Abstract
Understanding how environmental and ecological factors shape variability in soil-associated microbial communities is a complex problem, particularly on islands, which contain a wide range of diverse and unique geology, fauna, and flora. The island of Crete features sharp altitudinal gradients, diverse landscapes, and distinct ecological zones shaped by its complex geological history making it an ideal natural laboratory for studying how environmental variation influences soil microbial communities. In this study, we characterized the soil microbial communities across Crete’s ecozones and identify environmental factors associated with their diversity and composition. We performed a single-day, island-wide soil microbiota investigation, the first of its kind, to address this challenge by eliminating sources of variability including seasonality, weather conditions, anthropogenic or land use changes over time, and ecological succession of microbial communities. This island collection event (Island Sampling Day, ISD) was conducted in conjunction with the annual meeting of the Genomic Standards Consortium, on the island of Crete, and utilized standard data and metadata collection protocols. We generated amplicon sequences (V3-V4 regions of the 16 S ribosomal RNA gene) and a metadata-enriched dataset from 435 soil samples across 72 sites and four distinct ecozones for future whole-island microbiome studies. Here we report on the study design and sample collection process along with our initial examination of the ecological drivers of soil microbial community variability (e.g., elevation, soil types, soil pH, soil moisture, vegetation type, land use) across the Crete ecozones (defined by elevation and distinct habitats).
Supplementary Information
The online version contains supplementary material available at 10.1186/s40793-025-00752-z.
Keywords: Soil metagenome, Island, Crete, Metadata standards, Citizen science
Introduction
Crete is the fifth largest Mediterranean continental island measuring over 260 km long with a breadth ranging from 12 to 60 km dominated by three mountains rising over 2,000 m. Characterized by sharp altitudinal gradients up to 2,500 m, it contains diverse and distinct assemblages of animal and plant species [1–3]. The Cretan landscape is diverse, with 39% of the island above 400 m, plateaus, small valleys, other mountains and hills, along with tens of gorges, rivers, streams, lakes, wetlands and beaches. In addition, there are fertile plains and areas with high anthropogenic impact (e.g., towns, agriculture and tourist attractions). Soil-associated microbiota are fundamental to nutrient cycling, decomposition, and soil formation, and directly influence plant productivity, vegetation patterns, and ecosystem stability [4, 5]. Aside from those associated with agricultural crops such as grains and grapes, the soil microbiota of Crete has never been surveyed [6, 7].
Advances in metagenomics and standardized methods like those from the Earth Microbiome Project have improved our understanding of soil microbial diversity across regions and time [8–12]. However, variability in sampling conditions along with geographic and seasonal differences can obscure the distinction between global and local environmental drivers of soil microbial communities [13–15]. To reduce these effects, the Genomic Standards Consortium (GSC, www.gensc.org) [16], the Institute of Marine Biology Biotechnology and Aquaculture (IMBBC, http://www.imbbc.hcmr.gr) and the Hellenic Centre for Marine Research (HCMR) conducted a one-day, full-island soil collection event in June 2016. This inaugural Island Sampling Day (ISD) pioneered a coordinated, island-wide, citizen-science effort to explore the diversity and structure of surface soil microbial communities across four out of Crete’s five ecozones, defined as geographic areas with distinct biodiversity of flora and fauna while limiting the influences of environmental confounding factors (e.g., weather, rainfall, temperature, sunshine, humidity). The scale of this one-day, whole-island sampling event, by necessity, was conducted by a large, coordinated team.
Herein, we present the first description of soil microbial diversity across crete’s distinct ecozones. Given that relationships between microbial community composition and soil moisture [15], organic nutrients [17], and elevation [18, 19] have been previously described, we sought to characterize these relationships among crete’s distinct ecozones. While global soil Microbiome studies have made significant progress, challenges remain in disentangling local versus global drivers due to Temporal and geographic variability. Our study contributes to this effort by providing a Temporally controlled, island-scale dataset that complements existing large-scale initiatives
Methods
Study design and sample collection
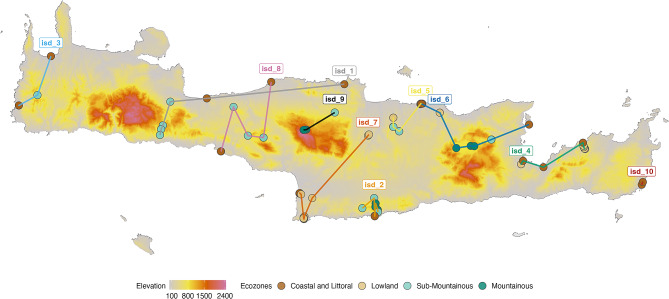
ISD was held on the final day of the GSC’s annual meeting which was hosted at the HCMR (15th June 2016, https://www.gensc.org/pages/meetings/GSC18/GSC18.html). ISD team members (scientists attending the GSC18 meeting, HCMR staff and a group of local volunteers) were trained on the sampling protocols on the previous day to facilitate standardized data collection including identification of soil types, local flora and pertinent metadata to be collected. Sampling teams were provided with an overview of Crete flora (https://github.com/GenomicsStandardsConsortium/ISD/tree/master/Methods/SampleCollection). Samples were collected in a single day to control for variations in environmental factors that may impact the abundance and diversity of microbes such as season, temperature and humidity. Sampling sites were chosen that included the dominant flora species. The sites were selected across Crete to encompass Crete’s distinctive elevation gradient and transects. In summary, ISD sampling was performed by 26 participants, organized in 10 teams, collecting soil samples from 72 distinct sites (2 subsites per site = 144 sample subsites) (Fig. 1). Sampling site Google Map: https://tinyurl.com/ISD.
Fig. 1.
Ten sampling routes were selected to examine the diverse and unique habitats and plants across the elevations on the island of Crete (white labels). Collection sites are indicated by dots colored by ecozones. Google Map: https://tinyurl.com/ISD
Prior to the study, a material transfer agreement (MTA) was established between the GSC and HCMR, to formally recognize and declare the samples and any products of the biological study were the sole ownership of Greece. The MTA, methods and associated scripts are archived in the GSC’s GitHub repository (https://github.com/GenomicsStandardsConsortium/ISD).
