Abstract
Serogroup A Neisseria meningitidis has repeatedly caused widespread epidemics of meningitis and septicemia throughout the 20th century. Recently, in a limited collection of strains, epidemic serogroup A isolates were found to have elevated mutation rates that was caused by defects in mismatch repair pathways. To ascertain the role of these mutators in the epidemic spread of this serogroup, the prevalence of hypermutability in a collection of 95 serogroup A N. meningitidis invasive isolates was determined. Overall mutability in Neisseriae can be described by measuring both missense mutation rates as well as phase variation frequencies of “contingency loci.” Fifty-seven percent of serogroup A isolates possessed elevated mutability, which could be divided into two classes: intermediate and high level. Eleven of 20 high-level mutators, with phase variation rates >100-fold higher than wild-type isolates, were defective in mismatch repair. Ten of the 34 intermediate mutators possessing >10-fold increases in phase variation rates could be partially complemented by a wild-type mutL allele. A high prevalence of mutators in epidemic isolates indicates that hypermutability may play a major role in the transmission of this pathogen. The added diversity derived from increased phase variation rates may allow fixation of mutator alleles more frequently during epidemic spread.
Keywords: meningitis‖evolution‖mismatch repair‖transmission‖virulence
The Gram-negative bacterium Neisseria meningitidis can be isolated from up to 10% of the world population as a commensal inhabitant of the human nasopharynx (1). In rare instances, this asymptomatic carriage can progress to meningococcal disease exemplified by life threatening meningitis and/or septicemia. However, it is known that not all isolates of N. meningitidis are equally invasive. N. meningitidis can be classified into 13 serogroups, based on the antigenicity of a polysaccharide capsule (2). Over 90% of invasive disease has been caused by organisms belonging to just 5 of these serogroups (A, B, C, W-135, and Y) (3). Furthermore, within these 5 serogroups exist a few hyperinvasive lineages, which have been shown to be responsible for much of the meningococcal disease throughout the 20th century (4, 5). For instance, most of the recent endemic and epidemic disease in Western Europe and North America has been caused by lineages ET-5, A4, and ET-37, which consist of serogroups B and C organisms (5). Outbreaks of meningococcal disease caused by these organisms can have attack rates of between 5 and 50 per 100,000 (3, 6). However, the developed world before the World War II, and currently in the developing countries of Africa and Asia, isolates belonging to serogroup A N. meningitidis (menA) cause epidemic disease that can reach incidences of 1,000 per 100,000, with carriage rates as high as 30% (3, 6). These epidemics have primarily been caused by hyperinvasive lineages referred to as subgroups I, IV-1, and III. In 1967 alone, nearly three million cases of menA disease occurred in China and resulted in 166,000 deaths (7). More recently, in 1996 menA organisms caused more than 153,000 cases of meningococcal disease including over 16,000 deaths in sub-Saharan Africa (8). It appears that menA isolates are particularly prone to rapid spread through susceptible host populations, yet the specific genetic determinants for this are unknown.
Because N. meningitis can thrive only in humans, direct transmission from host to host is crucial for the maintenance of epidemics. Successful transmission may require rapid adaptation to changes encountered in the new host. N. meningitidis accomplishes this adaptation, in part, through stochastically altering surface composition by modulating the expression of 20–60 “contingency loci” throughout the genome (9, 10). Briefly, this organism has evolved short, simple DNA repeats within or near specific coding regions. The replicative instability of these repeat tracts can shift reading frames or alter promoter strength, thus oscillating gene expression in a process known as phase variation. N. meningitidis has evolved repeat tracts within genes encoding surface-exposed molecules that are in direct contact with the host environment (9, 10). Thus, N. meningitidis adapts to new challenges by means of a random mutational process that ensures survival at the population level.
