Abstract
Background
Litchi, an economically important fruit crop in Southeast Asia, is renowned for its distinctive flavor and nutrient value, but its genetic diversity and genetic basis underlying fruit quality regulation remain largely unexplored.
Results
We re-sequence 276 litchi accessions collected globally and identify 54 million high-quality biallelic SNPs. We then analyze the population structure and reveal four main subgroups within the population. By phenotypic profiling of 21 fruit-quality-related traits, followed with genome-wide association study, we identify a plethora of candidate genes and genomic loci that are responsible for regulating seed and fruit quality traits. In particular, we characterize and experimentally validate an invertase gene, LcSAI, encoding an enzyme catalyzing the conversion of sucrose into reducing sugars, as a key regulator of sugar composition in litchi fruit, affecting the sweetness of litchi fruits.
Conclusions
Our study demonstrates the genetic diversity and population structure of litchi. We delineate the genetic basis of fruit quality through comprehensive genomic analyses and phenotypic profiling. These findings provide valuable resources and knowledge for understanding the genetic basis of fruit quality in litchi, which contribute to the theoretical basis for further genetic improvement.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-025-03693-5.
Keywords: Genetic diversity, Genetic basis, Fruit quality, Litchi (Litchi chinensis Sonn.), Invertase gene
Background
Litchi (Litchi chinensis Sonn.), a member of the Sapindaceae family, holds significant economic importance as a subtropical fruit tree in the Southeast Asia region, especially in the southern China [1]. The origin of litchi can be traced back to Yunnan Province, China, where it has been cultivated for over 2000 years. Over time, a diverse collection of litchi germplasm resources has been developed through natural selection and artificial breeding in various growing conditions, providing opportunities for continuous genetic improvement [2]. Although research endeavors have been dedicated to examining and assessing the diversity of litchi through the analysis of observable characteristics [3, 4], the associated genetic variations within the central germplasm, which harbors a substantial wealth of genetic data, remain largely unexplored and underutilized. Recent years, with the rapid development of genomics and deep sequencing technologies, the application of association analysis between genotypes and phenotypes in natural populations has unveiled a multitude of natural allele variations and potential genes in many fruit crops such as peach, apple, citrus, grape, and pear [5–9], offering substantial resources for accelerated breeding. Nevertheless, investigations pertaining to genome-wide association analysis are limited in litchi.
Fruit is the main product of commercial value for litchi; therefore, the primary goal of research and breeding is to ensure high fruit quality and stable yield. For instance, smaller seed is a character highly favored by consumers in industry, and small-seed fruits possess greater commercial value in comparison to normal-seed fruits. Smaller seeds are often resulted from embryo abortion in the process of seed development, giving rise to aborted seeds with reduced size. The seed-abortion trait exhibits a negative correlation with fruit yield and may impose limitations on the selection of female parents in hybrid breeding. And the seed-abortion trait is unstable and susceptible to environmental influences [10, 11]. In addition to seed size, the quality of litchi fruit is significantly influenced by sweetness, which is determined by the content and composition of soluble sugars [12]. It is worth noting that different fruits exhibit varying compositions of soluble sugars. For instance, fructose is the predominant soluble sugar in apples and pears [13, 14], whereas sucrose serves as the primary soluble sugar in citrus, bananas, peaches, and mangoes [15]. The majority of litchi varieties contain reducing sugars, namely glucose and fructose, as the primary soluble sugars in the pulp, while certain varieties predominantly consist of sucrose. Recently, more and more genes related to sugar metabolism have been identified, and they play important roles in regulating sugar composition, content, and quality in fruits. Malek et al. [16] identified two invertase copies are associated with dry fruit sucrose content in data palm fruit. The heterologous expression of the apple hexose transporter MdHT2.2 in tomato fruit increases cell wall invertase activity, leading to the increase of both fructose and glucose content and meanwhile the significant decrease of sucrose [17]. The vacuolar invertase PbrvacInv1 was found to be involved in sucrose decomposition during pear development [18]. In litchi, the activities of cell wall invertases (CWIN) and vacuolar invertases (VIN) in the aril show a strong positive correlation with their hexose/sucrose ratios. Interestingly, this hexose/sucrose ratio is also positively correlated with seed weight, and LcCWIN5 was reported specifically expressed in anthers and pistils, then result in impaired liquid endosperm development, smaller seeds, and/or higher seed abortion rate [19, 20]. Additionally, the sucrose synthase (SuS) activities in litchi also show a significantly positive correlation with the hexose/sucrose ratio. Five SuS genes are identified in litchi, only the expression profile of LcSuS1 was consistent with the trend of sugar accumulation during aril development, and LcSuS2, LcSuS4, and LcSuS5 show varied expression patterns between cultivars with different hexose/sucrose ratios [21]. Although several genes related to abortive seed development and sugar metabolism have been documented, the precise genetic mechanisms remain elusive [22], and their potential use in breeding, for example, molecular marker development, has not yet been tested.
Litchi is native to south China, in which there are profuse natural germplasm resources. The National Litchi Germplasm Resource Nursery (NLGRN) in Guangzhou, China, has amassed hundreds of litchi varieties sourced from litchi-producing nations across the globe [23]. In this study, we selected a collection of 276 representative accessions from NLGRN and profiled their genetic diversity and variations using deep sequencing. The abundance of genomic variation data has provided us with new insights into the population structure of litchi. In addition, we recorded as many as 21 phenotypic traits related to fruit quality and used them for genome-wide association study (GWAS). This led to the identification of a series of genomic loci and candidate genes associated with certain traits. The findings from our study have provided valuable knowledge for the understanding of the population structure of litchi and rich gene resources that can be potentially used for targeted breeding and improvement efforts in litchi.
Results
Genetic variation and population structure of litchi
Previously, our investigations have revealed that wild litchi species mainly belong to two groups, YNG (Yunnan Group) and HNG (Hainan Group) [2]. To gain a more comprehensive view of the population structure of litchi, in this study, we newly collected 276 litchi accessions (Additional file 2: Table S1) (Fig. 1A). In combination with 38 wild litchi samples from the previous study [2] (Additional file 2: Table S1), a total of 314 litchi samples were used for genetic variation profiling.
Fig. 1.
