Abstract
Comparative phylogeography has proved useful for investigating biological responses to past climate change and is strongest when combined with extrinsic hypotheses derived from the fossil record or geology. However, the rarity of species with sufficient, spatially explicit fossil evidence restricts the application of this method. Here, we develop an alternative approach in which spatial models of predicted species distributions under serial paleoclimates are compared with a molecular phylogeography, in this case for a snail endemic to the rainforests of North Queensland, Australia. We also compare the phylogeography of the snail to those from several endemic vertebrates and use consilience across all of these approaches to enhance biogeographical inference for this rainforest fauna. The snail mtDNA phylogeography is consistent with predictions from paleoclimate modeling in relation to the location and size of climatic refugia through the late Pleistocene-Holocene and broad patterns of extinction and recolonization. There is general agreement between quantitative estimates of population expansion from sequence data (using likelihood and coalescent methods) vs. distributional modeling. The snail phylogeography represents a composite of both common and idiosyncratic patterns seen among vertebrates, reflecting the geographically finer scale of persistence and subdivision in the snail. In general, this multifaceted approach, combining spatially explicit paleoclimatological models and comparative phylogeography, provides a powerful approach to locating historical refugia and understanding species' responses to them.
Phylogeography seeks to reveal biogeographical history of species and the habitats they occupy via (i) qualitative spatial association of divisions between monophyletic clusters of alleles with biogeographic barriers, and (ii) quantitative estimates of historical population size (1–4). Much of this work has focused on mitochondrial DNA; however, stochastic variance limits our confidence in reconstructions of history from a single gene. One approach solving this limitation is to sample more genes (5). A more common approach is comparative phylogeography (6) in which sequence variation is surveyed at a single gene for multiple species across the same landscape. A limitation here is that histories of local extinction and recolonization may vary among species despite a common history of habitat fluctuation.
To improve inference of historical biogeography, we need to incorporate spatially explicit evidence from paleoecology into interpretation of species' phylogeography. Some recent studies have promoted the use of fossil evidence along with phylogeography to estimate historical distributions (7) or have examined sequence variation in the fossils themselves (e.g., refs. 8 and 9) However, appropriate fossils are sparse or nonexistent for most taxa. We explore a novel and more widely applicable approach that uses paleoclimatological models of species' distributions in conjunction with phylogeography.
Bioclimatic modeling (10) predicts potential distributions for species by deriving an environmental envelope from known distribution points and projecting this onto a spatially interpolated climate surface of an area. For paleomodeling, the same climate envelope for a species is mapped onto inferred paleoclimatic surfaces, these being based on analysis of local paleopalynology or other indices (11, 12). This method assumes that the species' physiological limits are constant over the time period concerned, an assumption that may not hold in some cases (8, 13). This approach is therefore most likely to be effective for species with climatically defined range limits in landscapes with steep climatic gradients, so that errors because of physiological evolution or from estimates of paleoclimate have less effect on predicted spatial distribution. These conditions hold for the fauna endemic to the Wet Tropics rainforests of north east Queensland, Australia.
The Wet Tropics (WT; Fig. 1) is the largest and most intact remnant of Gondwanan-derived wet forests that dominated the continent of Australia to the mid Miocene, declining in the Tertiary to its current state of fragmentation and isolation (12, 14, 15). This long-term decline, and particularly the Pleistocene climate oscillations, appears to have driven cycles of contraction and expansion of rainforest, probably structured around montane remnants surrounded by dry sclerophyll woodlands (16). These montane blocks are centers of endemism and have been proposed as Pleistocene refugia (17–19), and form the basis of the recognized subregions within the Wet Tropics (see Table 1 and Fig. 1). However, analysis of subfossil charcoal records have cast doubt on the integrity and existence of several putative refugia (20).
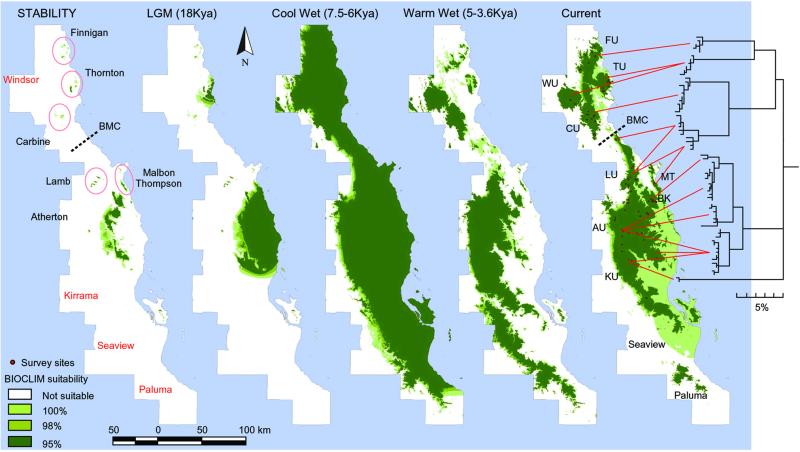
Figure 1.