Recently, N. meningitidis isolates with elevated rates of phase variation have been described (11–13). Particularly, strains defective in postreplicative mismatch repair possess phase variation rates greater than 100-fold higher than that of wild-type N. meningitidis. In a limited collection of serogroup A, B, and C isolates, mismatch repair defects were only seen in epidemic menA strains (13). A link between hypermutation and hypervirulence has been difficult to establish despite a number of investigations. Hypermutable bacterial isolates can be found in nature with frequencies of a few percent, with exceptions correlating with very specific environments (i.e., chronic lung infections of Pseudomonas aerugenosa in cystic fibrosis patients) (14–16). In general, mutators (hypermutable isolates) do not reach high frequencies in nature, probably because of the burden of excess deleterious mutations associated with high mutation rates (17). The present study investigates the prevalence of mutators in epidemic isolates of serogroup A N. meningitidis, an organism that primarily uses “directed mutations” to adapt to its natural environment. Because hyperinvasive lineages within serogroup A seem to be uniquely capable of transcontinental spread and disease, the frequency of mutators in this population could yield insight into the effects of elevated phase variation rates on the transmissibility and virulence of this organism (4, 18, 19).
Materials and Methods
Bacterial Strains and Media.
N. meningitidis isolates were cultured in GC Media (GIBCO/BRL) supplemented with 20 mM Na2CO3 and with Kellog's supplements I and II at 36.5°C. Escherichia coli strains were cultured in Luria Medium at 37°C. Antibiotic selection was performed in N. meningitidis (E. coli) by using 3 μg/ml (300 μg/ml) erythromycin, 750 μg/ml (100 μg/ml) streptomycin, 100 μg/ml (50 μg/ml) kanamycin, and 100 μg/ml (100 μg/ml) spectinomycin. Kanamycin selection in N. meningitidis was performed on Brain-Heart Infusion Agar (Difco) supplemented with 2.5% heat inactivated FBS (GIBCO/BRL).
A total of 128 serogroup A N. meningitidis isolates were chosen from a collection at the Meningitis and Special Pathogens Branch, Centers for Disease Control and Prevention, Atlanta, GA. Most serogroup A isolates can be categorized into nine subgroups based on Multilocus Enzyme Electrophoresis (MLEE) data (18). This collection of strains consists of invasive isolates from eight of the nine subgroups within the serogroup A lineage. Each strain was passaged once, then hmbR and hpuB were inactivated in transformable isolates as previously described (12). Derivatives of isolates with at least one active phase OFF Hb receptor were used for subsequent analysis. A total of 74 of the 128 clinical isolates met both of the above criteria (transformable and possessing at least one phase OFF Hb receptor). A total of 25 of the isolates were nontransformable, and the remaining 29 isolates had both Hb receptors in the phase ON configuration.
Phase OFF alleles of hmbR were used to replace alleles in 21 of the 29 transformable isolates in which both Hb receptors were phase ON. Spontaneous streptomycin (rpsL) mutants were selected in such wild-type isolates, then hmbR was inactivated by using pFLOB4300 conferring erythromycin resistance and streptomycin sensitivity (20). This insertionally inactivated allele was replaced by a phase OFF hmbR allele by transformation with XbaI-linearized pARR2055 (cloned hmbR gene of strain IR4048 containing seven consecutive G residues), and selection for streptomycin resistance followed by screening for erythromycin sensitivity. The hmbR poly(G) tracts were sequenced for verification as described (12). In total, 95 isolates could be used for analysis and are described in supporting information, which is published on the PNAS web site (www.pnas.org).
Mutant Frequency Assays.
Spontaneous mutation rates to rifampicin resistance were determined as described (13). Briefly, ≈1010 colony-forming units (cfu) were serial diluted and plated on selective media (GC media supplemented with rifampicin, 3 μg/ml). Rifampicin-resistant cfu per total viable cfu were calculated, and medians of at least three independent measurements were used for analysis. Spontaneous rifampicin resistance rates were classified as being either HIGH or LOW based initially on visual inspection. Both groups were normally distributed (P < 0.005), and the distinction between the classifications was confirmed by observation of the 95% confidence interval (HIGH = 8.8 × 10−9 to 6.0 × 10−8, LOW = 1.6 × 10−9 to 4.3 × 10−9).