Sampling sites
Ten sampling routes (Fig. 1), across Crete, were chosen from a set of twenty potential sites. In total, 72 sampling sites were chosen and in each site two subsites were sampled for soil cores thus leading to 144 subsite locations. The routes were selected to cover the different ecological zones (ecozones) of the island. Ecological zones were characterized based on elevation as follows: coastal and littoral (0–19 m), lowland (20–339 m), sub mountainous (340–799 m), mountainous (800–2199 m) and alpine (above 2200 m). For this study, given the challenges of high elevation sampling, samples were not collected from the highest ecozone (alpine, above 2200 m). Each ecological zone has distinct on dominant (most abundant) vegetation: lower elevations: oaks, carob, junipers, and tree-spurge (Euphorbia dendroides); Imbros Gorge (upper reaches: Cypresses, Kermes oaks, Creten maples; lower gorge: Calabrian pines (Pinus brutia), olive and Plane trees, Oleander and Chaste Tree shrubs; middle elevations: pine and holly oak forests; and higher elevations: cypress woodlands, evergreen Cretan maple.
Sample collection
Each team followed a standard sampling procedure for each sampling site (Table S1) including a ram (a narrow tube with a plastic head for driving the soil borer into the soil and to push the soil out of the tube after sampling) (Figure S1).
Metal soil borer (15 cm long, 2 cm wide aluminum tube), constructed by Dimitris Tsaparis (HCMR)
On each of the 72 sites, teams selected two specific sampling subsites that were at least 3 m from the edge of the road and 0.6 m from the base of the identified plant. Soil was collected from the two subsite locations (1.5–3 m apart). At each subsite, three soil cores were extracted as replicates, one inch apart from each other. Prior to sampling, any organic material (e.g., leaves, seeds, branches) were swept away from the surface of the soil. Wearing latex gloves, the soil borer tube and ram were rinsed with water and wiped with a paper towel to remove any dirt. Both were sterilized by spraying with 15% hydrogen peroxide solution followed by three minutes of air drying [20, 21]. Soil was collected by: [1] placing the bottom of the borer tube on the ground and pushing the tube in the soil up to the 8 cm ring marked on the tube; and [2] removing the tube from the ground and placing the lower edge of the borer tube inside the 50 mL Falcon tube resulting in ca. 40 cc of collected soil. When the soil was extremely hard, a peroxide sterilized metal spoon was utilized to loosen soil prior to sampling. This was repeated to collect a total of three replicates of 50 mL Falcon tubes at each of the two subsite locations. Replicate Tube 1 was for metagenomic analysis, Tube 2 for soil chemistry analysis and Tube 3 for permanent storage at HCMR. The soil sample tubes were stored immediately in dry-ice containing coolers during the sampling event and stored at -20 °C at HCMR prior to shipping. In total, 432 soil cores were collected (3 per 144 subsites).
In addition, in each site, teams recorded metadata on ISD sheets prepared with GSC MIxS standard metadata terms (https://www.gensc.org/pages/standards/all-terms.html). The ISD sheets metadata included collection date and time, air temperature (°C), humidity, elevation, latitude and longitude, city (nearest town), plant type/species, land use, place name, environmental feature (habitat), soil type, litter type and on site measurements of soil moisture (%), soil temperature (°C), pH and litter depth (approximation in cm). The pH, soil moisture and temperature were measured within the same area of soil (6 cm, to the right of the spot where the soil was sampled). Where possible, metadata was also collected via the GIS cloud app (https://www.giscloud.com/). Proximity to urban, forested, and agricultural land was also documented and the flora located at the sampling subsite was identified, collected and photographed.
Controlled vocabularies were utilized for environmental features, soil type and land use. Soil types were identified (shale, conglomerates, clay deposits, limestone, limestone & flysch, mari, alluvial fan, flysch, clay, sandy clay, silty clay, clay loam) and recorded. Environmental feature terms included: agricultural land, alpine, beach, botanical garden, boulder field, cave, crevice, cultivated habitat (crop production), desert, farm, field, forest (e.g. oak forest), gravel field, grassland, greenhouse, lacustrine beach, meadow, national forest, nature reserve, olive grove, orchard, park, pasture, planted forest, plateau, sandy beach, valley, vineyard. Land use terms included: agricultural, botanical garden, cultivated, pristine, beach.
Soil chemistry analysis
Two replicate sets were shipped on dry ice to the University of Maryland Soil Lab for RNA and DNA extraction and soil chemistry analysis (USDA PERMIT NUMBER: P330-16-00090) [22–24]. Prior to DNA extraction, the moisture, total organic Carbon and Nitrogen (CN) weights and CN analysis were determined. Soil moisture was determined following the USDA Soil Survey Laboratory Methods Manual (https://www.govinfo.gov/content/pkg/GOVPUB-A57-PURL-gpo93947/pdf/GOVPUB-A57-PURL-gpo93947.pdf), with the moisture value calculated by subtracting the weight of the dry soil from the weight of the moist soil and then this value is divided by the weight of the dry soil. The CN analysis was conducted on October 4th, 2016. Combustion capsules of soil were submitted for C&N analyses using LECO CN628 (LECO Corporation, Saint Joseph, MI, USA).
