-actin-β-gal construct, and pCB-6 (+) (empty vector). Cells were harvested 30–48 h after transfection, and luciferase activity was measured by using the Luciferase Assay System (Promega). β-galactosidase activity was measured as previously described (20).
Expression of Recombinant mOvo1a Polypeptide.
A movo1 cDNA fragment encoding a truncated mOvo1a protein missing the first 29 aa (polypeptide 30-267) was cloned into pQE32 (Qiagen) or the pFastBac vector (GIBCO/BRL). The production and purification of this His-6-tagged mOvo1a polypeptide from bacteria or baculovirus-infected insect cells was performed according to manufacturers' instructions.
Preparation of Epidermal Nuclear Extract and Electrophoretic Mobility-Shift Assays (EMSA).
Isolation of newborn mouse skin epidermis by dispase was done as previously described (21). Epidermal nuclear extract, prepared from isolated epidermis as described (22), or recombinant mOvo1a polypeptide 30-267 (≈25 ng per reaction or as indicated) was incubated with a 32P-labeled, double-stranded version of the following oligonucleotide (oligo): 5′-GTTCCTTTTACAGTTACATAGCAATCGTC-3′. The binding reactions contained 20,000 cpm of 32P-labeled oligo (≈4 fmol) that was incubated at room temperature for 30 min with mOvo1a in 20 mM Hepes (pH 7.9), 75 mM KCl, 2.5 mM MgCl2, 2 mM DTT, 1 mM EDTA, 12% glycerol, and 1 μg of poly(dI-dC).
DNase I Footprinting.
An MluI fragment containing the putative LEF/TCF binding site from the movo1 promoter was end-labeled by using T4 polynucleotide kinase, followed by cleavage with SphI. The MluI-SphI fragment labeled with 32P at the MluI site was gel-purified, and 20,000 cpm of labeled fragment were incubated with different concentrations of partially purified recombinant LEF1 protein in DNase I footprinting assays as described (19).
Immunofluorescence, Immunoblotting, and Immunohistochemistry.
The procedures for indirect immunofluorescence, immunoblotting, and the production and purification of peptide antibodies were as described (10). Immunohistochemistry was performed by using the Vectastain ABC kit according to the manufacturer's recommendations (Vector Laboratories). Rabbit α-mOvo1a polyclonal antibodies were raised against the His-6-tagged recombinant mOvo1a polypeptide 30-267 (Covance Research Products, Denver, PA). Anti-β-catenin antibody raised against a C-terminal peptide was kindly provided by W. Birchmeier (Max-Delbrueck Center for Molecular Medicine, Berlin) (23) and anti-E-cadherin antibody was from BD Transduction Laboratories.
Results
movo1 Is a Homolog of dovo and a Member of a Family of ovo Genes Conserved in Multicellular Organisms.
Three different transcripts are produced from the movo1 locus (10). The two smaller transcripts are abundantly expressed in skin and encode mOvo1a, a protein of 267 aa that contains a C-terminal zinc-finger region bearing extensive sequence homology to that of dOvo. The homologous C-terminal region of mOvo1a and dOvo appear complex: besides the existence of four classic C2H2 zinc fingers, a region bearing resemblance to a FYVE finger domain (24) and a PHD-finger domain (25) are present (Fig. 1A). Moreover, a putative nuclear localization sequence is located within the zinc finger domains. These characteristic motifs define a family of Ovo-related proteins (Fig. 1B), including mOvo2 (ref. 9; B.L. and X.D., unpublished results) and mOvo3, predicted from mouse expressed sequence tag sequences (accession nos. BF714064 and BF715622). By using fluorescence in situ hybridization analysis, we mapped movo1 to chromosome 19, close to the centromere (Fig. 1C), a region syntenic to human chromosome 11q13 where a human homolog, hovo1, resides (8). Database searches revealed that the three human ovo genes reside in distinct chromosomes, suggesting that movo1, movo2, and movo3 are distinct, unlinked genes. Despite the existence of three ovo genes in the mouse genome, movo1 appears to carry out the ovo function in mouse epidermal appendage differentiation (10).