BIOCLIM distribution models with mtDNA haplotype phylogeny, showing the geographical association of the major clades. Region names to the left, with codes used throughout on the right. Light to dark shades of suitability denote models by using respectively 100, 98, and 95% observed distribution climate parameter limits. Distribution sites for modeling shown in red. Eastern boundary is current coastline; the remainder, spatial data limits. Sequence divergence scale under phylogeny. Right to left: current climate model; then the three paleoclimate models warm-wet, cool-wet, and LGM; far left, the STABILITY surface—intersection of the other four models. On the STABILITY surface refuges for FU, TU, CU, LU, and MT are circled, and areas going extinct are named in red. The central part of the WT is shown again in Fig. 2, in more detail.
Table 1.
Definition and attributes of nine Wet Tropics subregions
| Region code | FU | TU | WU | CU | LU* | BK† | AU‡ | KU | MT |
|---|---|---|---|---|---|---|---|---|---|
| Nucleotide diversity (π) | |||||||||
| G. bellendenkerensis | 0.54 | 0.43 | 0.46 | 0.60 | 1.86 | 1.71 | 3.29 | 0.09 | 0.40 |
| C. laevis | 0.12 | 0.11 | 0.11 | 0.44 | 0.47 | 0.27 | 0.37 | 0.14 | 0.00 |
| G. queenslandicus | 0.71 | 0.26 | 0.19 | 0.19 | 0.46 | 0.00 | 0.13 | 0.16 | 0.43 |
| Vertebrate average π§ | 0.36 | 0.12 | 0.13 | 0.25 | 0.33 | 0.24 | 0.19 | 0.24 | 0.10 |
| C.l.-G.q. average π¶ | 0.42 | 0.19 | 0.15 | 0.32 | 0.47 | 0.14 | 0.25 | 0.15 | 0.22 |
| Snail/vert. π ratio | 1.30 | 2.34 | 3.07 | 1.91 | 4.01 | 12.67 | 13.15 | 0.59 | 1.88 |
| Rainforest area‖ | |||||||||
| Recent | 28,090 | 23,770 | 25,620 | 39,430 | 20,824 | 29,260 | 170,489 | 59,093 | 6,192 |
| LGM | 2,399 | 31,504 | 0 | 1,869 | 2,034 | 29,260 | 334,099 | 27 | 6,192 |
| STABILITY | 2,400 | 3,121 | 0 | 1,870 | 2,034 | 26,957 | 76,975 | 27 | 4,484 |
| Sampling | |||||||||
| Genetics | |||||||||
| Individuals | 7 | 8 | 2 | 15 | 10 | 10 | 57 | 8 | 4 |
| Haplotypes | 5 | 5 | 2 | 10 | 7 | 5 | 30 | 2 | 3 |
| Locations | 4 | 4 | 2 | 6 | 6 | 5 | 20 | 3 | 3 |
| Modeling | |||||||||
| Locations | 10 | 13 | 3 | 14 | 12 | 15 | 29 | 3 | 3 |
Includes Mcallister Range.
Bellenden Ker and Bartle Frere.
Includes Herberton and Walter Hill Ranges, and BK.
Average of the six species in ref. 22.
Average of C. laevis and G. queenslandiae.
BIOCLIM model, hectares.
Congruence among vertebrate phylogeographies across these subregions support long-term restriction of rainforest-dependent fauna to at least two long-standing refugia, separated by the Black Mountain Corridor (BMC), across which rainforest was absent until ≈8 thousand years ago (Kya; refs. 21 and 22; Fig. 1). In addition, some vertebrate species show idiosyncratic disjunctions, perhaps indicating additional refugia in which these species persisted whereas the others did not (22). A more detailed record of historical rainforest habitat structuring might be preserved in the genetic pattern of taxa showing finer spatial scales of persistence and low vagility (6, 7). Such characteristics make land snails good candidates for recovering details of historical biogeography (23, 24).
Here, we use paleoclimatological modeling of the endemic land snail, Gnarosophia bellendenkerensis (Brazier 1875) to predict the location of refugia and to estimate the magnitude of change in the extent of suitable habitat in each biogeographic subregion and then use a mtDNA phylogeography of the snail to evaluate these predictions. We also compare the snail phylogeography to those reported previously for rainforest-restricted vertebrates endemic to the wet tropics (22, 25). The WT paleoclimates and modeling cover the last glacial maximum-Holocene period (last 20 thousand years). The palynological record also shows similar fluctuations back at least 100 Kya (16), whereas the vertebrate data suggests much older refugial patterns, more in keeping with Pliocene ages. Therefore, the modeling should allow direct quantitative comparison to within population genetic patterns (at the tips of the genealogy) and qualitative comparison to older spatial patterns. In particular, we are interested in evidence of multiple Pleistocene refugia and Holocene patterns of expansion from these.
Materials and Methods
Species and Modeling.
G. bellendenkerensis is a moderately large (height = 33.08 mm, width = 40.19 mm, n = 97 adults) globose Camaenid land snail endemic to Wet Tropics rainforests. It is a leaf litter/log generalist and occurs predominantly in the upland (above 400 m) mesothermal rainforests extending from the Finnegan uplands (FU) in the north (15° 43′ 10′′ S) to the end of the Kirrama uplands (KU) in the south (18° 14′ 6′′ S; Fig. 1). We used either specimens from the Queensland Museum (stored in 70% ethanol) or live-caught animals. The museum database and field collections provide 102 records, accurate to 3 decimal degree places, for distribution modeling. Extensive molecular systematic surveys by the authors (unpublished data) show G. bellendenkerensis to be monophyletic. G. bellendenkerensis has recently been removed from the genus Hadra (26). Our definition of geographic subregions within the Wet Tropics (Table 1) follows previous biogeographical studies (18, 19, 21, 22, 27).