DNA extraction, amplicon sequencing, and sequence quality filtering
DNA and RNA extraction followed the Earth Microbiome Project (EMP) standard protocols (www.earthmicrobiome.org/protocols-and-standards/16s) [11] utilizing the MoBio RNA extraction kit and the MoBio DNA elution kit to go with RNA kit (Qiagen, Hilden, GER). DNA quantification was performed using Qubit (Thermofisher). When the first extraction did not yield sufficient DNA, a second DNA extraction was attempted using the MoBio PowerMac Kit. Sequencing was conducted at the Institute for Genome Sciences, Genomics Resource Center (Maryland Genomics, https://marylandgenomics.org/) at the University of Maryland School of Medicine. The total DNA extracted in 100 µL was calculated by multiplying the concentration (µg/mL) by 0.1. This value was then scaled up to estimate the DNA content per gram of soil by multiplying by 1/0.25 and converted to milligrams by dividing by 1000. Sequencing libraries were prepared using a 2-Step PCR method [25], where the first PCR used short, target-specific primers with heterogeneity spacers and Illumina sequencing primer sequences, and the second PCR added dual-index barcodes and flow cell adaptors. Amplicon libraries targeting the 16S ribosomal RNA gene V3-V4 region of 16S rRNA gene were sequenced (Primers: 338F 5’-ACTCCTACGGGAGGCAGCAG-3’; 806R 5’-GGACTACHVGGGTWTCTAAT-3’) as described elsewhere [25]. Libraries were sequenced on an Illumina HiSeq2500 alongside four positive in-house and four negative controls representing each step of library preparation (extraction, amplification, barcoding, sequencing). Sequencing data were processed as described by Holm et al. [25] using DADA2 [26] to produce amplicon sequence variants (ASVs). Briefly, raw paired-end reads were processed using the DADA2 pipeline (v1.6.0). Reads were filtered and trimmed using filterAndTrim() with parameters truncLen = c(255,225), maxN = 0, maxEE = c [2], and truncQ = 2, and PhiX reads were removed. Quality profiles were inspected post-trimming. Error rates were learned from 1 million reads per direction. Amplicon sequence variants (ASVs) were inferred using the DADA algorithm and paired-end reads were merged. Chimeric sequences were removed using the default consensus method. An ASV was defined as a unique, non-chimeric sequence inferred after denoising and merging, representing a biological sequence variant at single-nucleotide resolution. Taxonomy was assigned using the RDP Classifier [27] trained on the SILVA NR99 v138 reference dataset and v128 (September 25, 2016) [28]. Counts from ASVs with the same taxonomic assignment were summed. ASVs assigned to d_Bacteria or d_NA and taxa present in any negative control with > 100 sequences were removed from downstream analyses (Escherichia/Shigella).
Descriptive and statistical analyses
Analyses were performed and figures generated using R Statistical Software (v4.4.0) [29]. Ecozones were summarized by elevation (m), soil moisture (log10-transformed), pH, organic carbon, and organic nitrogen content and each were compared between ecozones using Kruskal-Wallis tests. Shannon’s H was calculated for each sample using the diversity function of the vegan package (v2.6-6.1) [30]. Wilcoxon rank-sum tests were performed within each ecozone to compare Shannon diversity differences between spatially proximate (< 1 m) and distant (> 1 m) soil samples. To determine if soil features were associated with Shannon’s H, a linear regression model was fit using elevation, soil moisture, pH, organic carbon, and organic nitrogen content as predictors and Shannon’s H as the response. Significance was detected via p-values < 0.05.
β-diversity was evaluated through Bray-Curtis dissimilarities using the vegdist function of the vegan package and visualized using PCoA (package ape v5.8) [31]. PERMANOVA tests (adonis2 function, vegan) were performed to identify associations with Bray-Curtis dissimilarities. PCoA analyses were employed to identify how sampling site characteristics, including elevation, ecozone, geographic region, pH, soil type, total organic nitrogen, and total organic carbon and their microbial communities related to one another. A heatmap was also constructed using pheatmap v1.0.12 to visualize the 75 most abundant taxa. Taxa-specific associations with soil physical and chemical features were performed using count data normalized using the “poscounts” method and dispersions were estimated with a local fit type using the estimateSizeFactors and estimateDispersions functions from the DESeq2 package [32]. Normalized counts were extracted from the DESeq2 object and transformed to log2-counts per million (logCPM) using the voom function, which also estimated the mean-variance relationship [33]. A linear model was fitted, and empirical Bayes moderation was applied to the standard errors of the estimated coefficients. The results were visualized using ggplot2 [34, 35], with significant taxa labeled based on adjusted p-values.
Results
The ISD teams collected 432 soil samples (3 replicates/subsite) from 46 coastal or littoral, 36 lowland, 38 submountain and 24 mountainous sampling sites (Table S2). One replicate from each subsite was used for amplicon sequencing analyses (n = 144). Three other samples, all collected in sand, had no detectable DNA following extraction (isd_10_site_1_loc_1, isd_10_site_1_loc_2, isd_10_site_2_loc_2), and two samples were excluded from downstream analyses because they had fewer than 1,000 sequences (isd_3_site_2_loc_2_repl_2_DNA, isd_7_site_10_loc_1_repl_2_DNA). For 139 remaining samples, an average of 270,439 quality-filtered sequences per sample were obtained (range: 41-406k) from 2,248 total taxa. A total of 179,903 amplicon sequence variants (ASVs) were detected with an average 5,268 ASVs per sample detected (range: 797-8,130).
The elevations of sampling sites ranged between 1 and 1733 m, with the highest elevation sampled at approximately 1730 m (sample: isd_9_site_3) located at Lakos of Migero on Psiloritis Mountain (municipality of Mylopotamos). Four ecological zones (ecozones) significantly differed by elevation (p < 0.001), soil moisture (p < 0.001), and organic nitrogen (p < 0.001, Table 1). Samples with the highest moisture and nitrogen content were the from the Coastal and Littoral ecozone specifically isd_4, known as the Richtis Gorge (Table S2). Total organic carbon was also greatest in the Coastal and Littoral ecozone, specifically isd_1_site_2_loc_2 sample, collected from Askiyfou- Sfakia at a lacustrine beach, with the local vegetation composed of grasses and dwarf shrubs. The highest percent of total organic Nitrogen was identified from the isd_4_site_8_loc_1 sample, which was collected in the Richtis Gorge, in a woodland area with Tamarix parviflora, known by the common name smallflower tamarisk. The highest DNA concentration was extracted from the isd_7_site_1_loc_2 sample, collected from an olive grove on a hillside with strawberry plants nearby near Dafnes village (Heraklion municipality).