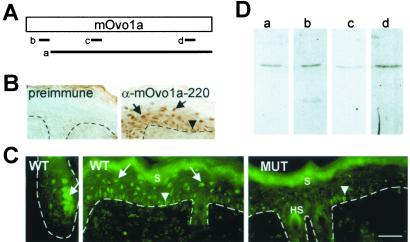
Figure 1.
movo1 and its family members. (A) Sequence and features of mOvo1a. Zinc fingers are underlined. The internal PHD/FYVE finger domains are shown in bold. The predicted nuclear localization sequence is boxed. (B) Amino acid sequence alignment of the zinc finger regions of Ovo family members. cOvo, C. elegans Ovo. *, Indicates amino acid identity. Positions of the zinc fingers are indicated above the alignment. Exon boundaries conserved among all species are indicated (S1 and S2). (C) Mapping of the movo1 gene to chromosome 19. Red (arrows) and green dots represent hybridization signals obtained with the λ clone containing movo1 genomic sequence and a chromosome 19-specific probe, respectively. The centromeres are highlighted in blue.
mOvo1a Is a Nuclear Protein That Binds to Similar but Not Identical DNA Sequences as dOvo.
Previously, we reported that a peptide antibody (α-mOvo1a-220, d in Fig. 2A) detected nuclear mOvo1a protein in transiently transfected COS cells (10). This antibody was used to determine the subcellular localization of mOvo1a in epidermis. Consistent with the suprabasal and precortex location of movo1 RNAs (10), strong staining was seen in the nuclei [as indicated by 4′,6-diamidino-2-phenylindole (DAPI) staining; data not shown] of the epidermal suprabasal cells and the hair follicle precortex cells (arrows in Fig. 2 B and C). Nuclear staining of basal cells was also observed (arrowheads in Fig. 2 B and C). These nuclear staining patterns were not detected with preimmune serum (Fig. 2B) or when an excess of the immunizing peptide was present (data not shown). Staining of the stratum corneum and hair shafts was observed (Fig. 2C); however, these signals persisted in peptide competition experiments and are therefore not peptide specific (not shown). The suprabasal nuclear stain was absent in skin from mutant mice in which the zinc finger-coding region (where this peptide antigen resides) was deleted from the movo1 locus (10), whereas a few basal cells in these mice still stained positive (Fig. 2C Right). These results indicate that the suprabasal nuclear stain indeed arises from mOvo1a protein, whereas the basal staining might be due to crossreactivity toward another mouse Ovo or unrelated proteins expressed in epidermis.
Figure 2.
Multiple antibodies detect nuclear mOvo1a protein in skin epidermis and hair follicles. (A) Diagram showing the positions of antigens used for anti-mOvo1a antibody production: a, polypeptide corresponding to amino acids 30-267 (mOvo1a); b, peptide CKRNWSELPDEERGE starting at position 15 (mOvo1a-15); c, peptide TDPQSRDQGFLRTK starting at position 91 (mOvo1a-91); d, peptide CTSESQEGHVLHLKERHPDS starting at position 220 (mOvo1a-220). (B) Immunohistochemistry of newborn skin by using preimmune serum or α-mOvo1a-220. (C) Indirect immunofluorescence of 6-day postnatal wild-type (WT) or movo1 mutant (MUT) skin by using α-mOvo1a-220. (D) Immunoblot analysis of epidermal nuclear extract with anti-mOvo1a antibodies: a, α-mOvo1a; b, α-mOvo1a-15; c, α-mOvo1a-91; d, α-mOvo1a-220. S, stratum corneum; HS, hair shafts. Dotted lines indicate the basement membrane. Arrows and arrowheads point to suprabasal/precortex and basal staining, respectively. Bar = 30 μm in B and C.
Three additional antibodies (Fig. 2A), all recognizing nuclear mOvo1a in transiently transfected COS cells (data not shown), were subsequently used to confirm the nuclear localization of mOvo1a in epidermis. All four anti-mOvo1a antibodies detected a single 43-kDa protein in epidermal nuclear extracts (Fig. 2D), suggesting that this nuclear protein is mOvo1a. Moreover, α-mOvo1a detected only suprabasal nuclear staining, but with lower signal intensity than that observed by using α-mOvo1a-220 (not shown).