Climate-based distribution modeling followed the BIOCLIM procedure (10) and used spatially interpolated estimates (anuclim, ref. 28) of annual mean temperature (AMT), annual mean precipitation (AMP) and precipitation of the driest quarter (PDQ) on an 80-m resolution digital elevation model for the wet tropics. These layers are the only ones for which paleoclimate scenarios are available; however, they are adequate to provide a good fit to current rainforest (12) and snail distributions (see below). The BIOCLIM procedure sets upper and lower boundary limits for each climate layer based on the upper and lower limits of the observed distribution points, allowing for trimming of outliers. Because the current distribution of the snail reaches the extremes of minimum temperature and maximum rainfall found in the wet tropics, we considered that these limits may not be intrinsic to the snail and therefore did not invoke these boundaries in the paleomodels. Boundary limits (100%) for the snail BIOCLIM model are AMT of 24°C (unused min. 15.5°), AMP of 1,398 mm (unused max. 6644 mm), and PDQ of 83 mm (unused max. 672 mm). Modeled distributions are restricted to the current coastline by the available digital elevation model. Whereas some refugia could have extended further east, the coastal platform was more likely gallery forest/savanna when exposed (29).
Paleoclimates have been estimated for the WT for late Pleistocene through the Holocene by examining overlap of current bioclimatic limits for species abundant in the respective sections of pollen cores (11, 12); these climate estimates have been used recently to model geographic shifts in vegetation types across the wet tropics (30). We predicted potential distributions of the snail for the three paleoclimates, corresponding to a cool-dry climate at the last glacial maximum (LGM, e.g., 18 Kya), a cool-wet period extending from ≈8–6 Kya, and warm-wet periods from about 5–3.6 Kya. Estimated shifts of the three climate parameters relative to present are as follows: LGM AMT −3.5°C, AMP 50%, PDQ 60%; Cool-Wet AMT −2.0°C, AMP 120%, PDQ 200%; and Warm-Wet AMT +2.0°C, AMP 125%, PDQ 150%. The STABILITY surface represents the intersection of all scenarios: presence in the current and three paleoclimate models. Areas included in the STABILITY surface therefore represent predicted refugia. For quantitative comparison with genetic estimates of expansion, we used the ratio of areas within the STABILITY surface vs. the CURRENT model as surrogates for historical fluctuation in population size.
Molecular Analyses.
For most specimens, DNA was extracted by using the Chelex (Bio-Rad) method. All specimens were sequenced by Applied Biosystems automated sequencing, most with both strands. The final dataset used here comprises 121 individuals from 53 localities scored for 460 bases of COII giving 67 haplotypes. COII primers are coding strand: 5′-AAA TAA TGC TAT TTC ATG AYC AYG C-3′; H strand: 5′-GCT CCG CAA ATC TCT GAR CAY TG-3′. The snail sequences are lodged with GenBank, accession nos. AY048376–AY048422.
All analyses used the Tamura-Nei model to estimate sequence divergence. A phylogeny of haplotypes was inferred with paup* (31) by Neighbor-Joining, and nucleotide diversity and net nucleotide divergences within and among populations estimated with reap (32). Simultaneous optimization and estimation of likelihood surfaces for theta and population growth rate parameters were done with the Metropolis-Hastings Markov Chain Monte Carlo genealogy sampler fluctuate of Kuhner et al. (33). Number and length of chains run in the Metropolis-Hastings sampler accorded with guidelines by using a maximum likelihood starting tree. Quantification of population size and size change from theta and growth parameters estimated from sequence data requires estimates of mutation rate and time. The relationship of effective population size (Ne), diversity (Θ), growth parameter (g), time (t) in generations, and mutation rate μ per generation is given by Ne = (Θ/2μ)e(−gμt) (33).
Results
BIOCLIM Modeling of Current and Paleodistributions.
The bioclimatic modeling of the snail (Figs. 1 and 2) predicts a substantial reduction in range during the arid and cool conditions of the LGM, particularly south of the Atherton Uplands (AU), and severe fragmentation north of AU. Specifically, it predicts small discrete refugia for three northern subregions, Finnigan (FU), Thornton (TU), and Carbine (CU) Uplands, another on the central coast (Malbon-Thompson Range, MT), a major refugial area across the eastern AU and adjacent coast, and smaller areas just to the north (Lamb Uplands, LU) and the west at Mt Fisher (see Fig. 2 for additional detail). Conversely, the LGM model predicts the snail to be absent from the Windsor Uplands (WU) and nearly so from the KU. The modeled range is maximally expanded through the cool-wet period of the early Holocene (6–7.5 Kya), highlighting the potential for recolonization of all areas. During the warmer wetter period of the late Holocene (3.6–5 Kya), the modeling predicts further contractions, especially from the lowlands and eastern uplands (e.g., TU and AU in Fig. 1). Overall, these predicted ranges are very similar to fluctuations in mesothermal rainforest predicted by using similar methods (refs. 12 and 30). The STABILITY surface is essentially the LGM model with the Thornton and Atherton uplands (TU and AU) reduced by limitations of the late Holocene warm-wet scenario. These models predict changes in range across subregions varying from relatively minor to drastic [e.g., BK (Bellenden Ker and Bartle Frere) vs. others in Table 1].