Table 1.
Physical and chemical characteristics of ecozones. Kruskal-Wallis (K-W) p-values are reported
| Coastal and Littoral (N = 42) |
Lowland (N = 35) |
Sub-Mountainous (N = 38) |
Mountainous (N = 24) |
K-W p-value |
|
|---|---|---|---|---|---|
| Elevation (m) | |||||
| Mean (SD) | 6.60 (4.89) | 132 (91.8) | 586 (165) | 1160 (346) | < 0.001 |
| Median [Min, Max] | 5.00 [1.00, 18.0] | 132 [20.0, 309] | 581 [340, 798] | 1030 [805, 1730] | |
| Soil Moisture (wfv) | |||||
| Mean (SD) | 2.23 (3.88) | 10.4 (28.7) | 2.61 (1.97) | 6.42 (4.00) | < 0.001 |
| Median [Min, Max] | 0.766 [0.0405, 21.8] | 1.77 [0.0584, 141] | 2.28 [0.0956, 10.4] | 5.51 [0.876, 12.5] | |
| pH (Corrected) | |||||
| Mean (SD) | 7.05 (0.319) | 6.95 (0.578) | 7.22 (0.510) | 7.25 (0.463) | 0.2 |
| Median [Min, Max] | 7.00 [6.50, 8.00] | 7.00 [5.80, 9.00] | 7.00 [6.60, 8.50] | 7.00 [7.00, 8.00] | |
| Missing | 14 (33.3%) | 4 (11.4%) | 12 (31.6%) | 16 (66.7%) | |
| Organic C (mg/kg soil) | |||||
| Mean (SD) | 59.3 (47.1) | 63.2 (37.6) | 49.2 (38.2) | 63.6 (48.6) | 0.3 |
| Median [Min, Max] | 53.1 [1.60, 238] | 59.1 [16.5, 173] | 45.3 [3.00, 141] | 40.5 [15.5, 175] | |
| Organic N (mg/kg soil) | |||||
| Mean (SD) | 1.60 (2.20) | 2.19 (1.65) | 1.72 (1.32) | 3.86 (2.14) | < 0.001 |
| Median [Min, Max] | 1.00 [0, 12.3] | 1.80 [0.100, 6.90] | 1.40 [0.100, 6.00] | 2.90 [1.30, 7.90] | |
| Total Estimated DNA (ug / g Soil) | |||||
| Mean (SD) | 6.36 (5.22) | 6.16 (5.62) | 7.30 (4.67) | 8.45 (4.61) | 0.04 |
| Median [Min, Max] | 5.18 [0.436, 20.0] | 5.00 [0.988, 32.0] | 6.54 [0.928, 23.7] | 7.42 [2.95, 17.8] | |
| Shannon Diversity Index | |||||
| Mean (SD) | 4.78 (0.253) | 4.84 (0.195) | 4.79 (0.213) | 4.74 (0.227) | 0.4 |
| Median [Min, Max] | 4.80 [4.07, 5.18] | 4.87 [4.42, 5.29] | 4.79 [4.27, 5.32] | 4.75 [4.23, 5.17] |
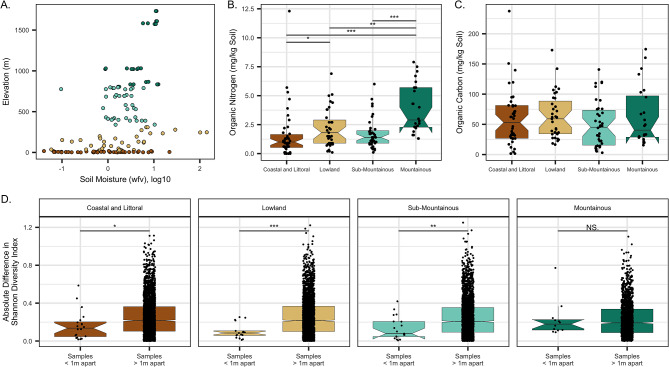
Mountainous ecozone samples featured significantly more organic nitrogen than other ecozones on average (Fig. 2A, B). Soil carbon content did not differ by ecozones (Fig. 2C). α-diversity of the soil microbiota was generally high with Shannon diversity indices (H) ranging from 4.1 to 5.2. Within all ecozones, Shannon’s H was more similar from samples collected 1 m apart compared to those from different collection sites, though the two nearby samples from mountainous isd_2_site_5 had the greatest difference in α-diversity than all other comparisons (Fig. 2D,|H1-H2| = 0.8). Shannon’s H did not differ between ecozones (Table 1) but was significantly associated with specific physical and chemical features: greater diversity was associated with lower elevations (p = 0.02, Table 2), higher moisture contents (p < 0.001), or when more organic carbon was present (p < 0.001).
Fig. 2.
(A) Elevation and soil moisture of samples colored by ecozone. (B) Soil nitrogen content was significantly higher in samples from the mountainous ecozone (and see Table 2). (C) Soil carbon content did not differ between ecozones (see Table 2). (D) α-diversity, as measured by Shannon’s H index, was more similar between samples from sites within 1 m compared to those from sites > 1 m apart in most ecozones. The difference was not significant among sites from the mountainous ecozone
Table 2.