Phase variation rates were similarly determined by a described method (12, 13). The medians of at least three independent measurements of Hb+ cfu per total viable cfu were used for analysis. All rates were corrected for the length of the poly(G) tract involved (see supporting information). Phenotypes of Hb receptor phase variation were assigned based on statistical analysis (see supporting information).
Determining the Effect of Repeat Tract Length on Phase Variation Frequencies.
The hmbR gene of a streptomycin resistant wild-type isolate (IR4048) and a mutL mutator isolate (IR4027) was insertionally inactivated by pFLOB4300 (ErmRrpsLS) (20). For the IR4048 background, this hmbR∷ErmRStrS allele was replaced with alleles with differing poly(G) tract length (IR2781 = g8, IR2860 = g10, IR4971 = g11, IR3277 = g13, and IR4975 = g14). In the IR4027 background, the hmbR∷ErmRStrS allele was replaced with alleles (IR4048 = g7, IR2860 = g10, IR4971 = g11, IR3277 = g13, and IR4975 = g14). hmbR phase variation frequencies were calculated as described in each engineered background (13).
Complementation of Mismatch Repair-Deficient Isolates.
MutS and MutL were used to complement any strain demonstrating a mutator phenotype (i.e., elevated phase variation and missense mutation rates). pARR2062 or pARR2063 containing a wild-type mutL or a mutS locus, respectively, was cointegrated into the chromosome of mutator isolates by means of homologous recombination (13). Full complementation was defined as restoring both phase variation rates and spontaneous rifampicin resistance rates to wild-type levels. Partial mutL complementation was defined as restoring wild-type phase variation rates without significant reductions in spontaneous rifampicin resistance frequencies. Dam methylase phenotypes were determined for all 95 isolates by digestion of 1 μg of chromosomal DNA with either DpnI or MboI for electrophoresis.
Results
Hb Receptor Phase Variation Frequencies Are Greatly Influenced by the Poly(G) Tract Length.
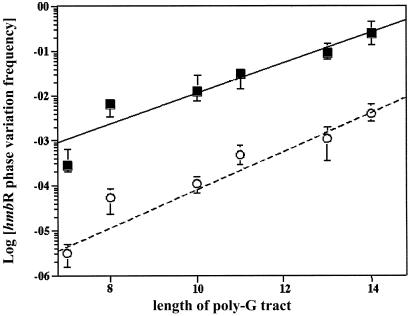
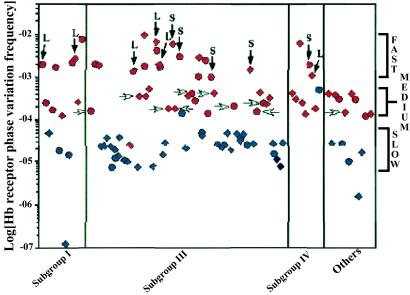
The sequence surrounding the poly(G) tracts of phase OFF hpuA and hmbR was determined for each of the 95 invasive isolates (104 phase OFF receptors in total). Although the majority (82 of 104) of tracts were between 7 and 9 consecutive G residues, many (22 of 104) had significantly longer tracts, the longest being 17 G residues (hpuA of IR4678). To determine the effects of different repeat tract lengths on phase variation rates, the hmbR gene in both a wild-type and a mutator menA isolate was replaced by alleles possessing poly(G) tracts of varying lengths (e.g., 7, 8, 10, 11, 13, or 14 consecutive G residues). Fig. 1 shows the effects of mononucleotide repeat tract length on phase variation rates in both wild-type and mutator backgrounds. Phase variation frequencies rise linearly with tract length in both backgrounds with similar slopes (mWT = 2.5, and mMutator = 2.1). However, in the mutator isolate, phase variation rates are nearly 3 orders of magnitude higher than in the wild-type strain (bWT = 9.3 × 10−9 per cfu, and bMutator = 7.7 × 10−6 per cfu). By using the relationship in Fig. 1, all Hb receptor phase variation rates were normalized by linear regression (see supporting information). Fig. 2 shows the 104 normalized receptor phase variation frequencies for all 95 isolates classified by subgroup. Three distinct classifications can be seen: SLOW, MEDIUM, and FAST, which corroborate previous results (see supporting information for definitions) (13). Most isolates (56 of 95) belong to either the FAST or MEDIUM categories, and nearly all of these isolates are global mutators (Fig. 2, red symbols, see below).