Litchi population structure and genetic diversity. A Geographical distribution of 277 litchi accessions; orange: YNG including YNW (Yunnan wilds), VNW (Vietnam wilds), GXDXW (Da Xin wilds in Guangxi), and EEMC/EMC (extremely-early/early maturing cultivars); purple: HNG including GXBBW (Bo Bai wilds in Guangxi) and HNW (Hainan wilds); green: FGG1 represents the class I accessions from Fujian, Guangdong, and Guangxi; and red: FGG2 represents the class II accessions from Fujian, Guangdong, and Guangxi. Accessions from neighboring locations were combined and displayed using pie charts, with the size of each pie representing the percentage of each group. B A maximum likelihood tree was constructed using fourfold degenerate SNP sites used HMD, LYLY, LL, and R267 as outgroup. HMD: Nephelium lappaceum, LYLY: Dimocarpus yunnanensis, R267 and LL: Dimocarpus confinis. C The tenfold cross-validation supported the population into four subpopulations, which had the lower CV error and a gradually increasing logarithmic likelihood. D The Fst values and nucleotide diversity (π) between the four subpopulations. E The decay of linkage disequilibrium of the four litchi subpopulations. F Estimated population structure of 277 litchi accessions using STRUCTURE. G PCA 3D-scatter plot of the 277 accessions. The dot colors of the scatter plot are the same as those in the ML tree
We sequenced all these new accessions and obtained 2.65 Tb data, with an average of coverage 20 ×. A set of 3.54 million high-quality biallelic SNP sites were detected, including 74.6k fourfold degenerate sites (4DTVs). Based on these 4DTVs, a maximum likelihood tree was constructed, and it is observed that all these litchi accessions could be classified into four groups: Yunnan Group (YNG, including wild litchi from Yunnan, Vietnam, and Daxing county in Guangxi Province), Hainan Group (HNG, including wild litchi from Hainan and from Bobai county in Guangxi, and consisting of accessions mainly from southern Guangdong and Guangxi), FGG1 Group, and FGG2 Group (both including accessions from Fujian, other parts of Guangdong and Guangxi) (Fig. 1A, B).
A subset of 439,759 SNPs was employed for further population analysis after pruning by PLINK. Tenfold cross-validation supported a K = 4 classification, which showed a low CV error (0.40187) and a steadily increasing logarithmic likelihood value (Fig. 1C). Although K = 5 had the smallest CV error (0.40098), it further subdivided FGG2 into two categories, which is conflicted with both the phylogenetic tree structure (Fig. 1B) and the geographic distribution of accessions. Nucleotide diversity was significantly higher in FGG1, FGG2, and HNG (π ≥ 1.73e − 3) compared to YNG (π = 7.2e − 4) (Fig. 1D). The lowest fixation index (Fst) was observed between FGG2 and HNG (Fst = 0.042), with Fst values of 0.047 and 0.060 between these two and FGG1, respectively. In contrast, YNG exhibited great genetic distance to HNG, FGG2, and FGG1 (Fig. 1D).
LD decay varied among subgroups, with YNG showing the longest decay distance of 1862 bp, followed by FGG2, FGG1, and HNG with distances of 447 bp, 422 bp, and 138 bp, respectively (Fig. 1E). Population admixture results indicated that YNG and HNG were primarily distinguished when K = 2, while FGG1 and FGG2 were discerned at K = 4 (Fig. 1F). A minor amount of FGG2 component was detected in HNG, and vice versa, while FGG1 exhibited a mixture of YNG, HNG, and FGG2 components (Fig. 1F). Population principal component analysis demonstrated significant differentiation between YNG and FGG2/HNG along PC1 (t-test, p < 0.05), and between FGG1/FGG2 and YNG/HNG along PC2 (t-test, p < 0.05) (Fig. 1G; Additional file 1: Fig. S1).
Taken together, based on these results, categorizing the litchi population into four groups may be more accurate than the previous classification of two major groups previously [2]. It is worth noting that incomplete lineage sorting could potentially hinder the finer classification of FGG1/FGG2, and more representative samples may clarify this.
Comprehensive characterization of fruit phenotypic traits in litchi germplasm
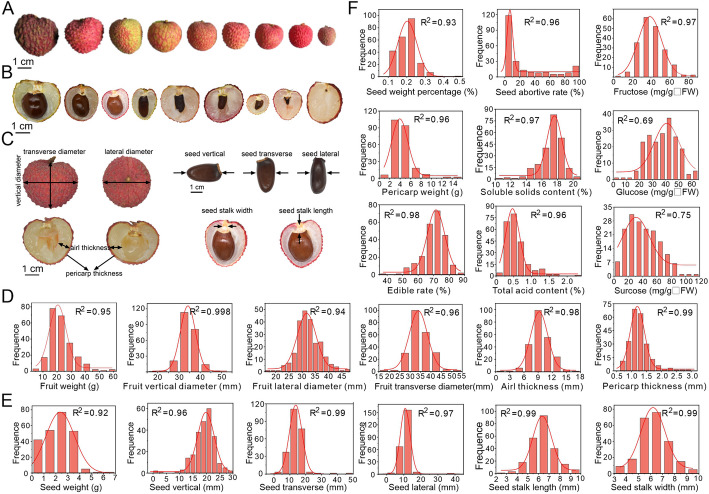
Fruit is the main commercial organ of economic importance for litchi. For the 276 litchi germplasm we studied here, their fruits were of great diversity in many aspects, for instance, the fruit size and the seed size (Fig. 2A, B). We conducted a broad collection of as many as 21 fruit phenotypic traits in 2020 and 2021 (Fig. 2; Additional file 1: Fig. S2). These traits encompass various aspects of fruit phenotypes, including overall fruit features (fruit weight, fruit longitudinal diameter, fruit transverse diameter, fruit lateral diameter, fruit peel thickness, fruit peel weight, fruit aril thickness, Fig. 2C, D), seed features (seed weight, seed longitudinal diameter, seed transverse diameter, seed lateral diameter, seed stalk length, and seed stalk width, seed weight percentage, Fig. 2C and E), and other quality-related traits (seed abortive rate, edible rate, TSS, contents of total acid, sucrose, glucose, and fructose, Fig. 2F). It was observed that almost all the 21 traits displayed wide variation in the 276 litchi varieties (Additional file 2: Table S2), for example, the fruit weight ranged from 5 to 60 g, with the majority of fruits being 20–30 g (Fig. 2D). But for the seed abortive rate, most of the accessions born normal (non-abortive) seed, with only a few varieties producing fully abortive seed (100% abortive rate), which is preferred by consumers (Fig. 2F). Most of these phenotypic traits represented a continuous distribution that fits the normal distribution very well, with only two traits (the content of glucose and sucrose) showing a relatively low conformity (R2 > 0.69, Fig. 2D–F). Therefore, all these fruit phenotypic traits can be used for the association study with the genotypic variations in our large collection of litchi germplasm.
Fig. 2.