Significant linear relationships exist between physical and chemical features and Shannon diversity index (H)
| Coefficient (β) | Std. Error | 95% Confidence Interval | F-statistic | p-value | |
|---|---|---|---|---|---|
| (Intercept) | 4.62E + 00 | 2.91E-01 | 4 to 5.2 | ||
| Elevation (m) | -7.94E-05 | 4.86E-05 | 0 to 0 | 5.4188 | 0.022 |
| Soil Moisture (wvf) | 4.17E-03 | 1.16E-03 | 0 to 0 | 16.2659 | < 0.001 |
| pH (corrected) | 1.17E-02 | 4.19E-02 | -0.1 to 0.1 | 1.3277 | 0.252 |
| C (mg/kg soil) | 2.25E-03 | 5.90E-04 | 0 to 0 | 13.3092 | < 0.001 |
| N (mg/kg soil) | -1.66E-02 | 1.25E-02 | 0 to 0 | 1.7744 | 0.186 |
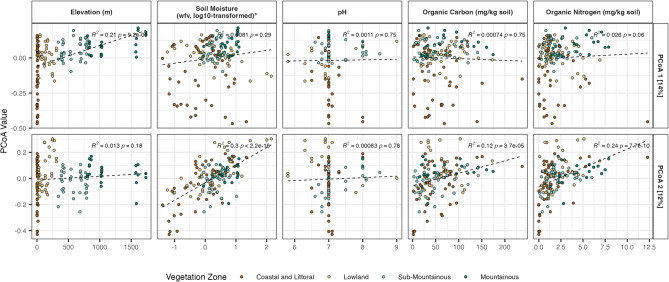
Ecozones were significantly associated with β-diversity of the soil microbiota, accounting for 14% of the variance (PERMANOVA F = 7.1, p < 0.001). When stratified by ecozone, 43% of β-diversity variance was explained by physical features of the soil (Fig. 3). A total of 26% of the variance in β-diversity was explained with PCoA axes 1 and 2. Higher values of PCoA 1 (14% of composition variance) were found in the coastal and littoral ecozone and was largely driven by elevation (R2 = 0.2, p < 0.001, Fig. 3, top row). The highest PCoA 2 values (12% of the composition variance) were observed in the lowland ecozone and correlated with higher soil moisture (R2 = 0.3, p < 0.001), organic carbon (R2 = 0.2, p < 0.001) and nitrogen content (R2 = 0.1, p < 0.001, Fig. 3, bottom row).
Fig. 3.
Microbial β-diversity represented by PCoA axes 1 (top row) and 2 (bottom row) differently associate with soil physical and chemical features
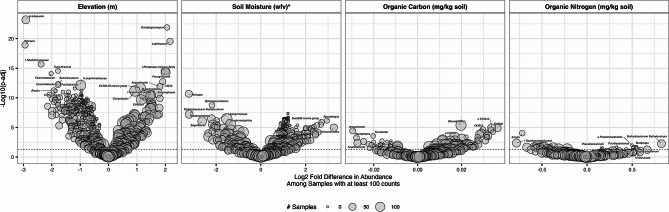
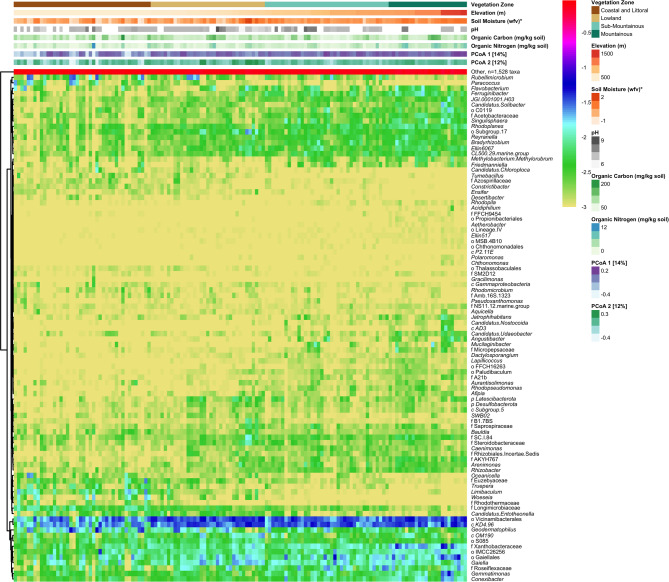
The most prevalent taxa in this study included Solirubrobacter, Nocardioides, and Blastococcus are globally prevalent [12]. On Crete, many microbial taxa were uniquely associated with specific physical and chemical soil features (Fig. 4 and see Table S3 for all results). In this analysis, log fold changes represent the change in taxon abundance per unit increase in each environmental variable, modeled as continuous predictors. Numerous ASVs from taxa within the Actinomycetota were associated with higher elevations including Angustibacter, Lapillicoccus, Candidatus Nostocoida, Candidatus Udaeobacter, Xanthobacteraceae, and Dactylosporangium, while Limibaculum and Woesia (Proteobacteria) were associated with lower elevations. ASVs from Methylobacterium methylorubrum was also associated with elevation, as well as drier soils, and higher organic nitrogen content. Soil moisture was positively associated with Algoriphagus and order Actinomarinales. The greatest associations with organic carbon content were with ASVs from SWB02, while those from Sediminibacterium had the strongest coefficients of association with organic nitrogen content. Ensifer, Geodermatophilus, and Desertibacter were each associated with drier soils. No significant observations were made regarding pH, likely due to the limited range of soil pH values sampled. The distribution of taxa across all physical and chemical features are presented in Fig. 5.
Fig. 4.
Microbial taxa significantly associated with physical and chemical soil features. Dashed line indicates adjusted p-value = 0.01. See Table S3 for all results
Fig. 5.
Heatmap of log10-transformed proportions of taxa. Featured taxa were significantly associated with physical or chemical features. All others were grouped into “Other”. Samples (columns) are sorted by elevation
Discussion
This study described the soil microbiota of the island of Crete and addressed the challenges of controlling for confounding environmental factors such as seasonality, anthropogenic or land use changes over time, and ecological succession of microbial communities to reveal the relationships between local ecological drivers of soil and its associated microbial diversity [30]. This whole island soil microbiome sampling event, enabled sampling across the breadth of ecoregions, and demonstrated capacity for a coordinated effort to collect the breadth of samples using consistent methods. Devising a systematic sampling plan facilitated sampling of diverse types of sites while also collecting enriched metadata describing the ecological context. Here, we have demonstrated ‘proof of principle’, that it is reasonably feasible to conduct single-day, large-scale collection studies, in a collaborative and standards compliant fashion.