Figure 1.
The linear relationship between the length of the hmbR poly(G) tract and phase variation frequency. Alleles with varying lengths of repeat tracts were used to replace the hmbR locus in wild-type strain IR4048 (○) and a mutator isolate IR4027 (■). The least squares fit lines describing the two isolates are: yWT(x) = 2.5x + 9.3 × 10−9 (P < 0.01), and yMutator(x) = 2.1x + 7.7 × 10−6 (P < 0.01). Each data point represents the medians of at least 10 independent measurements with error bars depicting plus/minus quartiles.
Figure 2.
Distribution of corrected Hb receptor phase variation frequencies in serogroup A N. meningitidis. The phase variation rates of hmbR (diamonds) and hpuAB (circles) were linearly regressed by using the data from Fig. 1 (see supporting information). The data are divided into subgroups: epidemic subgroups I, III, and IV, as well as geographically isolated subgroups II, V, VI, VII, and VIII (OTHERS). Nearly all of the isolates with elevated Hb receptor phase variation frequencies also possess HIGH rifampicin resistance rates (red symbols), whereas strains with LOW rifampicin resistance rates (blue symbols) generally have SLOW phase variation rates. Mutator isolates that could have wild-type mutability restored by complementation with either MutS or MutL are indicated by vertical dark arrows. MEDIUM switching strains that could be partially complemented by wild-type MutL (see text) are indicated with horizontal white arrows. Each data point represents the median of at least three independent measurements.
The Majority of Isolates Possess a Mutator Phenotype.
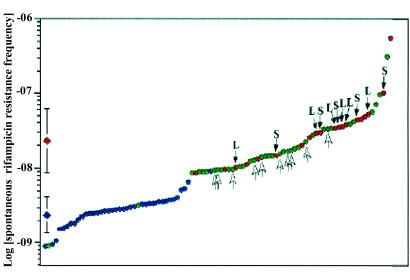
The rate of spontaneous resistance to rifampicin was determined for each of the 95 serogroup A isolates. These data could be divided into two classes: a HIGH class made up of the majority of isolates (55 of 95) with rates above 8.8 × 10−9 per cfu, and a LOW class of 40 strains possessing rifampicin resistance rates below 8.8 × 10−9 per cfu (Fig. 3). The rates of rifampicin resistance in the isolates belonging to the HIGH class were significantly greater, by nearly an order of magnitude, than those in the LOW class (meanHIGH = 2.3 × 10−8, meanLOW = 2.6 × 10−9, P < 0.01). Comparisons of missense mutation rates (rifampicin resistance) with phase variation rates (Hb utilization) revealed that the majority of isolates (57%, 54 of 95) possess elevated global mutability (both elevated phase variation and missense mutation rates). Furthermore, the rifampicin resistance rate and the phase variation rate are correlated within a given isolate. For instance, nearly all (54 of 56) isolates with elevated phase variation frequencies also possessed HIGH rifampicin resistance rates (Fig. 2). At the same time, of the 55 isolates with HIGH rifampicin resistance rates, all but 1 possessed elevated phase variation frequencies (Fig. 3).
Figure 3.