Phenotypic trait assessment of litchi fruit. A Litchi fruit size displays extensive variation. B Litchi seed size exhibits a wide range of variations. C Diagram illustrating the phenotypic traits of litchi fruit. D to F Histogram of all 21 fruit traits showed a normal or approximate normal distribution in 2020 year
Phenotype-driven shaping of population structure
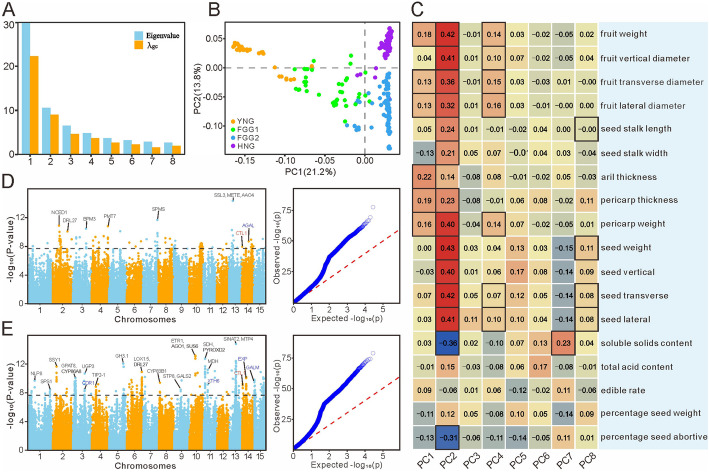
Natural selection is the driving force of evolution. To identify genomic loci under selection, EigenGWAS was applied in our population analysis [24]. The eigenvalues of population structure for all 314 samples all exceeded λgc, indicating natural selection acting across multiple principal components (Fig. 3A). Principal component analysis revealed that PC1 and PC2 accounted for 21.2% and 13.8% of the total variation in population genotypes, respectively (Fig. 3B). Intriguingly, PC2 notably divided the litchi populations into two groups: FGG1/FGG2 and HNG/YNG (Fig. 3B). The latter group encompassed a substantial number of wild litchi individuals, suggesting that artificial selection for fruit and seed size could be involved in the domestication of wild litchi into cultivated ones (Additional file 1: Fig. S3). In contrast, PC1 distinguished the YNG group from the FGG2/HNG group, which inhabit distinct ecological niches, with YNG usually exhibiting early maturation and FGG2/HNG later maturation [2, 25] (Fig. 3B).
Fig. 3.
Phenotype-driven shaping of population structure. A Distribution of eigenvalues of population structure. B PC2 clearly separates the litchi populations into two groups, HNG/YNG, including most wild accessions, and the FGG1/FGG2, mostly containing cultivated varieties. C Heatmap displayed Person correlation coefficient between 18 phenotypic traits and different PCAs. Square with a dark frame shows correlation of significant difference (p < 0.01, t-test). The Manhattan and QQ plot for EigenGWAS association analysis using the PC2 (D) and PC4 (E) as traits
Correlation analysis between eigenvector and 18 fruit-related traits revealed that PC2 was remarkably positively correlated with fruit weight and size as well as seed weight and size, while showing a strong negative correlation with TSS and seed abortion rate (Fig. 3C; Additional file 1: Figs. S4, S5). PC1 and PC4 also exhibited noticeable correlations with fruit weight and size, albeit with lower coefficients. PC1 also demonstrated moderate associations with pericarp and aril thickness (Fig. 3C). For these traits, PC2 represented strong associations with the majority of them, encompassing 77.8% (14/18), followed by PC4 and PC1 at 38.9% (7/18) and 33.3% (6/18), respectively (Fig. 3C).
GWAS was conducted using the PC2 eigenvector as the phenotype, revealing 124 significantly associated loci distributed across chromosomes 2, 7, and 13 (Fig. 3D). Similarly, the GWAS involving PC4 detected 336 significantly associated loci (Fig. 3E). Within the ± 50 kb of loci associated with PC2 and PC4, there were 534 and 1273 genes, respectively, with an overlap of 37 genes (Additional file 1: Fig. S6; Additional file 2: Table S3). Notably, many associated loci were also detected on other principal components, such as PC1 and PC3 (Additional file 1: Fig. S7). For the genes associated with PC2, predominant involvement was observed in hormone synthesis and transport, such as 9-cis-epoxycarotenoid dioxygenase (NCED1) implicated in ABA biosynthesis, spermine synthase (SPMS) in spermine biosynthesis, and choline transporter-like protein 1 (CTL1) in PIN trafficking (Additional file 2: Table S4; Additional file 1: Fig. S8).
Genes potentially involved in seed abortion
Litchi fruits with no or small seeds are favorable in the market. Small seeds often come from the embryo abortion in the process of seed development. Abortive-seed fruit exhibits notable benefits, like a thicker aril (the flesh of the fruit) and a higher edible rate, and consequently, abortive-seed has become an important desirable trait in the cultivation and breeding of litchi (Fig. 4A).
Fig. 4.
Profiling of candidate genes associated with seed abortion in litchi. A Fruits of abortive-seed (above) and normally developed large-seed (below) varieties. B Morphological changes during seed development in abortive-seed type (NMC: Nuomici, HZZ: Hongzhenzhu) and normal large-seed type (YS: Yeshenglizhi #10, HY: Heiye). DAA: days after anthesis. Bars (“-”) represent 0.2 mm at 0–20 DAA and 2 mm at 25–70 DAA for all four accessions. C Venn diagram showing the overlap among the PC2-associated genes, PC4-associated genes, and differential expressed genes (DEGs) identified by RNA-seq at 10–25 DAA. D to E, I to J Expression levels of candidate genes validated by qRT-PCR. H The Manhattan plot for GWAS analysis on seed transverse diameter. The dashed line represents the significance threshold, and statistical analysis was conducted using a two-tailed Wald test
In the seed development process of litchi varieties prone to early embryonic abortion, “Nuomici” (NMC) and “Hongzhenzhu” (HZZ), seed abortion was noted 10–15 days after anthesis (DAA) (Fig. 4B). With the seed development, endosperm in these varieties progressively diminished and vanished at approximately 25 DAA, leading to embryo development deficiency and eventually the formation of smaller aborted seeds (Fig. 4B). In contrast, for normal large-seed varieties, “Yeshenglizhi #10” (YS) and “Heiye” (HY), they commenced endosperm production at 10 DAA and started to develop cotyledonary embryo after 25 DAA, ultimately into plump, large seeds. Hence, the crucial period for early seed abortion in litchi is identified from 10 to 25 DAA (Fig. 4B).
Therefore, we conducted transcriptome analysis of the early seed development process (0–35 DAA) for these four litchi varieties and identified a total of 2136 differential expression genes (DEGs) (p < 0.01 and fold-change > 2) for the period of 10–25 DAA when the seed abortion occurs (Additional file 2: Table S5). Among them, one gene (LcCTL1, LITCHI004494), encoding a choline transporter-like protein (CTL1), was also significantly associated with both the PC2 and PC4 eigenvectors (Figs. 3D, E and 4C). LcCTL1 is of higher expression at 10–25 DAA of abortive-seed varieties, especially in HZZ (Fig. 4D). It is reported that AtCTL1 regulates intracellular trafficking of PIN-type auxin transporters and affects seedling growth in Arabidopsis [26]. Twenty-one other PC2-associated genes and 44 PC4-associated genes were also found to be differentially expressed (Fig. 4C; Additional file 2: Table S6), and some of them are likely involved in seed development. For instances, the PC2-associated gene (LITCHI006014), encoding the alpha-galactosidase (AGAL1) important for cell wall loosening and expansion [27], expressed at a remarkably higher level in abortive-seed varieties at 5–15 DAA (Fig. 4E). Two PC4-associated genes (LITCHI005289 and LITCHI027369) showed higher expression in large-seed varieties in most stages of seed development (Fig. 4F- G), especially for the latter which encodes constitutive disease resistance 1 (CDR1) protein with aspartic protease activity. The LITCHI005289 is a homolog of Arabidopsis expansin-A25 (EXPA25), which plays a potential role in seed development [28].