The four ecological zones aligned to those categorized by previous biodiversity studies in Crete [36, 37]. Although the ecozones varied significantly in elevation, soil moisture, and nitrogen content, these differences did not correspond to significant variation in microbial α-diversity, which remained consistently high across zones (HISD >4). However, overall α-diversity in the ISD samples (H = 4–5) was lower than that reported in a recent European soil microbiota study (H = 6–8) [38]. This disparity may be because the latter soil study covered a relatively wider region over a longer time (multiple seasons) compared to the ISD project which was specifically designed to complete all sampling within a single day. These differences likely also contribute to the relatively higher percent variance in β-diversity explained by the PCoA analysis of the ISD project (26% variance explained by PCoA axes 1 and 2) compared to the European soil project (18% variance explained by dbRDA axes 1 and 2). Soil bacterial biodiversity is very complex, even sites a few meters apart can differ significantly in their community composition (Fig. 2D). This is a fact that is sometimes left unmentioned in worldwide studies [38]. Islands like Crete can be useful to perform denser soil samplings to exclude variables such as latitude to explain the microbial diversity.
Elevation emerged as a strong associate of β-diversity, with higher elevations characterized by increased organic nitrogen content. Taxa such as Xanthobacteraceae and Candidatus Udaeobacter, both known nitrogen fixers and recently proposed bioindicators of soil health [39–41], were enriched at higher elevations, consistent with their ecological roles in nutrient-limited environments. Additionally, several members of the phylum Actinomycetota—including Angustibacter, Lapillicoccus, and Dactylosporangium—were also associated with high-elevation sites, aligning with previous reports of their prevalence in oligotrophic, montane soils [42]. Soil moisture was also a major driver of β-diversity. Associations between soil moisture and microbial diversity has been previously reported and taxa such as Geodermatophilus, Desertibacter isolated from desert soils were unsurprisingly associated with low-moisture settings [43–47]. Conversely, Algoriphagus and members of the order Actinomarinales were associated with wetter soils, consistent with their aquatic or semi-aquatic origins. Interestingly, Methylobacterium methylorubrum was associated with both higher elevation and drier soils, suggesting a potential role in nitrogen cycling under xeric conditions [48]. In contrast, soil pH did not significantly influence microbial composition, likely due to the narrow pH range observed across samples. Overall, the taxa-environment associations observed in this study largely align with known ecological traits, reinforcing the robustness of the ISD dataset and its potential for identifying microbial indicators of environmental conditions.
The findings presented here hold promise for benchmarking bacterial community composition while controlling for variation across seasons, temperature and humidity. Our open data approach will serve future repetitions of the experiment both in the island of Crete as well as other islands, for interisland community comparison purposes. Through this first Island Sampling Day study, our team demonstrated that it is not only feasible but fruitful to conduct an entire island microbiome study in a single day, to provide novel insights of an understudied ecosystem, by controlling for environmental variables which vary over time and season [49]. This snapshot of the soil microbial diversity across the island of Crete: [1] put metadata into action through a citizen science project to demonstrate their value in future samplings; [2] described the first full island microbial community assessment and [3] shed light on microbial phylotypes variability across Crete’s distinct terrestrial habitats.
Our selection of Crete as the first island microbiome study presented an opportunity to study a unique ecosystem with diverse ecoregions, an established model for traditional biodiversity research [50]. We invite and encourage reuse of this rich metadata dataset. More recently, samples have been collected, following the protocols devised for this work, for a second Crete ISD #2 and Tahiti ISD in 2022, to further this work through a JGI initiative to conduct full metagenomic sequencing of the Crete and Tahiti datasets.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 2: Metal soil borer (15 cm long, 2 cm wide aluminum tube), constructed by Dimitris Tsaparis (HCMR).
Acknowledgements
We greatly thank alumnae and current Genomic Standards Consortium board members, who supported the GSC initiating the study of an island microbiome through a single day, all island sampling event: Phil Hugenholtz, Nikos Kyrpides, Folker Meyer, Susanna Sansone, Linda Amaral-Zettler, Jim Cole, George Garrity, Rob Knight, Dave Ussery, Frank Oliver Glockner, Bonnie Hurwitz, Chris Hunter, Chris Meyer, Jack Gilbert, Maria Chuvochina, Emiley Eloe-Fadrosh, Rob Finn, Alice McHardy, Scott Jackson, João Setubal, Kasthuri Venkateswaran, Ruth Timme, Ramona Walls and Tanja Woyke. We thank the HCMR for their event coordination and Dimitra Manou at the HCMR for establishing the ISD material transfer agreement and Iraklis Vretzakis for his assistance in the compilation of the metadata from sampling log sheets.
Author contributions
EP, LS, MS, DT, GK, AM, GC: designed and organized the research. EP, LS, MS, DT, MP, PK, SD, AO, ZK. PP, PH, WHCFK, JS, BS, IL, PG, ND, CL, LS, GS, ST, IM, DP, PY, CC, PB; PG: Conducted the field sampling, metadata annotation.DM, AM, IV: Sample shipment, processed legal framework arrangements.MH, AC: conducted sequencing and bioinformatics analysis.SY, JG: processed the soil samples, extracted the DNA and conducted the soil chemistry analyses. JH, SP, EP and LS: Analyzed the data and wrote the paper; andLS, ND, GS, ST, IKM, PY, PG: GSC Board members on sampling team.
Funding
The field work was supported by the Genomic Standards Consortium and the HCMR. SP was supported by the 3rd H.F.R.I. (Hellenic Foundation for Research and Innovation), Scholarships for PHD Candidates (no. 5726). [CL] The National Ecological Observatory Network is a program sponsored by the U.S. National Science Foundation and operated under cooperative agreement by Battelle. This material is based in part upon work supported by the U.S. National Science Foundation through the NEON Program. WHCFK received funding for travel from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 730984, ASSEMBLE Plus project. The work of IKM was supported by the National Center for Biotechnology Information of the National Library of Medicine (NLM), National Institutes of Health.