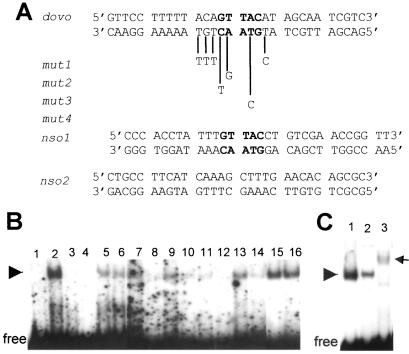
The nuclear localization of mOvo1a is consistent with the possibility that it acts as a DNA-binding transcription factor. We next examined whether mOvo1a recognizes dOvo cognate DNA sequences, identified by using a truncated dOvo protein containing the zinc finger domain (26). A recombinant, truncated mOvo1a protein containing the zinc finger domain was used for EMSA with an oligonucleotide (dovo) containing a dOvo binding site and including the core consensus sequence (GTTAC, Fig. 3A). Addition of mOvo1a polypeptide resulted in a single gel-shift band (Fig. 3B, lane 2), suggesting that mOvo1a indeed binds to this oligo. Whereas the presence of unlabeled dovo oligo completely abolished the gel-shift band (Fig. 3B, lanes 3 and 4), oligos containing mutations at the GTTAC core and/or flanking sequences displayed reduced ability to compete for binding, especially when present at a low excess (Fig. 3B, lanes 5–14). The ability to compete ranks as follows: mut1 = mut3 > mut2 > mut4 > nso1. In contrast, a nonspecific oligo (nso2) bearing no resemblance to dovo oligo did not compete for mOvo1a binding (Fig. 3B, lanes 15 and 16). Therefore, both the GTTAC core sequence in dovo oligo and sequences flanking the core are important for optimal mOvo1a binding.
Figure 3.
A zinc finger-containing mOvo1a polypeptide binds to an oligonucleotide sequence (dovo) containing a dOvo consensus binding site. (A) Sequence of the dovo oligo is shown at the top. The core consensus sequence, GTTAC, for dOvo binding is in bold. Base changes in mutant oligos (mut 1, 2, 3, and 4) used as unlabeled competitors are shown below dovo sequence. The sequences of two nonspecific oligos (nso 1 and 2), one containing the GTTAC core in a non-homologous context, and the other containing a completely unrelated sequence, are also indicated. (B) EMSA by using purified recombinant mOvo1a polypeptide 30-267 (see Materials and Methods). Lane 1, unbound labeled dovo oligo; lane 2, plus recombinant mOvo1a; lanes 3–16, competition of protein–DNA complex (arrowhead) by 20-fold (3, 5, 7, 9, 11, 13, and 15) or 100-fold (4, 6, 8, 10, 12, 14, and 16) excess of unlabeled oligos: 3 and 4, dovo; 5 and 6, nso1; 7 and 8, mut1; 9 and 10, mut2; 11 and 12, mut3; 13 and 14, mut4; 15 and 16, nso2. (C) EMSA by using recombinant mOvo1a (1) or epidermal nuclear extract (2, 3) in the absence (1, 2) and presence (3) of α-mOvo1a. The arrowhead and arrow indicate the protein–oligo complex and antibody–protein–oligo complex, respectively.
A complex of similar size was observed when epidermal nuclear extract was used (Fig. 3C). This complex was competed away by α-mOvo1a, indicating that it arises from binding of endogenous mOvo1a to the dovo oligo. These results, taken together with prior studies (10), suggest that (i) mOvo1a is a sequence-specific DNA binding protein, (ii) the DNA sequence specificities of mOvo1a and dOvo are similar but not identical, and (iii) mOvo1a and dOvo are biochemically homologous proteins with similar genetic functions in epidermal appendage differentiation.
An Active movo1 Promoter Fragment Contains a LEF1 Binding Site.
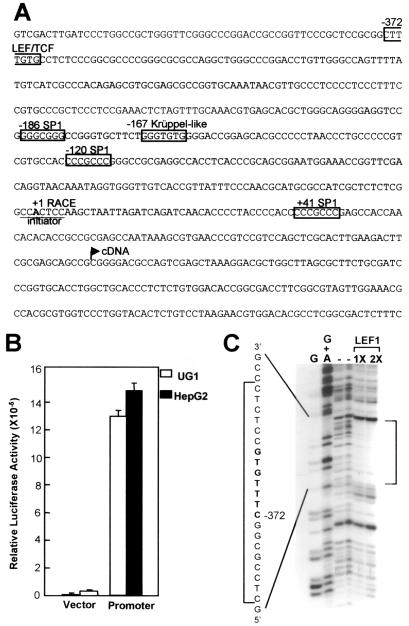
To understand how movo1 transcription is regulated, we analyzed the gene regulatory region of movo1. By using 5′ rapid amplification of cDNA ends (RACE), the 5′ end of movo1 mRNAs was identified as an A residue (+1 in Fig. 4A), 132 nucleotides upstream of the longest movo1 cDNA clone. No TATA consensus sequence is present upstream of the +1 site, indicating that movo1 promoter is TATA-less, much like its fly counterpart (5, 26). Instead, the sequence encompassing the +1 site, CCA+1CTCC, matches the consensus determined for an initiator element in eukaryotic cells, PyPyA+1NT/APyPy (27).