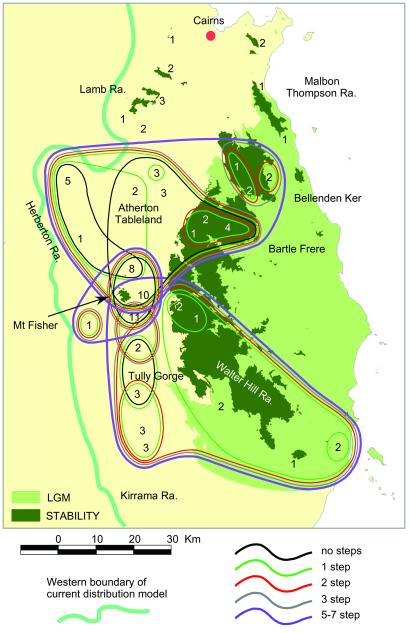
Figure 2.
MtDNA nested clade map of the Atherton Tableland and adjacent regions overlaying BIOCLIM LGM and STABILITY paleodistribution models (light and dark shades, respectively). Drawn for haplotypes shared across sites, sample sizes indicated for localities. The three 5- to 7-step clades (outermost lines) require another 8 steps to coalesce. Broad band to left indicates western boundary of current predicted distribution.
Molecular Phylogeography and Comparison with Bioclimate Models.
The 67 haplotypes detected fell into six divergent and geographically exclusive lineages; four with low and unstructured diversity (FU, TU/WU, CU, KU), one structured into two sub lineages (LU/MT), and the last a large complex (AU) of three sub lineages (Table 1, Fig. 1; all bootstraps >90%). Maximum sequence divergence among lineages was 12–15% and net divergence (Da) among areas (summarized in Fig. 3) ranged from 6.5% to 15.4% with average of 10.7%. These six major lineages showed almost complete geographical restriction to a single biogeographic subregion. Exceptions were identical or closely related (one step) haplotypes shared between WU and TU, AU and BK, and MT and LU.
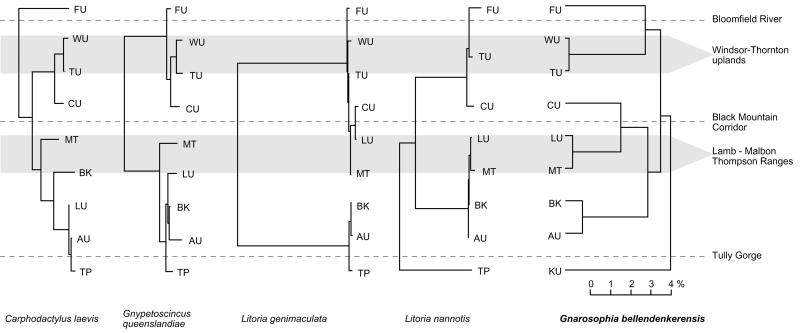
Figure 3.
Neighbor-joining trees of mtDNA net nucleotide divergence (Da) for snail and four vertebrate species, all drawn to the same nucleotide divergence scale, Tamura-Nei model. Snail; COII gene, lizards Carphodactylus and Gnypetoscincus: Cyt b, and Litoria frogs: COI. The southern subregion defined here as Kirrima Uplands (KU; Fig. 1) is to be compared with TP of Schneider et al. (22). L. nannotis not sampled in WU. Relationships between WU-TU and between LU-MT highlighted, dashed lines mark major genetic breaks in the vertebrates.
This phylogeographic pattern can be compared qualitatively with the predictions of the paleomodeling: each of the predicted discrete LGM/STABILITY refugia of FU, TU, CU, LU, and AU hosted a discrete genetic lineage, with the latter showing greatest internal complexity (see below). The absence of a distinct genetic lineage for the WU matches the predicted elimination of suitable environment at the LGM; rather, the WU samples were a subset of the adjacent TU lineage, suggesting colonization from that source. However, the LU samples included, in addition to a locally endemic lineage, another shared with the adjacent coastal refuge (MT). The modeling also failed to clearly predict a refuge to match the highly divergent lineage found south of the Tully Gorge (KU, Table 1).
The relatively large AU-BK region is the best sampled for both genetic and spatial modeling analyses. Fig. 2 shows the geographical spread of nested haplotype clades superimposed on the LGM model and STABILITY surface (100% bounds). Again, the combination of molecular data and paleomodeling is highly informative. Key features are as follows: (i) the northern and southern limits of the AU/BK area represent contact zones with divergent lineages; (ii) several haplotypes are widespread (i.e., spread across at least half the area); (iii) there is substantial diversity in (western) areas predicted to be largely unsuitable at LGM but occupied in later phases; and (iv) diversity is structured into three sublineages (with bootstrap support >90%), one widespread with closely related haplotypes (0–1 step) across the Atherton Tableland (AT) and Bartle Frere and a sister group confined to Bellenden Ker, a second widespread with closely related haplotypes (1 step) across the Walter Hill Range and adjacent western areas, and the third confined to the western AT and centered around Mt. Fisher. Thus, the range of each lineage includes at least one predicted refuge (which equals STABILITY areas in Fig. 2), and all three lineages overlap in the vicinity of the predicted Mt. Fisher refuge.