Data availability
The ISD Metadata is freely available in the GSC/ISD/Data archive https://github.com/GenomicsStandardsConsortium/ISD/tree/master/Data. Amplicon sequencing data are available through the BioProject PRJEB21776. The raw soil chemistry results are available in the GSC/ISD GitHub repository https://github.com/GenomicsStandardsConsortium/ISD/tree/master/Data.
Declarations
Competing interests
Lynn M. Schriml and Ilias Lagkouvardos are Associate Editors on Environmental Microbiome.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Johanna B. Holm and Savvas Paragkamian contributed equally to this work.
References
- 1.Rackham O, Moody J. The making of the Cretan landscape. Manchester University; 1996.
- 2.Tsantilis D. Crete: a continent in an Island. Natural History Museum of Crete; 2015.
- 3.Vogiatzakis I, Mannion A, Sarris D. Mediterranean Island biodiversity and climate change: the last 10,000 years and the future. Biodivers Conserv. 2016;25:2597–627. [Google Scholar]
- 4.Kovacs ED, Kovacs MH. Global change drivers impact on soil microbiota: challenges for maintaining soil ecosystem services. Vegetation dynamics. Changing Ecosystems and Human Responsibility: IntechOpen; 2023. [Google Scholar]
- 5.Banerjee S, van der Heijden MGA. Soil microbiomes and one health. Nat Rev Microbiol. 2023;21(1):6–20. [DOI] [PubMed] [Google Scholar]
- 6.Kalamaki MS, Angelidis AS. High-Throughput, Sequence-Based analysis of the microbiota of Greek Kefir grains from two geographic regions. Food Technol Biotechnol. 2020;58(2):138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papadopoulou E, Bekris F, Vasileiadis S, Papadopoulou KK, Karpouzas DG. Different factors are operative in shaping the epiphytic grapevine Microbiome across different geographical scales: biogeography, cultivar or vintage? J Sustainable Agric Environ. 2022;1(4):287–301. [Google Scholar]
- 8.Bakker P, Berendsen RL, Van Pelt JA, Vismans G, Yu K, Li E, et al. The Soil-Borne identity and Microbiome-Assisted agriculture: looking back to the future. Mol Plant. 2020;13(10):1394–401. [DOI] [PubMed] [Google Scholar]
- 9.Cowan DA, Lebre PH, Amon C, Becker RW, Boga HI, Boulange A, et al. Biogeographical survey of soil microbiomes across sub-Saharan africa: structure, drivers, and predicted climate-driven changes. Microbiome. 2022;10(1):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karimi B, Terrat S, Dequiedt S, Saby NPA, Horrigue W, Lelievre M, et al. Biogeography of soil bacteria and archaea across France. Sci Adv. 2018;4(7):eaat1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, et al. A communal catalogue reveals earth’s multiscale microbial diversity. Nature. 2017;551(7681):457–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-Gonzalez A, Eldridge DJ, Bardgett RD, et al. A global atlas of the dominant bacteria found in soil. Science. 2018;359(6373):320–5. [DOI] [PubMed] [Google Scholar]
- 13.Zhang K, Delgado-Baquerizo M, Zhu Y-G, Chu H. Space is more important than season when shaping soil microbial communities at a large Spatial scale. mSystems. 2020;5(3). 10.1128/msystems.00783. [DOI] [PMC free article] [PubMed]
- 14.Gupta VVSR, Tiedje JM. Ranking environmental and edaphic attributes driving soil microbial community structure and activity with special attention to Spatial and Temporal scales. mLife. 2024;3(1):21–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li SP, Wang P, Chen Y, Wilson MC, Yang X, Ma C, et al. Island biogeography of soil bacteria and fungi: similar patterns, but different mechanisms. ISME J. 2020;14(7):1886–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, Amaral-Zettler L, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol. 2011;29(5):415–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganzert L, Lipski A, Hubberten H-W, Wagner D. The impact of different soil parameters on the community structure of dominant bacteria from nine different soils located on Livingston island, South Shetland archipelago, Antarctica. FEMS Microbiol Ecol. 2011;76(3):476–91. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Heal KV, Shi M, Chen W, Zhou C. Decreasing molecular diversity of soil dissolved organic matter related to microbial community along an alpine elevation gradient. Sci Total Environ. 2022;818:151823. [DOI] [PubMed] [Google Scholar]
- 19.Tang M, Li L, Wang X, You J, Li J, Chen X. Elevational is the main factor controlling the soil microbial community structure in alpine tundra of the Changbai mountain. Sci Rep. 2020;10(1):12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linley E, Denyer SP, McDonnell G, Simons C, Maillard J-Y. Use of hydrogen peroxide as a biocide: new consideration of its mechanisms of biocidal action. J Antimicrob Chemother. 2012;67(7):1589–96. [DOI] [PubMed] [Google Scholar]
- 21.Magiopoulos I, McQuillan J, Burd C, Mowlem M, Tsaloglou M-N. A multi-parametric assessment of decontamination protocols for the subglacial lake Ellsworth probe. J Microbiol Methods. 2016;123:87–93. [DOI] [PubMed] [Google Scholar]
- 22.Kepler RM, Epp Schmidt DJ, Yarwood SA, Cavigelli MA, Reddy KN, Duke SO et al. Soil microbial communities in diverse agroecosystems exposed to the herbicide glyphosate. Appl Environ Microbiol. 2020;86(5). [DOI] [PMC free article] [PubMed]
- 23.Kepler RM, Schmidt DJE, Yarwood SA, Cavigelli MA, Reddy KN, Duke SO et al. Erratum for Kepler Soil Microbial Communities in Diverse Agroecosystems Exposed to the Herbicide Glyphosate. Appl Environ Microbiol. 2020;86(22). [DOI] [PMC free article] [PubMed]
- 24.Yarwood SA. The role of wetland microorganisms in plant-litter decomposition and soil organic matter formation: a critical review. FEMS Microbiol Ecol. 2018;94(11). [DOI] [PubMed]
- 25.Holm JB, Humphrys MS, Robinson CK, Settles ML, Ott S, Fu L et al. Ultrahigh-Throughput multiplexing and sequencing of > 500-Base-Pair amplicon regions on the illumina HiSeq 2500 platform. mSystems. 2019;4(1). [DOI] [PMC free article] [PubMed]
- 26.DADA2: High-resolution sample inference from Illumina amplicon data. (2016). [DOI] [PMC free article] [PubMed]
- 27.Lan Y, Wang Q, Cole JR, Rosen GL. Using the RDP classifier to predict taxonomic novelty and reduce the search space for finding novel organisms. PLoS ONE. 2012;7(3):e32491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The SILVA. ribosomal RNA gene database project: improved data processing and web-based tools., (2013). [DOI] [PMC free article] [PubMed]
- 29.R Core Team R. R: A language and environment for statistical computing. 2024.