We also conducted GWAS analysis directly using the six seed phenotypes. A total of 66 significantly associated SNP signals were identified (Additional file 1: Figs. S9, S10; Additional file 2: Table S7), and eight SNP loci were specifically associated with seed transverse diameter (Fig. 4H). A few protein-coding genes were found close to these associated loci and are likely involved in seed development (Additional file 1: Figs. S11, S12), such as LITCHI000638 and LITCHI001708 in chromosome 5 (Fig. 4H) and LITCHI021007 in chromosome 12 (Fig. 4H). LITCHI000638 encodes wall-associated receptor kinase gene 22 (LcWAK22), belonging to the WAKs/WAKLs family of roles in plant cell elongation and expansion and female gametophyte formation [29]. LITCHI001708 encodes UDP-glucosyltransferase 76E1 (LcUGT76E1), one of whose homologs (GhUGT71C4) regulates the seed size by affecting phenylpropanoid metabolism in cotton [30]. LITCHI021007 encodes a NAC transcription factor, and it is close to the AtNAC41 gene which is crucial in regulating the development of endosperm cells in Arabidopsis [31]. Transcriptome and qRT-PCR analysis revealed that the expression of LcWAK22 was significantly higher in abortive embryos compared to large-seed during 5–15 DAA (Fig. 4I). In contrast, expression levels of LcUGT76E1 and LcNAC41 were notably lower in abortive materials in the whole process of seed development (Fig. 4J - K). These observations indicate that these genes are likely critical factors regulating the early development of litchi seed embryos and affecting the seed formation ultimately.
LcSAI gene controls sugar composition in litchi fruit
The sugar content serves as a significant determinant in assessing the taste profile of fruits. According to the composition of main sugars in the flesh, litchi fruit can be classified into three types: sucrose-accumulated (SA) type which preferentially accumulate sucrose (sucrose/reducing sugar > 1), reducing sugar (glucose and fructose) accumulated (RA) type with predominant reducing sugar (reducing sugar/sucrose > 2), and intermediate type (IA) with the ratio of reducing sugar to sucrose ranging from 1 to 2 [32]. In this study, we determined the sugar content and composition of fruits for 190 litchi varieties by high-performance liquid chromatography (HPLC) (Additional file 2: Table S8). Subsequently, with the content of sucrose and glucose as traits, GWAS analysis detected a strong association peak on chromosome 7 (Fig. 5A; Additional file 1: Fig. S13), and it was situated within the gene body region of the gene LITCHI009111 (Fig. 5B). This gene encodes a beta-fructofuranosidase, also referred to as invertase (Fig. 5B), which catalyzes degradation of sucrose into glucose and fructose. Hence, we considered this gene as the best candidate gene controlling sugar composition and content in litchi, and named it as LcSAI (soluble acid invertase).
Fig. 5.
LcSAI is essential for the sugar accumulation in litchi fruit. A The Manhattan plot of GWAS analysis on fruit sucrose content. Dashed line represents the corrected p value cutoff with the Bonferroni method. B Three significantly associated SNPs (red points) all fell within an intron of the LITCHI009111 gene (upper). The LD heatmap of the 109 SNP loci around the significant peaks (bottom). C The reducing sugar and sucrose content of varieties with different genotypes at the SNP site Chr7:22,496,380. Box plots show the median, box edges represent the interquartile range, and whiskers represent the maximum and minimum data points. D The expression of the LcSAI gene in litchi sucrose accumulation (A:A) and reducing sugar accumulation (A:G/G:G) types. E The expression levels of LcSAI gene in wild and T1 transgenic tomato fruits. Error bars represent the standard deviation of three replicates. F The enzyme activity of LcSAI in wild and T1 transgenic tomato fruits. G Sucrose, fructose, and glucose content in wild and T1 transgenic tomato fruits. H Fruits of wild and transgenic tomato
Through phylogenetic analysis of LcSAI with invertases from other species, it is observed that LcSAI was clustered with two vacuolar invertases (VI1/VI2) from Arabidopsis thaliana (Additional file 1: Fig. S14). In Arabidopsis, VIs can hydrolyze sucrose into reducing sugars (glucose and fructose), thereby altering the ratio of sucrose to reducing sugar and impacting plant growth and development in multiple ways [33]. It was found that the SNP with the highest associated significance was at the position 22,496,380 bp of chromosome 7, which is located in the second intron of the LcSAI gene. It can be either adenine (A) or guanine (G) in litchi population. The homozygous “A:A” genotype was associated with comparable level of sucrose and reducing sugar content (Fig. 5C). In contrast, varieties carrying heterozygous “A:G” or homozygous “G:G” at the position were of a much higher level of reducing sugar content, but a lower level of sucrose content (Fig. 5C), with the “G:G” genotype represented the highest level of reducing sugar content and the lowest level of sucrose content (Additional file 1: Fig. S15). These composition trends suggested that the “G” allele is positively associated with the reducing sugar content in litchi aril, and mainly present in the RA/IA type of litchi varieties.
Next, we compared the expression level of LcSAI in the two classes of litchi varieties and found that LcSAI was expressed at a significantly higher level in the RA/IA-type (“A:G” or “G:G”) accessions than that SA-type (“A:A”) accessions, a few of which were even of almost no expression detected (Fig. 5D). These observations suggested that the “G” allele of the LcSAI gene in the RA/IA type enabled a high-level expression of LcSAI, which subsequently stimulated the conversion of sucrose to reducing sugars. To verify the function, the LcSAI was expressed in Escherichia coli, and the enzyme activity of LcSIA-GST was significantly higher compared to the control (Additional file 1: Fig. S16A).
To further confirm the function of the identified LcSAI gene, we overexpressed a copy of it in the tomato (Micro-Tom). Compared to the wild type, the vegetative status of transgenic plants was not affected. It was observed that expression of the LcSAI gene and the enzyme activity of LcSAI were significantly higher in the overexpressed tomato fruits (OE1, OE2, and OE3) compared to the wild type (Fig. 5E, F, Additional file 1: Fig. S16B, C). Consequently, the content of reducing sugars was greatly increased in the transgenic fruits (Fig. 5G; Additional file 1: Fig. S16D), demonstrating the enzymatic function of LcSAI catalyzing the conversion of sucrose to reducing sugars. Noticeably, the size of fruit from transgenic lines was smaller than that of the wild type (Fig. 5G). Taken together, the identified invertase gene LcSAI is a key regulator of sugar content accumulation in litchi fruits.