Distribution of the rates of spontaneous resistance to rifampicin in serogroup A N. meningitidis. Spontaneous rifampicin resistance rates were determined for each of the 95 menA isolates and sorted from the lowest to the highest. The division between LOW and HIGH rifampicin rates was determined to be 8.8 × 10−9 per cfu (see text). Each division is normally distributed and significantly different from the other (P < 0.01). The 95% confidence intervals about the mean for each group are shown at the left (red diamond for HIGH and blue diamond for LOW). Data points are coded for Hb receptor phase variation rate phenotypes: SLOW (blue), MEDIUM (green), and FAST (red) (see supporting information for definitions). Mutator isolates that could have wild-type mutability restored by complementation with either MutS or MutL are indicated (dark arrows). MEDIUM switching strains that could be partially complemented by wild-type MutL (see text) are indicated with white arrows. Each data point represents the median of at least three independent measurements.
Many Mutator Isolates Possess Defects in the Post Replicative Mismatch Repair System.
N. meningitidis isolates lacking a fully functional mismatch repair system have been shown to be global mutators (13). A functional copy of mutS or mutL was provided for each of the 54 established serogroup A mutator isolates in an attempt to complement the mutator phenotype. MutS was able to restore wild-type mutability in 5 mutator strains, and MutL complemented 6 isolates. All 11 mismatch repair defective strains displayed the HIGH/FAST phenotype (Figs. 2 and 3). A group of 10 MEDIUM/HIGH strains could be partially complemented by a wild-type mutL allele. In these isolates, SLOW phase variation rates could be restored by the presence of wild-type MutL, but the HIGH spontaneous rifampicin resistance rates did not change significantly (Figs. 2 and 3). Finally, 33 global mutators could not be complemented by either MutS or MutL (9 FAST/HIGH isolates and 24 MEDIUM/HIGH isolates). Clearly more than one pathway exists which generate mutator phenotypes in N. meningitidis. Specifically, 39% (21 of 54) of mutator phenotypes are linked to defects in the neisserial DNA mismatch repair system. Conflicting results have been reported on the role of Dam methylase in the neisserial mismatch repair system (11, 13). All 95 menA isolates in this collection lacked Dam activity, including the 40 isolates displaying wild-type mutability (data not shown).
Discussion
Serogroup A N. meningitidis has caused the major epidemics of meningococcal disease throughout the 20th century (3, 6). Particularly, a group of closely related menA isolates (subgroup III) has caused the pandemic spread of disease through Asia, Africa, Northern Europe, and South America (6, 19). MenA organisms generally account for ≈90% of disease during these epidemics despite the underlying presence of endemic nonserogroup A cases (3, 6). Indeed, of the five major disease-causing N. meningitidis serogroups, menA seems to exploit susceptible human populations to the greatest extent. The source of menA epidemic potential is not known; however, it is clear that these organisms must rapidly adapt to changes that occur on transmission into a new host. The large number of phase-variable surface antigens in the N. meningitidis genome no doubt aids in this adaptation process. However, both sequenced N. meningitidis genomes (serogroup B isolate MC58 and serogroup A isolate Z2491) possess between 20 and 60 simple repeats that may be substrates for phase variation (10, 21, 22). In fact, the serogroup B isolate, MC58, appears to contain more putative phase-variable genes than the serogroup A Z2491 (10, 21, 22). Therefore, merely possessing an extensive repertoire of phase-variable antigens cannot account for the unique epidemic nature of serogroup A organisms. Rather, elevation of phase variation rates might increase the epidemic potential of a strain by allowing for even more heterogeneity within a finite inoculum. This heterogeneity would enhance the adaptability of an organism living under dynamic selection such as successive transmissions into new hosts.