- 30.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, et al. The vegan package. Community Ecol Package. 2007;10:631–7. [Google Scholar]
- 31.Paradis E, Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35(3):526–8. [DOI] [PubMed] [Google Scholar]
- 32.Love MI, Huber W, Anders S. Moderated Estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Law CW, Alhamdoosh M, Su S, Smyth GK, Ritchie ME. RNA-seq analysis is easy as 1-2-3 with limma, glimma and edger. F1000Res. 2016;5:1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hadley W. Ggplot2. New York, NY: Springer Science + Business Media, LLC; 2016. pages cm p.
- 35.Wickham H. Ggplot2: elegant graphics for data analysis. New York: Springer; 2009. viii, 212 p. p.
- 36.Trigas P, Panitsa M, Tsiftsis S. Elevational gradient of vascular plant species richness and endemism in Crete–the effect of post-isolation mountain uplift on a continental Island system. PLoS ONE. 2013;8(3):e59425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chatzaki M, Lymberakis P, Markakis G, Mylonas M. The distribution of ground spiders (Araneae, Gnaphosidae) along the altitudinal gradient of crete, greece: species richness, activity and altitudinal range. J Biogeogr. 2005;32(5):813–31. [Google Scholar]
- 38.Labouyrie M, Ballabio C, Romero F, Panagos P, Jones A, Schmid MW, et al. Patterns in soil microbial diversity across Europe. Nat Commun. 2023;14(1):3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilhelm RC, Amsili JP, Kurtz KSM, van Es HM, Buckley DH. Ecological insights into soil health according to the genomic traits and environment-wide associations of bacteria in agricultural soils. ISME Commun. 2023;3(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oren A. The family Xanthobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: Alphaproteobacteria and Betaproteobacteria. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. pp. 709–26. [Google Scholar]
- 41.Brewer TE, Handley KM, Carini P, Gilbert JA, Fierer N. Genome reduction in an abundant and ubiquitous soil bacterium ‘candidatus Udaeobacter copiosus’. Nat Microbiol. 2016;2(2):16198. [DOI] [PubMed] [Google Scholar]
- 42.Wang DS, Xue QH, Ma YY, Wei XL, Chen J, He F. Oligotrophy is helpful for the isolation of bioactive actinomycetes. Indian J Microbiol. 2014;54(2):178–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hackl E, Zechmeister-Boltenstern S, Bodrossy L, Sessitsch A. Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl Environ Microbiol. 2004;70(9):5057–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romanowicz KJ, Freedman ZB, Upchurch RA, Argiroff WA, Zak DR. Active microorganisms in forest soils differ from the total community yet are shaped by the same environmental factors: the influence of pH and soil moisture. FEMS Microbiol Ecol. 2016;92(10). [DOI] [PubMed]
- 45.Han JR, Li S, Lu CY, Lian WH, Shi GY, Feng CY et al. Rubellimicrobium arenae sp. nov., isolated from desert soil. Int J Syst Evol Microbiol. 2023;73(7). [DOI] [PubMed]
- 46.Liu M, Dai J, Liu Y, Cai F, Wang Y, Rahman E, et al. Desertibacter roseus gen. Nov., sp. Nov., a gamma radiation-resistant bacterium in the family rhodospirillaceae, isolated from desert sand. Int J Syst Evol Microbiol. 2011;61(Pt 5):1109–13. [DOI] [PubMed] [Google Scholar]
- 47.Castro JF, Nouioui I, Sangal V, Trujillo ME, Montero-Calasanz MC, Rahmani T, et al. Geodermatophilus Chilensis sp. nov., from soil of the Yungay core-region of the Atacama desert, Chile. Syst Appl Microbiol. 2018;41(5):427–36. [DOI] [PubMed] [Google Scholar]
- 48.Leducq J-B, Seyer-Lamontagne É, Condrain-Morel D, Bourret G, Sneddon D, Foster James A, et al. Fine-Scale adaptations to environmental variation and growth strategies drive phyllosphere Methylobacterium diversity. mBio. 2022;13(1):e03175–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chase AB, Weihe C, Martiny JB. Adaptive differentiation and rapid evolution of a soil bacterium along a climate gradient. Proceedings of the National Academy of Sciences. 2021;118(18):e2101254118. [DOI] [PMC free article] [PubMed]
- 50.Vogiatzakis I, Rackham O, Crete. Mediterranean Island landscapes: natural and cultural approaches. Springer; 2008. pp. 245–70.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 2: Metal soil borer (15 cm long, 2 cm wide aluminum tube), constructed by Dimitris Tsaparis (HCMR).
Data Availability Statement
The ISD Metadata is freely available in the GSC/ISD/Data archive https://github.com/GenomicsStandardsConsortium/ISD/tree/master/Data. Amplicon sequencing data are available through the BioProject PRJEB21776. The raw soil chemistry results are available in the GSC/ISD GitHub repository https://github.com/GenomicsStandardsConsortium/ISD/tree/master/Data.