Development of molecular marker associated with sugar composition
Next, we checked the genomic variations along the LcSAI gene locus, especially potential structural variations, which have not been examined thoroughly in this study. Our analysis revealed that there was a 524-bp deletion (Chr7:7,985,725-7986251bp) in the second intron region proximal to the GWAS peak of LcSAI gene mainly in the SA type materials, and the deletion and the GWAS peak was in strong linkage (Fig. 6A). The genomic read coverage in the deletion region is significantly lower in the A:A genotype which was associated with the SA-type accessions than the A:G/G:G genotypes linked with RA/IA type (Fig. 6B, C). The accessions with the deletion (heterozygous or homozygous) had higher level of sucrose and lower level of reducing sugar (Fig. 6D). A molecular marker consisted of two PCR primers designed based on the deletion could accurately distinguish the sugar accumulation types of different litchi varieties (Fig. 6E, F; Additional file 2: Table S9). Specifically, the SA type predominantly yielded a single band of approximately 700 bp, while the RA type a single band of approximately 1150 bp, and the IA type exhibited both bands concurrently. The results indicated the molecular marker developed from the sugar-type associated deletion has the potential to be used in the breeding of litchi varieties for desired sugar composition.
Fig. 6.
Development of a molecular marker for sugar signal-associated loci. A Mapping data reveals a 524-bp deletion in the second intron of the LITCHI0009111 gene in sucrose accumulation type. p1 and p2 marked the PCR primer locations for genotyping. GWAS peak shows the associated SNP loci (SNP9 in GW genome, also corresponds to Chr7:22,496,380 A:G in FZX genome, Additional file 2: Table S10). B The AA genotypes exhibited significantly lower read coverage in the deleted region compared to the AG and GG genotypes. Box plots show the median, box edges represent the interquartile range, and whiskers represent the maximum and minimum data points. Statistics were performed with Student’s t-test. C Both the sucrose accumulation type (SA) and intermediate type (IA) showed significantly lower coverage compared to the reducing sugar accumulation type (RA) in the 524-bp deletion region (light blue region). D The boxplot of reducing sugar (fructose and glucose) and sucrose content among −/−, +/−, and +/+, which corresponds to values 0–0.5, 0.5–1, and > 1 according to the coverage of deletion region divided by genome-wide median coverage. E The 524-bp deletion can be used as a molecular marker to distinguish between SA, IA, and RA accessions. Expression of LcSAI was included above or below the gel graphs. “NA” marks accessions in which the expression of LcSAI was unknown. F The band type was significantly correlated with the sugar accumulation type (SA, IA, and RA). Fisher’s precise test was used for statistics analysis
Discussion
In this study, a comprehensive investigation of genetic variations in litchi germplasm resources was carried out through large-scale resequencing and population analysis. We obtained genomic data from a large natural population comprising 276 litchi germplasm accessions [25] (Additional file 2: Table S1), comprising nine wild germplasms and 267 landraces or cultivars, and many of them were collected from eastern and northern Guangxi, coastal areas of Fujian, and Taiwan (Fig. 1A), which were not covered previously [2]. This population contains representative varieties from all litchi-producing regions in China as well as some foreign accessions. Litchi is a perennial fruit tree generally propagated vegetatively in cultivation. After excluding 37 clonal samples identified in our clonal and pedigree analysis [34, 35], we detected no parent-offspring (PO) pairs and only a few full-sibling (FS) relationships among the remaining accessions. This is in marked contrast to similar collections in apples and grapes, where PO, FS, and clonal relationships are routinely observed [36, 37]. The scarcity of close kinship in our dataset is unexpected and remains an intriguing topic for future investigation.
Using this large population, as many as 3.54M high-quality SNPs were identified, comprising to date the most comprehensive dataset of genetic variations for litchi. In our previous study, the curated 72 accessions were divided into two major clusters: I and II, with cluster I including YNW, VNW, GXDXW, and EEMC (extremely-early maturing cultivars), and cluster II comprising HNW, GXBBW, and LMC (late maturing cultivars) [2]. Taking advantage of the larger population with more representative litchi accessions, this study categorized litchi germplasms into four subgroups: HNG, FGG2, FGG1, and YNG (Fig. 1B). YNG corresponds to the I cluster; HNG contains wild and cultivated litchi from Hainan and includes many accessions from adjacent areas of Guangxi and Guangdong, including GXBBW; FGG1 and FGG2 predominantly contain EEMC/EMC (early maturing cultivars) and LMC cultivars, respectively (Additional file 1: Fig. S17). The new classification shows that the majority of landraces and cultivars are included in FGG1/FGG2, which exhibit a mixture of genetic components from HNG and YNG (Fig. 1F). This phenomenon is consistent with previous studies suggesting that YNG overlaps with HNG in the border area between Guangxi and Guangdong provinces, which served as the center of litchi domestication [2]. Subsequently, cultivated litchi spread to the entire provinces of Guangdong and Fujian, before disseminating to Southeast Asia and the rest of the world [38]. These new knowledges gained from population classification and demographic history analysis will facilitate the further understanding of litchi genetic diversity and the genomics-assisted breeding.
In combination with the profiling of 21 fruit phenotypic traits, this study identified a total of 460 SNPs significantly associated with fruit traits and 1807 candidate genes (Fig. 3E; Additional file 2: Table S3). A total of 66 significantly SNPs and seven candidate genes were identified to be associated with abortive-seed formation (Fig. 4C; Additional file 2: Table S7). Their homologous genes play important roles in plant seed development (such as AGAl1 [27], WAK22 [29], UGT76E1 [30], NAC41 [31]). Small abortive-seed or seedlessness is a desirable trait in many fruit crops. A few genes related to seed development have been identified in citrus and grape, such as CitRWP [7], VvMADS39 [39], and VvAGL11 [40]. Seed abortion in litchi is a complex genetic characteristic that can be influenced by environmental factors, including temperature and pollen sources [10], and only a few related genes have been documented [20, 41]. For instance, silencing of cell wall invertase genes (LcCWIN2 and LcCWIN5) results in smaller seeds in litchi.
Among the various factors affecting litchi fruit quality, sweetness is a key determinant of flavor. Previous studies have shown that different sugar components are major contributors to fruit sweetness [13–15, 17]. Among them, fructose, as the sweetest soluble sugar, plays a decisive role in determining the sweetness of the fruit [42, 43]. However, high sucrose content is a distinctive feature of date palm fruit and affects flavor as well as texture and water retention [16]. Soluble sugars in litchi fruit mainly consist of sucrose, glucose, and fructose. Fruits of most litchi germplasms mainly accumulate reducing sugars (fructose and glucose), which play a regulatory and balancing role in the flavor of litchi, resulting in a more delicate and delicious sweetness [44, 45]. Previous studies have reported on the accumulation of different types of sugars in litchi [19, 33, 46] and highlighted the role of sucrose cleavage enzymes and sucrose synthase [33] in influencing on sugar accumulation pattern in litchi flesh. In this study, we identified an invertase gene, LcSAI, which catalyzes the conversion of sucrose into glucose and fructose. The function of LcSAI in sugar conversion is consistent with that of its homologous genes reported in tomatoes and other plants [20, 33]. In addition, glucose is crucial for early embryo development in many plants, which serving as a stimulatory signal for cotyledon embryo formation [47]. In maize and rice, a higher ratio of glucose to sucrose can stimulate cell division during the early stages of grain development, leading to the production of more endosperm cells and reducing abortive-seed rates [48]. These results suggest that LcSAI may play a role in early seed development, similar to their homologous genes [20]. However, further investigation is needed to validate this speculation.