This study demonstrates that the majority (57% 54 of 95) of menA isolates display elevated mutability. Two distinct classes of mutators emerged from this collection of strains: intermediate (MEDIUM/HIGH) and high (FAST/HIGH) level mutators. Intermediate level mutators are the larger of the two classes, and have increased phase variation rates of the same magnitude as missense mutation rates, ≈10-fold. Ten of the 34 intermediate-level mutator isolates could be partially complemented by a wild-type mutL allele restoring SLOW phase variation rates while leaving missense mutation frequencies unchanged (HIGH). The molecular mechanism behind this partial complementation is not understood. High-level mutators have a larger increase in phase variation frequencies than in other types of mutations (>100-fold increase in phase variation frequencies compared with ≈10-fold increase in spontaneous rifampicin resistance rates). The majority of isolates in this category (11 of 20) possessed defects in mismatch repair. These types of mutators would benefit from a large increase in phase variation rate, and would suffer a relatively smaller increase in deleterious mutation accumulation. Furthermore, it has been shown that defects in mismatch repair lead to higher rates of recombination (23). Therefore, mismatch repair-deficient isolates may provide an evolutionary “jump start” to this species by creating extensive diversity through increased rates of recombination as well as generation of beneficial mutations. The fact that the majority of mutators (24 medium level and 10 high level) could not be complemented by either wild-type MutS or MutL indicates that all factors which can influence mutability in Neisseriae have not yet been elucidated.
It is possible that mutators in N. meningitidis provide the “high speed” adaptation required for the maintenance and spread of epidemic disease. The increased variability within a limited population of a mutator clone might increase the probability of establishing successful colonization in a new host. Thus, the successive transmission events during epidemic spread of N. meningitidis may select for clones with hypermutability, creating a bias for mutators in this collection of isolates. Indeed, it seems that not all disease isolates are equally likely to be mutators. Twenty-five percent of isolates from major epidemic/pandemic meningococcal disease (subgroups I, III, and IV) were high-level mutators, whereas none of the high-level mutators in this collection came from patients in nonpandemic disease (Fig. 2; P = 0.036, Fisher's Exact test). In addition, patients from the ends of the subgroup III pandemics seem to be more often infected with mutators than individuals from China, the origin of all three pandemics, indicating that mutators are enriched for over time during epidemic spread (data not shown). These data suggest that our collection of epidemic disease isolates may be biased toward high mutator prevalences caused by the increased transmissibility that mutators may possess.
An alternative view for the high menA mutator prevalence is that the selection for mutators occurs at the level of each individual host. Because N. meningitidis is primarily a commensal inhabitant of the nasopharynx, disease organisms isolated from blood or cerebrospinal fluid would have had to exist in highly selective and unusual environments. These clones would have benefited from the extra diversity provided by high frequency phase variation and, therefore, a collection of invasive isolates might be biased toward mutators. There is evidence for this bias in our study through analysis of subgroup III pandemics. Mutators within single pandemics of subgroup III arise because of mutations in different genes, namely mutS or mutL (see supporting information). Moreover, two independent mutS mutators from the first subgroup III pandemic have different mutations in otherwise identical alleles, an ATG → TTG in IR4561 and a single base pair insertion in a run of four C residues in IR4562. These data indicate that pandemics are not caused by single mutator clones sweeping through the population, but rather mutators arise independently in the N. meningitidis populations colonizing individual hosts. Thus, two invasive isolates from the same epidemic need not possess the same mutator genotype.
Clearly the two hypotheses, selection at the level of the host population vs. the individual, are not mutually exclusive. The increased transmission efficiency afforded mutator clones may lead to their fixation in the host population. In addition, these mutators may also prove to be more invasive. Therefore, the high prevalence of mutators in epidemic serogroup A isolates might then be attributed to bias from two sources: invasive isolates may have high mutator prevalences because of increased virulence, and epidemic isolates may have an over representation of mutators because of increased transmissibility. Comparison of results from this work with data from collections of non-disease causing “carriage” strains, or with other serogroups that are less prone to cause pandemic disease would be helpful in determining the relationship between these two sources of selection for mutator phenotypes. This work demonstrates that mutator phenotypes play an important role in the evolution of serogroup A N. meningitidis. A general link between hypermutability and hyperinvasiveness has not been clearly established in any organism to date (14–16). Here we investigate mutator isolates of a mucosal “pathogen” that uses phase variation, a form of “directed mutation”, rather than extensive environmental gene regulation to adapt to new environments. It may be that in the evolution of these types of pathogens (i.e., N. meningitidis, Neisseria gonorrhoeae, Haemophilus influenzae, Helicobacter pylori, Campylobacter jejuni) mutators play pivotal roles which have yet to be fully understood.