Figure 4.
The movo1 promoter contains a LEF1 binding site. (A) Nucleotide sequences encompassing the transcription start site of movo1. Putative binding sites for transcription factors are boxed. The starting points of the cDNAs obtained by RACE (+1 RACE) and by library screening (cDNA) are shown. (B) The fragment shown in A is able to direct transcription in epithelial cells. Luciferase activities are normalized for transfection efficiency by using a β-actin promoter driving lacZ as an internal control. An average of three experiments is shown. (C) DNase I footprinting revealing binding of recombinant LEF1 protein to the predicted LEF/TCF site (shown in bold). The DNase I cleavage patterns are shown next to the Maxam-Gilbert G and G + A sequencing reactions obtained on the same DNA fragment. The protected area is shown by brackets.
To identify potential regulatory sequences, we tested the ability of a 732-bp movo1 genomic fragment (from −431 to +303) to direct transcription of a luciferase reporter gene. In transient transfection assays, this fragment was able to confer a ≈60-fold increase in relative luciferase activity over a promoterless control vector in UG1 mouse keratinocytes [known to express movo1 transcripts (10)] or in HepG2 human epithelial cells (Fig. 4B), indicating that the movo1 promoter is active in these cells. Scanning sequences surrounding the +1 site (Fig. 4A) revealed putative binding sites for several transcription factors: three GC-rich stretches including SP1 binding sites at −186, −120, and +41, and a binding site for Krüppel-like factors at −167. Additionally, a putative binding site for LEF/TCF is present at −372.
The presence of a putative LEF/TCF site in movo1 promoter is exciting in light of the finding that dovo is genetically downstream of the wg signaling pathway in flies (2). We therefore asked whether this putative element is a bona fide recognition site. As shown in Fig. 4C, recombinant LEF1 protein indeed protected the predicted binding sequence. This result, together with the detection of movo1 mRNAs (10) as well as nuclear LEF1 and β-catenin proteins in the hair follicle precortex cells (16, 28), suggests a model that activated wnt signaling in the precortex cells causes stabilization of β-catenin and formation of LEF1/β-catenin complexes to activate the expression of movo1.
movo1 Promoter Is Activated by the LEF1/β-Catenin Complex.
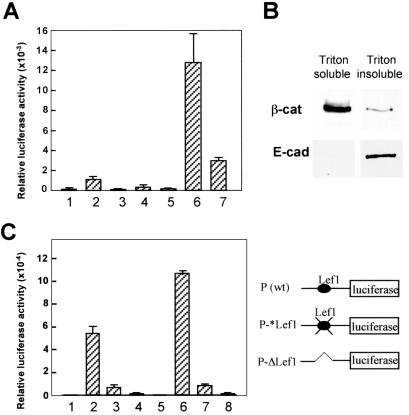
To test the notion that movo1 is directly regulated by the LEF1/β-catenin complex, we examined the effect of LEF1 and β-catenin expression on movo1 promoter activity. Since precortex-derived cell lines are not available, UG1 keratinocytes were used because (i) they are derived from mouse epidermis, which shares the same embryonic origin as precortex cells, (ii) they are responsive to wnt signaling (14, 28), and (iii) the movo1 promoter is active in these cells.