Conclusions
In this study, a comprehensive analysis of 276 litchi accessions refined the population structure into four distinct groups, providing deeper insights into the crop’s domestication history. Through the integration of large-scale genomic data with the profiling of 21 fruit-related phenotypes, we have delineated the genetic architecture of crucial quality traits. Notably, we identified and characterized a soluble acid invertase gene, LcSAI, as the key regulator of sugar composition in litchi fruit. A structural variation within this gene was found to be strongly associated with the accumulation of either sucrose or reducing sugars. Several candidate genes potentially involved in seed abortion were identified as well. Collectively, these findings provide valuable genomic resources and a foundational understanding of fruit quality genetics, which will greatly accelerate the genetic improvement and molecular breeding of litchi.
Methods
Plant material and growth conditions
A total of 314 litchi accession samples were used in this study, including 38 wild litchi samples from the previous study [2] (Additional file 2: Table S1), and a comprehensive collection of 276 litchi core germplasm resources was obtained from the National Litchi Germplasm Repository (Additional file 2: Table S1), Institute of Fruit Tree Research, Guangdong Academy of Agricultural Sciences, Guangzhou, Guangdong Province. The orchard, situated on mildly undulating topography, has nurtured these germplasm trees for a period exceeding 10 years. The consistent soil fertility and management practices implemented in the orchard guarantee an annual occurrence of flowering and fruiting.
Phenotypic data collection
All of the measurements for the agronomic traits were based on the Description and data standard for litchi (Litchi chinensis Sonn.) [49]. Data were collected in 2020 and 2021. Fifty fully mature litchi fruits were selected from each variety for phenotypic measurements. The weight of individual fruits (FW, g), pericarp (PW, g), and seeds (SW, g) was measured using a Mettler Toledo balance with a precision of 1/1000th of a gram. The longitudinal, transverse, and lateral diameters (mm) of both the fruit and seed were measured using a digital vernier caliper. The developmental stage of the embryo (normal seed/aborted seed) was observed and recorded. The Edible rate (%), Seed weight percentage (%), and Seed abortive rate (%) are calculated from the fruits and seed data. Edible rate = (FW − SW − PW)/FW*100, Seed weight percentage = SW/FW*100, Seed abortive rate = abortive-seed number/total seed number*100. The pulp of the fruit was juiced, and the content of soluble solids (TSS) and total acidity in individual fruits were measured using a digital refractometer (ATAGO, Japan). Sucrose and reducing sugars (glucose and fructose) were quantified using high-performance liquid chromatography (HPLC) on an Agilent 1260 Infinity HPLC system following the methods described in a previous report [32]. Each sample was measured three times, and the average value was recorded. The statistical analysis and data visualization of the phenotypic traits were performed using Origin software (version 2022). Statistical analysis was conducted using SPSS software (SPSS 27.0, SPSS Inc.). One-way analysis of variance (ANOVA) was employed for the statistical analysis. The significance level for the ANOVA was set at p = 0.05 or 0.01.
Resequencing and SNP calling
The genomic DNA was extracted from the fully expanded and green litchi tender leaves of each accessions using Super Plant Genomic DNA Kit (Polysaccharides & Polyphenolics-rich, TIANGEN). Then, the 350-bp whole-genome libraries were constructed for each accession and sequenced on the Illumina sequencing platform (Illumina Novaseq6000 sequencer) by Biomarker Biotechnology Company (Beijing), generated a total of 8G (average 20 ×) clean raw data with a read length of 150 bp per sample (Additional file 2: Table S1 for more details). The software with parameter settings of the quality control, adapter trimming, mapping, alignment, and SNP calling methods used for the raw resequencing data of 276 accessions, which were the same as resequencing data of 72 accessions as described in our previous study [2]. The trimmed reads were mapped to the reference litchi genome (Feizixiao), and the 9.48 million biallelic SNPs were kept with parameter “–maf 0.01 –geno 0.2” using PLINK2 [34].
Clonal and pedigree analyses
To ensure the accuracy of population analysis, the putative clones were examined and removed, as litchi is a perennial woody fruit tree of vegetative propagation. The 9.48 M SNPs were filtered using strict parameters (“–maf 0.05 –geno 0.05 –hwe 0.001”) in PLINK2 [34], resulting in 3.59 M SNPs. These were then analyzed using the KING software with the “–kinship” parameter to infer relatedness [35]. Samples with a kinship value of ≥ 0.354 were classified as putative clonal, leading to the removal of 37 clones. Following this, 3.54M SNPs were retained for subsequent population structure analyses.
Phylogenetic tree
Fourfold degenerate sites were obtained across the entire genome using custom scripts. These sites were then intersected with 3.54M SNPs, yielding 74.6k shared sites. Then, a maximum likelihood tree was constructed with the optimal model TVM + F + ASC + R6 using IQ-TREE (version 1.6.10) [50]. And ultrafast bootstrap with 1000 replicates was conducted using “-bb 1000 -bnni” parameters. The resulting tree was visualized using FigTree software (https://github.com/rambaut/figtree/releases/tag/v1.4.3) and evolview (version 2) [51]. The construction of the phylogenetic tree of LcSAI (Additional file 1: Fig. S13) proceeded as follows: First, proteins of 11 species, such as Vitis vinifera and Solanum lycopersicum, were retrieved from public databases as our study previously described [2]. The sequences of INVERTASE from Arabidopsis thaliana (AtVI1 ~ 2, AtCWI1 ~ 6) were subjected to blastp comparison against the aforementioned protein sequences, using an e value threshold of 1e–5. The resulting protein sequences were utilized for constructing maximum likelihood trees using MUSCLE (version 3.8.31) [52] and TrimAL (version 1.4.22) [53] with default parameters. Additionally, IQ-TREE [50] was employed with the same parameters as previously mentioned.
Population structure and genetic differentiation analysis
The hard-filtered 3.54M SNPs were pruned using the recommended parameters “-indep-pairwise 50 10 0.1” in ADMIXTURE software [54], resulting in 439.8k SNPs for population structure analysis. The optimal number of populations (K) ranged from 2 to 10 was determined using tenfold cross-validation with the 439.8k SNPs. Linkage disequilibrium (LD) in population groups were calculated by PopLDdecay (3.40) [55] software using the correlation coefficient (r2) of 3.54M SNPs. The resulting output was input into R, and a linear model was applied to the data using the “lm” function with log10 transformed distance vs. R2 value data, and the point where R2 dropped to 50% of the initial value was reported as the measure of decay. PCA was calculated for evaluating genetic structure using KING. Fixation statistics (FST) and nucleotide diversity (θπ) were calculated using VCFtools software (version 0.1.15) [56] in windows of 100 kb.