For most regions, nucleotide diversity is low (π < 0.6%; Table 1), three exceptions being LU, BK, and AU, each of which is structured into multiple sublineages. The AU region includes the three sublineages discussed above. The BK region is separated in the STABILITY surface into two areas corresponding to the two highest mountains in Queensland—Bartle Frere and Bellenden Ker, each of which hosts a separate three-step clade (Fig. 2). These observations indicate that the high diversity in each of these areas reflects admixture from separate refugia. The same reasoning can be applied to LU, which hosts two lineages, one shared with MT (another predicted refuge).
The distribution modeling predicts population expansion for some regions, defined as the area change between STABILITY and current models. This ratio can be compared with signals of population change in the genetic data, defined as the likelihood estimates of the exponential growth parameter, g (33). Only six of the nine subregions had sample sizes (n > 7) adequate for this purpose, including a sublineage of AU extending from Mt. Bartle Frere across the Atherton Tableland (AT-BF; see Fig. 3). This subset of the AU samples was included to represent a presumed expansion from a single source in keeping with the assumptions of the analysis. Table 2 gives the combined optimal Θ and g estimates, and likelihood support for g > 0.
Table 2.
Optimal g/θ values and ΔlnL support for g > 0
| Region | Area ratio* | θ | g | ΔlnL |
|---|---|---|---|---|
| AU | 2.2 | 0.054 | 34 | 0.7 |
| TU | 7.6 | 0.018 | 611 | 1.0 |
| LU | 10.2 | 0.023 | 105 | 0.6 |
| FU | 11.7 | 0.046 | 934 | 1.1 |
| AT-BF† | 14.5 | 0.056 | 2266 | 13.1 |
| CU | 21.1 | 0.050 | 1639 | 4.9 |
Ratio of modeled current distribution to STABILITY surface (100%).
Bartle Frere-Atherton Tableland clade, see Fig. 2.
There is a reasonable correlation across subregions between area ratio and estimates of g relationship (r = 0.76; P = 0.04): subregions with a large area increase also show substantial g, except LU being low. Likelihood ratio tests reject constant population size for CU and BF-AT. For CU, we get a relationship of g = 1,639 to area ratio = 21.1, with an overall area ratio/g ≈ 20/2000. Similar results apply by using the LGM areas. Snail effective generation time of about one year (34)—the LGM time frame and clear subfossil evidence of rainforest expansion by 8 Kya (22)—suggests a time scale for expansion of the order of 10–20,000 t. For the genetic growth to quantitatively match the amount of spatial growth then requires a μ of around 0.7–1.3 × 10−7, between phylogenetic and within population estimates of mtDNA mutation rate (26, 35). We note that some areas included here deviate from the assumptions of the coalescent expansion model in having substantial substructure (AU) or possible admixture (LU), and this result may underly the observed underestimate of g relative to the values predicted from the paleomodeling.
Comparison of Snail and Vertebrate Phylogeography.
Geographic patterns of molecular diversity for the snail and some previously studied rainforest-restricted lizards and frogs from the wet tropics (22, 25) are summarized in Table 1 and the Da area phenograms in Fig. 3. The snail phylogeography is essentially a composite of the patterns for individual vertebrate species, reflecting idiosyncratic as well as common genetic breaks among the former, as well as admixture zones and inferred patterns of recolonization.
Proceeding from north to south, the divergence between FU and TU in the snail reflects the pattern seen in the gecko Carphodactylus laevis, and to a lesser extent, the skink Gnypetoscincus queenslandiae. The phylogeographic connection between WU and TU in the snail is common to all vertebrate species sampled from both areas (Fig. 3) where distinction allows. For the coastal MT range, vertebrates were a mixture of endemic (C. laevis, G. queenslansdiae) and immigrant lineages, the latter derived from either the north (Litoria genimaculata, Carlia rubrigularis) or the south (Litoria nannotis). The particular MT-LU connection seen in the snail occurs also in C. rubrigularis and L. genimaculata. In the center of the range, the association between AU and the adjacent BK is present in all of the vertebrates assayed except for C. laevis (Fig. 3), and the substructuring within AU-BK (Fig. 3) reflects patterns of mtDNA diversity within G. queenslandiae (36). At the southern limit of the snail, the divergent KU lineage reflects the pattern seen in L. nannotis.
Zones of admixture between major phylogeographic lineages of the snail have been observed at sites on the northern Atherton Tableland (AU/LU) and also near the Tully Gorge (KU/AU clades). Each of these corresponds to admixture zones seen in vertebrates, the former region having secondary contacts between major northern and southern lineages for the skink C. rubigularis, the frog L. genimaculata, and the bettong Bettongia tropica (ref. 37; B. Phillips, unpublished data) and the latter for the frog L. nannotis (ref. 22; M. Cunningham, unpublished data).