When introduced alone, LEF1 modestly activated movo1 promoter activity, whereas β-catenin had little effect (Fig. 5A). That LEF1 alone activates transcription in the absence of exogenous β-catenin raises two possibilities: (i) LEF1 activation of movo1 promoter might be independent of β-catenin, but instead depends on other transcription factors (29, 30), or (ii) LEF1 activation of movo1 promoter is mediated through endogenous “free” β-catenin present in UG1 cells cultured under our conditions. To distinguish between these two possibilities, we first tested a mutant LEF1 protein (ΔN-LEF1) missing the β-catenin-interacting domain at the N terminus and found that it failed to activate movo1 promoter (Fig. 5A). Instead, this mutant LEF1 protein acts in a dominant negative fashion (repressing by 50–80%), as previously reported (19). This result supports the notion that LEF1 activation of movo1 promoter depends on its interaction with endogenous β-catenin, and in the absence of β-catenin, LEF1 can recruit corepressors to suppress promoter activity. When both LEF1 and β-catenin were introduced, a dramatic increase in activation (>90-fold) was observed. Again, this synergistic effect depends on the interaction between LEF1 and β-catenin, because substitution of wild-type β-catenin with Δ19-β-catenin, a mutant protein that lacks the LEF1-interacting domain but retains the ability to localize to nuclei (18), reduced the fold of activation. Δ19-β-catenin can still interact with axin and might therefore cause a stabilization of the endogenous β-catenin by competing for axin binding (19). When exogenous LEF1 was present, this increase in “free” endogenous β-catenin would lead to increased LEF1/β-catenin complex formation, accounting for the residual activation observed. Without exogenous LEF1, addition of wild-type β-catenin or Δ19-β-catenin resulted in little activation, suggesting that the endogenous LEF1 levels in UG1 cells are limiting.
Figure 5.
The movo1 promoter is activated by the LEF1/β-catenin complex. (A) UG1 keratinocytes (passages 19–28) were transfected with plasmids as indicated: 1, promoter; 2, plus LEF1; 3, plus ΔN-LEF1; 4, plus β-catenin; 5, plus Δ19-β-catenin; 6, plus LEF1 and β-catenin; 7, plus LEF1 and Δ19-β-catenin. Luciferase activities are normalized as in Fig. 4B. Each bar represents the average of three triplicates. Results shown represent those from one of several experiments. (B) Triton (1%)-soluble (1.5 mg/ml) and insoluble fractions (0.2 mg/ml) prepared from UG1 keratinocytes were subjected to immunoblot analysis by using anti-β-catenin or anti-E-cadherin antibodies. (C) Activities of wild-type (wt; 2 and 6) and mutant (3, 4, 7, and 8) movo1 promoters in the absence (1–4) or presence (5–8) of exogenous LEF1 1 and 5, no promoter;3 and 7, P-*Lef1; 4 and 8, P-ΔLef1. Promoter constructs are shown on the right.
The above results imply the existence of a cytoplasmic pool of endogenous β-catenin in UG1 cells that can translocate into the nucleus, bind LEF1, and activate transcription. Indeed, when examined by immunoblotting, a significant fraction of β-catenin protein was detected in Triton-soluble cell extracts in addition to that present in Triton-insoluble fractions (Fig. 5B Upper). As a control, E-cadherin proteins were detected only in the Triton-insoluble fractions (Fig. 5B Lower). Taken together, these results suggest the presence of non-cadherin-bound, “free” cytoplasmic β-catenin in these UG1 cells. Compared with cells used previously (14, 28), our cells had undergone extensive passaging, which might have led to cellular events that facilitated the accumulation of cytoplasmic β-catenin. In this context, it is interesting to note that the proliferation rate of UG1 cells increased with the number of passages (not shown). Furthermore, populations of human primary keratinocytes that display a higher proliferative potential contain higher levels of cytoplasmic β-catenin (31).
To test whether the LEF1 binding site at −372 is required for movo1 promoter activity and for activation by exogenous LEF1, we generated mutant promoters in which either two point mutations were introduced into the LEF1 binding site (Fig. 5C, P-*Lef1: CGTGGTG) that would abolish LEF1 binding (32), or a 67-bp sequence surrounding the LEF1 binding site was deleted (P-ΔLef1). Whereas P-*Lef1 displayed a >8-fold reduction in promoter activity in UG1 cells, the activity of P-ΔLef1 was reduced to almost background level. Furthermore, both mutant promoters could no longer be activated by the addition of exogenous LEF1 protein. Collectively, these results demonstrate that the LEF1/β-catenin complex activates movo1 promoter at least in part through the LEF1 binding site at −372 and implicate movo1 as a direct wnt/LEF1/β-catenin target.