GWAS analysis
To better utilize the phenotypic data, the clonal lines were included in our GWAS analysis. The 9.48M SNPs were pruned using “-indep-pairwise 50 5 0.5,” resulting in 4.43M SNPs. The 4.43 million SNPs were further filtered using “–maf 0.05,” and 38 wild litchi samples lacking phenotypic data were removed. Ultimately, 2,442,670 SNPs were retained from 276 litchi varieties and were used in association with 21 phenotypic traits. The EigenGWAS analyses were conducted using a linear mixed model [24] without covariate matrix in R package (version 3.5.2). When using the mixed linear model (MLM) in TASSEL [57] for association analysis, structure results (Q matrix from admixture analysis), kinship (K matrix), and PCA results were used individually or in combination to correcting for population structure. In the case of the MLMM model for six phenotypic data of seed trait, we conduct association analysis using both the Bayesian-information and linkage-disequilibrium iteratively nested keyway (BLINK) [58] and fixed and random model circulating probability unification (FarmCPU) [59] algorithms simultaneously with R-package GAPIT version 3 [60] (http://www.zzlab.net/GAPIT/GAPIT.library.R). The significance threshold for genome-wide association was determined based on a false discovery rate (FDR-adjusted p < 0.05).
Functional annotation of candidate genes in the target region
Access the Litchi genome database (http://www.sapindaceae.com/), enter the Genome Browser (Litchi genome) interface. The gene IDs of candidate genes were obtained through using a sliding window of + 50 kb upstream and downstream of the physically associated trait loci, and perform a BLAST search to compare the candidate region genes with functional databases such as NR, SwissProt, GO, COG, and KEGG for functional annotation. Further compare with the Arabidopsis genome (https://www.arabidopsis.org/) to preliminarily determine the biological functions of homologous genes in Arabidopsis within the candidate region. Concurrently, integrate transcriptome data to preliminarily identify genes with significant differences in expression levels as candidate genes.
Sampling of developmental seeds
In this study, two abortive-seed varieties, “Nuomici” and “Hongzhenzhu,” were selected because both of them have stable rates of seed abortion. “Nuomici” is an old cultivar well-known for its small seeds coming from stable seed abortion. “Hongzhenzhu” is a new cultivar we selected recently. Our field trials of multi-year (at least 3 years) and multi-location revealed that the abortive seed rate of “Hongzhenzhu” is consistently above 95%, indicating a high stability of seed abortion. When sampling, we also manually removed non-abortive seeds as aborted seeds are usually smaller, deformed, or lacking certain anatomical structures, which could be distinguished as early as 10 DAA (as shown in Fig. 4B). Two large-seed varieties, “Yeshenglizhi #10” (YS) and “Heiye” (HY), with normal seed development, were selected for comparison.
The cross section of seed development at 0, 5, 10, 15, 25, 30, and 35 days after anthesis (DAA) are from two large-seed (YS and HY) and abortive-seed varieties (NMC and HZZ), the images were taken under a stereomicroscope (Zeiss, Discovery.V20, Germany), and pictures of other fruits and seeds were all taken with a Nikon camera (Nikon, D850, Japan). All the fresh seeds of two type (large- and abortive-seed) varieties was collected described above from 0 to 35 DAA, which was frozen rapidly in liquid nitrogen and stored in a − 80 °C freezer for subsequent RNA sequencing and qRT-PCR analysis.
RNA-seq and data analysis
The transcriptomic data were generated from different seed development stages (0, 5, 10, 15, 20, 25, and 35 DAA) of two large-seed (YS and HY) and abortive-seed varieties (NMC and HZZ) by the majorbio company (www.majorbio.com). A total of 1 μg RNA was used for library construction following Illumina® Stranded mRNA Prep, Ligation from Illumina (San Diego, CA). RNA-seq sequencing was sequenced with the NovaSeq 6000 sequencer (2 × 150 bp read length). Each sample was sequenced with three biological replicates. Clean reads were separately aligned and mapped to the litchi genome using HISAT2 [61] software. The raw count of each gene was calculated by “prepDE.py” according to the StringTie method [62]. Both the raw count of large-seed and abortive-seed samples were as input to DESeq2 [63]. The DEGs were evaluated in |log2FC| ≧ 2 and adjusted p value < 0.01. The heatmap of expression level of DEGs were visualized by TBtools II (V2.025) [64].
Quantitative reverse transcription (qRT)-PCR expression analysis
Total RNA was extracted using the RNAprep Pure Kit (Polysaccharides & Polyphenolics-rich, TIANGEN). cDNA synthesis was performed using PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time, TakaRa). qRT-PCR was performed in a CFX96 real-time PCR system (Bio-Rad, CA) using iTaq Universal SYBR Green Supermix (Bio-Rad). Primers are shown in Additional file 2: Table S11. LcActin [65] or ubiquitin (UBI; serial number: Solyc01g056940) was used as reference gene to normalize the transcript levels of target genes for litchi and Micro-Tom tomato. Software was utilized to calculate the relative expression level based on the ratio of the target gene and the reference gene with three biological replicates. The statistical analysis and data visualization were using GraphPad prism 8.0 software (GraphPad Software).
Vector construction and genetic transformation
The coding sequence (CDS) of LITCHI009111 m3, obtained from the reference genome “Fuzixiao,” was synthesized by a commercial company (Tsingke Biotech Co., Ltd.). The CDS was designed with XbaI and pstI restriction enzyme sites and was constructed into the super 1300 vector, which carry chloramphenicol resistance gene and were under control of the 35S promoters. And transformed into Agrobacterium tumefaciens strains (GV3101), which was used for tomato (Micro-Tom) genetic transformation as reported previously [66]. The transgenic plants were selected on MS medium with antibiotic selection of the construction vector. The genetic transformation and planting of experiments were conducted in a greenhouse at 26 °C with a light cycle of 16 h light and 8 h darkness. Positive transgenic plants were screened using PCR amplification with genomic DNA; the sequence across the LITCHI009111 was amplified and sequenced. The primers used for amplification are listed in Additional file 2: Table S11 and T1 generation over-expression plants were selected for subsequent fruit analysis.
Enzyme activity determination
For the enzyme activity of LcSAI in E. coli strain Rosetta (DE3), the sequence of LcSAI (LITCHI00911m3 CDS, 643 aa) was constructed into the pET28a-GST and transformed into E. coli Rosetta (DE3) cells. The DE3 cells were harvested by centrifugation at 5000 × g for 30 min and then resuspended in a lysis buffer (phosphate‐buffered saline, pH 4.8, 1 mmo/L phenylmethanesulfonyl fluoride (PMSF), 0.2 mg/mL lysozyme, and 0.02 mg/mL DNase I), followed by disruption with a sonicator. Cell debris was removed by centrifugation at 14,000 × g for 30 min. The supernatant was then applied on an affinity (glutathione S‐transferase (GST)) column. The target protein was eluted using a 10 mmol/L GSH. The microplate reader (SpectraMax ABS plus, Molecular Devices, CA, USA) was used to measure the concentration of purified proteins by soluble protein BCA method. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis was performed to verify the purity protein.