Turning to quantitative estimates, we compared nucleotide diversity (π) in the snail with that in the two most highly structured lizards (C. laevis and G. queenslandiae) as well as the average for all frogs and lizards (Table 1). In most areas, diversity in the snail is about 2- to 3-fold higher than vertebrates. Taking CU as a discrete area with the best individual estimates of π for both vertebrates and snails, we get 0.60% for the snail and 0.25–0.32% for vertebrate averages, a 2-fold difference as expected for mtDNA Ne in a hermaphrodite. Relative to vertebrates, snail diversity is exceptionally high in BK and AU. For each of these areas, the comparison of modeling and phylogeography suggests admixture from multiple refugia in the snail. Across four common and major phylogeographic divisions (FU-TU, LU-CU, AU-KU, and AU-CU) estimates of net sequence divergence (Da) for the snail and the more highly subdivided vertebrates are comparable (Fig. 3): snail Da ranges from 6.5% to 15.4% (avg. 9.7%), vertebrates from 4.2% to 14.8% (avg. 7.9%). Overall, the similarity of snail and vertebrate divergences and diversities suggests similar substitution rates, rather than the 10- to 20-fold higher rate postulated recently (38–40).
Discussion
Two concerns for comparative phylogeography are stochastic variance of loci, the “gene tree-species tree” problem, and differing responses among taxa because of varying habitat requirements or vagility. Modeling of paleodistributions may help to address both of these issues and can provide spatially explicit hypotheses about historical distributions for taxa lacking evidence from fossils.
In most natural environments, current patterns result from an amalgam of historical and current processes. For the WT fauna, extensive fluctuations in rainforest area and connectivity have led to episodes of differential contraction and extinction, followed by range expansion and recolonization, from multiple refugia. Paleomodeling can provide the explicit spatial hypotheses needed to untangle such complex histories and resultant phylogeographies. In this sense, paleomodeling provides an alternative to gathering more genetic data (i.e., more loci), and one that provides an independent demarcation of historical population structure. The WT provides an optimal testbed for this approach because species distributions are structured by steep environmental gradients into discrete geographic zones (12), because there are multiple detailed analyses of pollen sequence from which recent local paleoclimates can be estimated (11, 16), and because there is extensive information on species' distributions (19, 27) and phylogeography (21, 22, 25). In the present study, the combination of modeled paleodistributions and mtDNA phylogeography for the snail is highly informative, providing a spatial template of the size and distribution of mesothermal rainforest refugia against which the evolutionary and biogeographic history of other taxa can be compared.
Implications for Biogeography and Evolution in the Wet Tropics Rainforest Fauna.
In general, attempts to locate and circumscribe historical refugia in tropical rainforest systems have proved contentious (41, 42), and the same is true for the Australian wet tropics where there is some discrepancy between inferences from paleoclimatic models (12), current phytogeography (17), and evidence for drier sclerophyll vegetation and burning within putative refugia (20). Although consistent phylogeographic divisions across the BMC in vertebrates support the division between the major northern and southern refugia predicted by modeling (22), further details are obscured for vertebrates by varying histories of local extinction and expansion, stochastic variance of gene trees, or both (43).
The BMC break evident in vertebrates is present in the snails (between LU/MT and CU clades), but, in the latter and the more subdivided of the vertebrates (Fig. 3), this result is just one of several deep divisions. For the snail, both the phylogeography and paleomodels indicate the presence of multiple refugia within the north (FU, TU and CU) and also across the eastern Atherton region (MT, LU, AU, and BK). Although the climate estimates necessary for paleomodeling are available for the LGM only through the Holocene, the substantial levels of sequence divergence seen among these areas for both the snail and some vertebrates are consistent with Kershaw's (16) suggestion that angiosperm-dominated rainforests were restricted to these refugial areas for much of the Quaternary and perhaps earlier.
Vertebrate phylogeographic analyses indicate some species survived the LGM in multiple refugia to the north and south of the BMC whereas others probably persisted in just one refuge in each area (Fig. 3; refs. 27 and 43). If we use the predicted refugia for the similarly distributed snail as a template, we can assess which areas were suitable for all vertebrate species and which for only some. The combined modeling and genetic analysis of the snail predict TU as the major northern refuge for vertebrates, with the related WU populations being recolonized from there. By contrast, FU and CU were probably too small or too fragmented (or both) to support refugial populations of most vertebrate species across the Pleistocene. Exceptions include C. laevis and perhaps G. queenslandiae, which appear to have persisted in the FU refuge (Fig. 3). Similarly, the AU/BK region was clearly the major refuge south of the BMC, although the same two lizard species may also have persisted with the snails in the small refuge on the coastal MT range. That most upland areas were too small for persistence of most endemic and rainforest-restricted vertebrates is also suggested in comparisons of local species richness and endemism (19, 44).
Although few vertebrate species seem to have survived within them, the presence of small, geographically disjunct refugia across the northern uplands (excluding WU) and on Atherton Tableland is consistent with analysis of taxa with a higher proportion of narrowly distributed species, e.g., low vagility insects and snails (27). For these species, areas such as CU, FU, KU, and MT have high species richness, including locally endemic species. Phylogenetic analysis of such groups may shed light on the extent to which long-term subdivision among the northern areas and across the Atherton region has contributed to speciation as well as phylogeographic structure within species. For example, sister species of flightless dung beetle (Temnoplectron) occur on adjacent refugial areas in the north (45).
In comparing taxa, a differing response because of ecology needs to be taken into account. This cross-ecology approach potentially can provide an additional perspective, but up to now has been done subjectively. In the present case, we have sufficient context to argue that the differences among taxa are largely due to different responses to a common history of changes in distribution of habitats. Paleodistribution modeling for each species may provide a more formalized way of expressing the ecological differences among taxa.