Discussion
Many of the genetic events governing hair morphogenesis remain to be elucidated. What are the putative wnts that trigger proliferating matrix cells at the base of a hair follicle to differentiate into the precortex? What are the genetic determinants for precortex cells to further differentiate and give rise to the hair shaft, a structure composed of terminal cells packed with cross-linked keratin filaments? Our previous genetic studies revealed that movo1 is a player in this morphogenic process (10).
Our molecular analyses indicate that movo1 is a homolog of the Drosophila ovo/svb gene. The ovo class of genes is conserved in multicellular organisms including worms, flies, mice, and human. ovo genes in mice and flies are required for the differentiation of epidermal appendages, whereas mutations in ovo of Caenorhabditis elegans (lin-48) led to no apparent epidermal defects (33). Although studies in Drosophila suggest that dovo gene products function as transcriptional regulators, it is important to test this possibility directly for movo1 gene products to validate movo1 as a good starting point to probe downstream and upstream genetic events during hair morphogenesis. It was this interest that stimulated the analyses presented here on the biochemical functions and the regulation of movo1. The results of our studies clearly demonstrate that mOvo1a is a sequence-specific nuclear DNA binding protein, produced as hair follicle and epidermal cells undergo differentiation, suggesting that it functions at least in part by regulating gene expression required for differentiation of hair follicles and the epidermis.
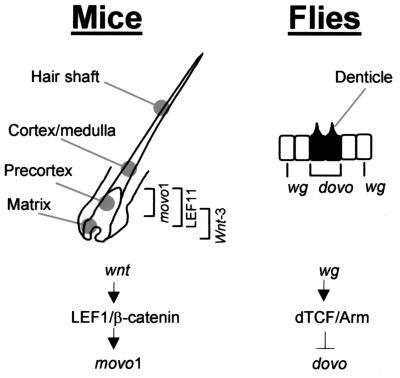
Previously, it was shown that an artificial LEF/TCF reporter promoter was activated in hair follicle precortex cells, suggesting active wnt signaling in these cells (28). Consistently, LEF1 and β-catenin proteins concentrate in the nucleus of precortex cells despite their presence in other compartments of the follicles (16, 28). Hair keratin genes have been implicated as targets of TCF/LEF in these precortex cells (13, 16, 34). Our results showing that the LEF1/β-catenin complex activates movo1 promoter, which is normally active in precortex cells, strongly suggest that movo1 is also a developmental target of activated wnt signaling pathway during hair morphogenesis (Fig. 6 Left). Further support of this notion comes from the observation of hair shaft defects in mutant mice lacking either LEF1 (12) or movo1 (10). Future work will focus on examining this LEF1/β-catenin-movo1 link in vivo.
Figure 6.
Models of wnt/wg–movo1/dovo pathways in mouse and fly epidermal appendage differentiation. (Left) Partial structure of a mouse hair follicle. Zones of cells expressing wnt-3 (11), LEF1, or movo1 are indicated by brackets. (Right) A simplified view of fly epidermis showing smooth cells (white) and denticle-producing cells (black). Cells that express wg or dovo are indicated.
In Drosophila, an activated wg signal leads to increased dTCF/arm activity in the smooth cells that do not produce denticles (Fig. 6 Right). Interestingly, in this case, the dTCF/arm complex inhibits rather than activates dovo expression and therefore prevents denticle formation. The exact location of dTCF expression in denticle-forming vs. smooth epidermal cells has not been reported. Therefore, it is unclear whether inhibition of ovo transcription by dTCF/arm is through intracellular repression (either directly or involving intermediate factors), or via a more indirect route (e.g., dTCF/arm regulates the expression of secreted factors that in turn act on smooth cells to inhibit dovo expression), albeit genetic data appear to be more consistent with the former possibility (2). A recurring theme in development is the use of a common set of molecular pathways to different effects. In our model, common regulatory pathways are used in two related developmental processes, collectively known as epidermal appendage differentiation, of different organisms to effect opposite morphological outcome.