Supplementary Material
Acknowledgments
We thank Andrea S. Robertson for statistical analysis and Drs B. Levin, L. Ancel, M. Achtman, and I. Matic for providing strains, invaluable insight, and suggestions. This work was supported by the Public Health Service Grant AI42870–01A1 and National Institutes of Health Training Grant 2T32 AI07470 (to A.R.R.).
Abbreviation
- menA
serogroup A N. meningitidis.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Stephens D S. Lancet. 1999;353:941–942. doi: 10.1016/S0140-6736(98)00279-7. [DOI] [PubMed] [Google Scholar]
- 2.Vedros N A. In: Evolution of Meningococcal Disease. Vedros N A, editor. Vol. 2. Boca Raton, FL: CRC Press; 1987. pp. 20–32. [Google Scholar]
- 3.Peltola H. Rev Infect Dis. 1983;5:71–91. doi: 10.1093/clinids/5.1.71. [DOI] [PubMed] [Google Scholar]
- 4.Maiden M C J, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, et al. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achtman M. In: Meningococcal Disease. Cartwright K, editor. New York: Wiley; 1995. pp. 159–175. [Google Scholar]
- 6.Caugant D A. APMIS. 1998;106:505–525. [PubMed] [Google Scholar]
- 7.Zhen H. In: Evolution of Meningococcal Disease. Vedros N A, editor. Vol. 2. Boca Raton, FL: CRC; 1987. pp. 19–32. [Google Scholar]
- 8.Tikhomirov E, Santamaria M, Esteves K. World Health Stat Q. 1997;50:170–177. [PubMed] [Google Scholar]
- 9.Moxon E R, Rainey P B, Nowak M A, Lenski R E. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 10.Saunders N J, Jeffries A C, Peden J F, Hood D W, Tettelin H, Rappuoli R, Moxon E R. Mol Microbiol. 2000;37:207–215. doi: 10.1046/j.1365-2958.2000.02000.x. [DOI] [PubMed] [Google Scholar]
- 11.Bucci C, Lavitola A, Salvatore P, Del Giudice L, Massardo D R, Bruni C B, Alifano P. Mol Cell. 1999;3:435–345. doi: 10.1016/s1097-2765(00)80471-2. [DOI] [PubMed] [Google Scholar]
- 12.Richardson A R, Stojiljkovic I. J Bacteriol. 1999;181:2067–2074. doi: 10.1128/jb.181.7.2067-2074.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson A R, Stojiljkovic I. Mol Microbiol. 2001;40:645–655. doi: 10.1046/j.1365-2958.2001.02408.x. [DOI] [PubMed] [Google Scholar]
- 14.LeClerc E J, Baoguang L, Payne W L, Cebula T A. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 15.Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Denamur E, Elion J. Science. 1997;277:1833–1834. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- 16.Oliver A, Cantón R, Campo P, Baquero F, Blazquez J. Science. 2000;288:1251–1253. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 17.Dawson K J. Theor Popul Biol. 1999;55:1–22. doi: 10.1006/tpbi.1998.1375. [DOI] [PubMed] [Google Scholar]
- 18.Wang J-F, Caugant D A, Li X, Hu X, Poolman J T, Crowe B A, Achtman M. Infect Immun. 1992;60:5267–5282. doi: 10.1128/iai.60.12.5267-5282.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu P, van der Ende A, Falush D, Brieske N, Morelli G, Linz B, Popovic T, Schuurman I G A, Adegbola R A, Zurth K, et al. Proc Natl Acad Sci USA. 2001;98:5234–5239. doi: 10.1073/pnas.061386098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston D M, Cannon J G. Gene. 1999;236:179–184. doi: 10.1016/s0378-1119(99)00238-3. [DOI] [PubMed] [Google Scholar]
- 21.Parkhill J, Achtman M, James K D, Bentley S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, et al. Nature (London) 2000;400:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 22.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K E, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, et al. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 23.Matic I, Rayssiguier C, Radman M. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.