Limitations and Developments.
The three key discrepancies between snail phylogeography and modeling (linkages between WU-TU and between LU-MT, and a KU refuge) are also evident in vertebrate phylogeographies, where resolution allows, indicating that the weakness lies with the modeling. In addition to the simplicity of the model used here, the climate estimates and spatial interpolations themselves have several limitations that may contribute to these discrepancies. Rainshadow effects may be behind the failure of the modeling to detect the WU-TU connection inferred from the genetic data, and whereas a KU refuge is not clearly within the BIOCLIM bounds, a larger area lays just below these limits. Other historical barriers such as rivers may also be important—MT currently is cut off from BK by the Mulgrave River. More sophisticated modeling systems that take into account climate parameter interactions, neighborhood size, and other nonlinear effects (reviewed in ref. 46) are effective in modeling snail current distribution (A.H. and A.M., unpublished results), and are being applied to modeling WT forest types (30); however, the main difficulty lies in developing paleoclimate estimates for these models. For reasons such as these, the combination of paleomodels and phylogeography is expected to be most informative at broad spatial scales and systems where simple parameters such as annual mean rainfall and temperature are highly informative.
Whereas our comparisons of modeling and genetics are largely qualitative, the results presented here are encouraging. Subregions predicted from the modeling (e.g., STABILITY surface, Fig. 1) to have contained refugia contained discrete phylogeographic lineages, whereas those from which the snail is predicted to have disappeared did not, but rather contained nested subsets of alleles as expected for recolonization from adjacent areas. There is also quantitative consistency between modeling and genetic estimates of the scale and tempo of population expansions from these refugia, in the correlation between area ratio and g, and plausible dependent parameters. However, each approach is laden with assumptions (e.g., area as a surrogate for Ne and panmixia and exponential growth in the genetic model), and the parameters obtained are not directly comparable. It would be far better to develop a framework in which the coalescent simulations on which genetic estimates of population parameters are based are combined directly with the spatially explicit predictions of the paleodistribution models.
Acknowledgments
We thank Michael Cunningham, Joanna Sumner, Conrad Hoskin, Chris Schneider, Keith McDonald, and Ben Phillips for collecting animals; Michael Cunningham, Stuart Baird, and Chris Schneider for discussion; and Catherine Graham, Leslie Rissler, Jim Patton, and David Wake for comments on the manuscript. This research was funded by grants from the Rainforest Cooperative Research Centre and the Australian Research Council.
Abbreviations
- WT
Wet Tropics
- BMC
Black Mountain Corridor
- Kya
thousand years ago
- AMT
annual mean temperature
- AMP
annual mean precipitation
- PDQ
precipitation of the driest quarter
- LGM
last glacial maximum
- MT
Malbon-Thompson Range
- BK
Bellenden Ker
- AT
Atherton Tableland
- FU
Finnegan Uplands
- KU
Kirrama Uplands
- AU
Atherton Uplands
- TU
Thornton Uplands
- CU
Carbine Uplands
- LU
Lamb Uplands
- WU
Windsor Uplands
Footnotes
References
- 1.Avise J C, Arnold J, Ball R M, Bermingham E, Lamb T, Neigel J E, Reeb C A, Saunders N C. Annu Rev Ecol Syst. 1987;18:489–522. [Google Scholar]
- 2.Slatkin M. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- 3.Templeton A R, Routman E, Phillips C A. Genetics. 1995;140:767–782. doi: 10.1093/genetics/140.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hewitt GM. Mol Ecol. 2001;10:537–549. doi: 10.1046/j.1365-294x.2001.01202.x. [DOI] [PubMed] [Google Scholar]
- 5.Edwards S V, DeBeerli P. Evolution. 2000;54:1839–1854. doi: 10.1111/j.0014-3820.2000.tb01231.x. [DOI] [PubMed] [Google Scholar]
- 6.Avise JC. Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard Univ. Press; 2000. [Google Scholar]
- 7.Cruzan M B, Templeton A R. Trends Ecol Evol. 2001;15:491–496. doi: 10.1016/s0169-5347(00)01998-4. [DOI] [PubMed] [Google Scholar]
- 8.Hadly E A, Kohn M H, Leonard J A, Wayne R K. Proc Natl Acad Sci USA. 1998;95:6893–6896. doi: 10.1073/pnas.95.12.6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leonard J A, Wayne R, Cooper A. Proc Natl Acad Sci USA. 2001;97:1651–1654. doi: 10.1073/pnas.040453097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nix HA. In: Atlas of Elapid Snakes of Australia. Longmore R, editor. Canberra, Australia: AGPS; 1986. [Google Scholar]