wnt/wg signaling has been implicated both in tumorigenesis by promoting proliferation and in development by influencing cell fate determination. In mammals, several downstream targets of LEF/β-catenin-mediated wnt signaling, including c-myc (35), cyclin D1 (36), WISP (37), matrix metalloproteinase-7 (MMP7; refs. 38 and 39), TCF-1 (40), LEF1 (19), and hair-specific keratin genes have been identified. movo1 represents the first mammalian transcription factor that likely functions downstream of wnt in postmitotic components of a normal developmental process. Our studies now allow a careful dissection of potential interactions/cross-talk between wnt signaling and other genetic pathways that play a role in differentiation of the hair follicle. In this context, it is interesting to note that (i) several CAGA- or GTCT-like sequences, to which Smad proteins, intracellular signaling mediators of transforming growth factor-β (TGF-β) pathways, bind (41), are present in the movo1 promoter, (ii) LEF1 and Smad have been shown to interact and regulate gene expression in a coordinated manner to integrate wnt and TGF-β signals (42, 43), and (iii) TGF-βs are known to play important roles in hair follicle development (44–46).
Acknowledgments
We thank Rob Steele and Haoping Liu for suggestions and critiques. This work was supported by National Institutes of Health Research Grant R01 AR47320, by University of California Cancer Research Coordinating Committee (CRCC) Grants 9-555112-36685 and 9-555112-37128, and by a University of California-Irvine–American Cancer Society-Institutional Research Grant to X.D.
Abbreviations
- LEF
lymphoid enhancer factor
- TCF
T cell factor
- EMSA
electrophoretic mobility-shift assay
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF487891, AF13804, and AF13805).
References
- 1.Fuchs E. Harvey Lect. 1998;94:47–77. [PubMed] [Google Scholar]
- 2.Payre F, Vincent A, Carreno S. Nature (London) 1999;400:271–275. doi: 10.1038/22330. [DOI] [PubMed] [Google Scholar]
- 3.Wieschaus E, Nusslein-Volhard C, Jurgens G. Wilhelm Roux's Arch Dev Biol. 1984;193:296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]
- 4.Andrews J, Garcia-Estefania D, Delon I, Lu J, Mevel-Ninio M, Spierer A, Payre F, Pauli D, Oliver B. Development. 2000;127:881–892. doi: 10.1242/dev.127.4.881. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Andrews J, Pauli D, Oliver B. Dev Genes Evol. 1998;208:213–222. doi: 10.1007/s004270050175. [DOI] [PubMed] [Google Scholar]
- 6.Eastman Q, Grosschedl R. Curr Opin Cell Biol. 1999;11:233–240. doi: 10.1016/s0955-0674(99)80031-3. [DOI] [PubMed] [Google Scholar]
- 7.Sharpe C, Lawrence N, Martinez Arias A. Bioessays. 2001;23:311–318. doi: 10.1002/bies.1045. [DOI] [PubMed] [Google Scholar]
- 8.Chidambaram A, Allikmets R, Chandrasekarappa S, Guru S C, Modi W, Gerrard B, Dean M. Mamm Genome. 1997;8:950–951. doi: 10.1007/s003359900620. [DOI] [PubMed] [Google Scholar]
- 9.Masu Y, Ikeda S, Okuda-Ashitaka E, Sato E, Ito S. FEBS Lett. 1998;421:224–228. doi: 10.1016/s0014-5793(97)01567-6. [DOI] [PubMed] [Google Scholar]
- 10.Dai X, Schonbaum C, Degenstein L, Bai W, Mahowald A, Fuchs E. Genes Dev. 1998;12:3452–3463. doi: 10.1101/gad.12.21.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Millar S E, Willert K, Salinas P C, Roelink H, Nusse R, Sussman D J, Barsh G S. Dev Biol. 1999;207:133–149. doi: 10.1006/dbio.1998.9140. [DOI] [PubMed] [Google Scholar]
- 12.van Genderen C, Okamura R M, Farinas I, Quo R G, Parslow T G, Bruhn L, Grosschedl R. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 13.Zhou P, Byrne C, Jacobs J, Fuchs E. Genes Dev. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]
- 14.Gat U, DasGupta R, Degenstein L, Fuchs E. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 15.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 16.Merrill B J, Gat U, DasGupta R, Fuchs E. Genes Dev. 2001;15:1688–1705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chong S S, Pack S D, Roschke A V, Tanigami A, Carrozzo R, Smith A C, Dobyns W B, Ledbetter D H. Hum Mol Genet. 1997;6:147–155. doi: 10.1093/hmg/6.2.147. [DOI] [PubMed] [Google Scholar]
- 18.Prieve M G, Waterman M L. Mol Cell Biol. 1999;19:4503–4515. doi: 10.1128/mcb.19.6.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hovanes K, Li T W, Munguia J E, Truong T, Milovanovic T, Lawrence Marsh J, Holcombe R F, Waterman M L. Nat Genet. 2001;28:53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 20.Eustice D C, Feldman P A, Colberg-Poley A M, Buckery R M, Neubauer R H. Biotechniques. 1991;11:739–740. , 742–743. [PubMed] [Google Scholar]
- 21.Wang X, Zinkel S, Polonsky K, Fuchs E. Proc Natl Acad Sci USA. 1997;94:219–226. doi: 10.1073/pnas.94.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leask A, Rosenberg M, Vassar R, Fuchs E. Genes Dev. 1990;4:1985–1998. doi: 10.1101/gad.4.11.1985. [DOI] [PubMed] [Google Scholar]
- 23.Hulsken J, Birchmeier W, Behrens J. J Cell Biol. 1994;127:2061–2069. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenmark H, Aasland R. J Cell Sci. 1999;112:4175–4183. doi: 10.1242/jcs.112.23.4175. [DOI] [PubMed] [Google Scholar]
- 25.Aasland R, Gibson T J, Stewart A F. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 26.Lee S, Garfinkel M D. Nucleic Acids Res. 2000;28:826–834. doi: 10.1093/nar/28.3.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo K, Smale S T. Gene. 1996;182:13–22. doi: 10.1016/s0378-1119(96)00438-6. [DOI] [PubMed] [Google Scholar]
- 28.DasGupta R, Fuchs E. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 29.Bruhn L, Munnerlyn A, Grosschedl R. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 30.Mayall T P, Sheridan P L, Montminy M R, Jones K A. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 31.Zhu A J, Watt F M. Development. 1999;126:2285–2298. doi: 10.1242/dev.126.10.2285. [DOI] [PubMed] [Google Scholar]
- 32.Waterman M L, Fischer W H, Jones K A. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 33.Johnson A D, Fitzsimmons D, Hagman J, Chamberlin H M. Development. 2001;128:2857–2865. doi: 10.1242/dev.128.15.2857. [DOI] [PubMed] [Google Scholar]
- 34.Dunn S M, Keough R A, Rogers G E, Powell B C. J Cell Sci. 1998;111:3487–3496. doi: 10.1242/jcs.111.23.3487. [DOI] [PubMed] [Google Scholar]
- 35.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 36.Tetsu O, McCormick F. Nature (London) 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 37.Pennica D, Swanson T A, Welsh J W, Roy M A, Lawrence D A, Lee J, Brush J, Taneyhill L A, Deuel B, Lew M, et al. Proc Natl Acad Sci USA. 1998;95:14717–14722. doi: 10.1073/pnas.95.25.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brabletz T, Jung A, Dag S, Hlubek F, Kirchner T. Am J Pathol. 1999;155:1033–1038. doi: 10.1016/s0002-9440(10)65204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford H C, Fingleton B M, Rudolph-Owen L A, Goss K J, Rubinfeld B, Polakis P, Matrisian L M. Oncogene. 1999;18:2883–2891. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- 40.Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Derynck R. Trends Cell Biol. 1999;9:274–279. doi: 10.1016/s0962-8924(99)01579-2. [DOI] [PubMed] [Google Scholar]
- 42.Labbe E, Letamendia A, Attisano L. Proc Natl Acad Sci USA. 2000;97:8358–8363. doi: 10.1073/pnas.150152697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishita M, Hashimoto M K, Ogata S, Laurent M N, Ueno N, Shibuya H, Cho K W. Nature (London) 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 44.Blessing M, Nanney L B, King L E, Jones C M, Hogan B L. Genes Dev. 1993;7:204–215. doi: 10.1101/gad.7.2.204. [DOI] [PubMed] [Google Scholar]
- 45.Foitzik K, Lindner G, Mueller-Roever S, Maurer M, Botchkareva N, Botchkarev V, Handjiski B, Metz M, Hibino T, Soma T, et al. FASEB J. 2000;14:752–760. doi: 10.1096/fasebj.14.5.752. [DOI] [PubMed] [Google Scholar]
- 46.Foitzik K, Paus R, Doetschman T, Dotto G P. Dev Biol. 1999;212:278–289. doi: 10.1006/dbio.1999.9325. [DOI] [PubMed] [Google Scholar]