- 11.Kershaw A P, Nix H A. J Biogeography. 1988;15:589–602. [Google Scholar]
- 12.Nix HA. In: Rainforest Animals: Atlas of Vertebrates Endemic to Australia's Wet Tropics. Nix H A, Switzer M, editors. Canberra, Australia: ANPWS; 1991. pp. 11–39. [Google Scholar]
- 13.Davis M B, Shaw R G. Science. 2001;292:673–679. doi: 10.1126/science.292.5517.673. [DOI] [PubMed] [Google Scholar]
- 14.Adam P. Australian Rainforests. Oxford: Clarendon; 1992. [Google Scholar]
- 15.Truswell E. Aust Syst Botany. 1993;6:533–557. [Google Scholar]
- 16.Kershaw A P. Palaeogeogr Palaeoclimatol Palaeoecol. 1994;109:399–412. [Google Scholar]
- 17.Webb L, Tracey J. In: Ecological Biogeography of Australia. Keast J A, editor. The Hague, The Netherlands: W. Junk; 1981. [Google Scholar]
- 18.Nix H A, Switzer M. In: Rainforest Animals: Atlas of Vertebrates Endemic to Australia's Wet Tropics. Nix H A, Switzer M, editors. Canberra, Australia: ANPWS; 1991. [Google Scholar]
- 19.Williams S E, Pearson R G, Walsh P J. Pacif Conserv Biol. 1996;2:327–362. [Google Scholar]
- 20.Hopkins M S, Ash J, Graham A W, Head J, Hewett R K. J Biogeogr. 1993;20:59–74. [Google Scholar]
- 21.Joseph L, Moritz C, Hugall A. Proc R Soc London Ser B. 1995;260:177–182. doi: 10.1098/rspb.1995.0077. [DOI] [PubMed] [Google Scholar]
- 22.Schneider C J, Cunningham M, Moritz C. Mol Ecol. 1998;7:487–498. [Google Scholar]
- 23.Thomaz D, Guiller A, Clarke B. Proc R Soc London Ser B. 1996;263:363–368. doi: 10.1098/rspb.1996.0056. [DOI] [PubMed] [Google Scholar]
- 24.Douris V, Cameron R A D, Rodakis G C, Lecanidou R. Evolution. 1998;52:116–125. doi: 10.1111/j.1558-5646.1998.tb05144.x. [DOI] [PubMed] [Google Scholar]
- 25.Schneider C J, Smith T B, Larison B, Moritz C. Proc Natl Acad Sci USA. 1999;96:13869–13873. doi: 10.1073/pnas.96.24.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanisic J. Mem Qld Mus. 2000;46:337–348. [Google Scholar]
- 27.Moritz C, Richardson K S, Ferrier S, Monteith G B, Stanisic J, Williams S E, Whiffin T. Proc R Soc London Ser B. 2001;268:1875–1881. doi: 10.1098/rspb.2001.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houlder D J, Hutchinson M F, Nix H A, McMahon J P. anuclim Users Guide (Australian National Univ., Canberra, Australia), Version 5.1. 2000. [Google Scholar]
- 29.Moss P T, Kershaw A P. Palaeogeogr Palaeoclimatol Palaeoecol. 2000;155:155–176. [Google Scholar]
- 30.Hilbert D W, Ostendorf B. Ecol Modell. 2001;146:311–327. [Google Scholar]
- 31.Swofford D L. paup* Sunderland, MA: Sinauer; 2000. , Version 4.0b5. [Google Scholar]
- 32.McElroy D, Moran P, Bermingham E, Kornfield I. J Hered. 1992;83:157–158. doi: 10.1093/oxfordjournals.jhered.a111180. [DOI] [PubMed] [Google Scholar]
- 33.Kuhner M K, Yamato J, Felsenstein J. Genetics. 1998;149:429–434. doi: 10.1093/genetics/149.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King H. MSc. thesis. Brisbane, Australia: Univ. of Queensland; 1975. [Google Scholar]
- 35.Denver D R, Morris K, Lynch M, Vassilieva L L, Thomas W K. Science. 2000;289:2342–2344. doi: 10.1126/science.289.5488.2342. [DOI] [PubMed] [Google Scholar]
- 36.Cunningham M, Moritz C. Biol Conserv. 1998;83:19–30. [Google Scholar]
- 37.Pope L C, Estoup A, Moritz C. Mol Ecol. 2000;9:2041–2053. doi: 10.1046/j.1365-294x.2000.01110.x. [DOI] [PubMed] [Google Scholar]
- 38.Chiba S. Evolution. 1999;53:460–471. doi: 10.1111/j.1558-5646.1999.tb03781.x. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi M, Chiba S. Biol J Linn Soc. 2000;70:391–401. [Google Scholar]
- 40.Thacker R W, Hadfield G. Mol Phys Evol. 2000;16:263–270. doi: 10.1006/mpev.2000.0793. [DOI] [PubMed] [Google Scholar]
- 41.Brown K S J. In: Biogeography and Quaternary History of Tropical America. Whitmore T C, Brown K S, editors. New York: Oxford Univ. Press; 1987. pp. 175–196. [Google Scholar]
- 42.Colinvaux P A, De Oliveira P E, Moreno J E, Miller M C, Bush M B. Science. 1996;274:85–88. [Google Scholar]
- 43.Schneider C J, Moritz C. Proc R Soc London Ser B. 1999;266:191–196. [Google Scholar]
- 44.Williams S E, Pearson R G. Proc R Soc London Ser B. 1997;264:709–716. doi: 10.1098/rspb.1997.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reid C A M, Storey R I. Mem Qld Mus. 2000;46:253–297. [Google Scholar]
- 46.Guisan A, Zimmermann N E. Ecol Modell. 2000;135:147–186. [Google Scholar]