All the activities of soluble acid invertase (including in DE3, the fruits of tomato in transgenic and WT lines) were measured using a biochemical reagent kit (NM-W-0223, Norminkoda Biotechnology Co., Ltd. Wuhan, China). The principle is that invertase catalyzes the decomposition of sucrose to produce reducing sugar, which further reacts with 3,5-dinitrosalicylic acid to produce a brownish red amino compound. It has characteristic light absorption at 540 nm, and within a certain range, the light absorption value at 540 nm is proportional to the amount of reducing sugar produced. The production of 1 μg reducing sugar per milligram of protein per minute is defined as one enzyme activity unit (U).
Marker development
Primer design is based on the deletion region in the second intron of the LITCHI009111 genomic sequence in two types of materials. PCR amplification was performed using PrimeSTAR® Max DNA Polymerase (Takara) with the following program: 98 °C for 10 s, 60 °C for 5 s, 72 °C for 1 min, for a total of 35 cycles. The amplification reaction was carried out in a T100 Therma Cycler (BIO-RAD), and all the amplification products were separated by 1.2% agarose gel electrophoresis.
Supplementary information
Additional file 1: Figures S1–S17. This file contains all supplementary figures. Fig. S1 PCA scatter plots of the 277 accessions. Fig. S2 Phenotypic data distribution for 21 fruit traits. Fig. S3 Differences of fruit trait phenotypic data in subpopulations. Figs. S4 and S5 Pearson correlation between 19 traits and different PCAs in 2 years. Fig. S6 Venn diagram of the overlapped genes between the PC2 and PC4 groups. Fig. S7 EigenGWAS of the PC1 and PC3 traits. Fig. S8 The expression level of the 35 seed-related genes assessed by RNA-seq. Figs. S9 and S10 Genome-wide association studies of seed traits by Blink and FarmCPU models in 2 years. Fig. S11 The expression of GWAS associated seed-related genes by MLM model. Fig. S12 Expression levels of candidate seed-related genes validated by qRT-PCR. Fig. S13 Genome-wide association studies of glucose and sucrose contents. Fig. S14 Phylogenetic tree of LcSAI in 11 plant species. Fig. S15 The sugar contents of varieties with different genotypes at the same SNP site. Fig. S16 The function of LcSAI validated in T0 transgenic tomato fruits. Fig. S17 Numbers of accessions related to maturity in four subpopulations
Additional file 2: Tables S1–S11. This file contains all supplementary tables. Table S1 Litchi samples and their origins and sequencing results. Table S2 Statistical traits and phenotypes of the 276 accessions. Tables S3 and S4 Genes in the regions under PC2 and PC4 by EigenGWAS. Table S5 Differential expression genes of the two-type seed development. Table S6 The overlap genes between PC2, PC4, and DEGs. Table S7 Genome-wide association signals of seed traits. Table S8 Sugar composition and contents under HPLC method. Table S9 Genotype and sugar accumulation type of different accessions under PCR amplification. Table S10 Linkage disequilibrium values between 7:22496380 G:A and their nearby sites. Table S11 The primers of qRT-PCRs and markers
Acknowledgments
Peer review information
David Edwards and Wenjing She were the primary editors of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. The peer-review history is available in the online version of this article.
Authors’ contributions
Q.Y., R.X. conceived the project. Q.Y, J.C., L.O., and Y.W. collected the samples and phenotypic data. J.F., Q.Y., K.H. Y.M., and H.L. performed the data analysis. H.L., F.S., Y.H., C.C., C. Y., and L.O. gave suggestions of the analysis and tested the functions. Q.Y., J.F. and R.X. prepared the figures and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work is also supported by the Key Area Research and Development Program of Guangdong Province (2022B0202070003), the National Science Foundation of China (#32102333, #32072547, and #32102320), the Guangxi Science and Technology Major Program (Grant No. GuikeAA23023007-2), the Natural Science Foundation of Guangdong (2021A1515011031), and National Litchi Longan Industrial Technology System (CARS-32–01).
Data availability
The raw sequence data have been deposited in the NCBI Sequence Read Archive (SRA) under accession PRJNA1107979 [25] and PRJNA747875 [2]. The litchi reference genome used in this study is available from the ‘Feizixiao’ of our previous study [2]. All other relevant data are contained within the paper and its Additional files. All other data generated or analyzed during this study are included in the article or its additional files. Specifically, Additional file 1 provides all supplementary figures, which present supporting data for the population structure analysis, GWAS results, and functional gene verification; Additional file 2 contains the comprehensive phenotypic data for 21 fruit traits, lists of candidate genes from GWAS and transcriptome analyses, significant SNP loci, sugar content measurements, and all primer sequences used.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qian Yan, Junting Feng and Jiezhen Chen contributed equally to this work.
Contributor Information
Liangxi Ou, Email: gdglxo2015@126.com.
Rui Xia, Email: rxia@scau.edu.cn.
References
- 1.Chen HB. OU LX, Li JG, SU ZX, Yang SN, WU ZX, Hu ZY. Fruit scientific research in new China in the past 70 years: litchi. J Fruit Sci. 2019;36:1399–413.
- 2.Hu GB, Feng JT, Xiang X, Wang JB, Salojärvi J, Liu CM, Wu ZX, Zhang JS, Liang XM, Jiang ZD, et al: Two divergent haplotypes from a highly heterozygous lychee genome suggest independent domestication events for early and late-maturing cultivars. Nat Genet. 2022; 54:73–83. NCBI Genome. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA747875/. [DOI] [PMC free article] [PubMed]
- 3.Wu JF, Zhang CY, Chen JZ, Cai CH, Wang LM, Fu DW, Ou LX. Morphological diversity within litchi (Litchi chinensis Sonn.) based on leaf and branch traits. Sci Hortic-Amsterdam. 2016;207:21–7. [Google Scholar]
- 4.Wu JF, Fu DW, Chen JZ, Cai CH, Yan Q, Ou LX. Pollen quantity and viability in 65 litchi (Litchi chinensis Sonn.) cultivars. Hortscience. 2017;52:1337–41. [Google Scholar]
- 5.Cao K, Zhou ZK, Wang Q, Guo J, Zhao P, Zhu GR, Fang WC, Chen CW, Wang XW, Wang XL, et al. Genome-wide association study of 12 agronomic traits in peach. Nat Commun. 2016;7:13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan NB, Bai Y, Sun HH, Wang N, Ma YM, Li MJ, Wang X, Jiao C, Legall N, Mao LY, et al. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat Commun. 2017;8:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Xu YT, Zhang SQ, Cao L, Huang Y, Cheng JF, Wu GZ, Tian SL, Chen CL, Liu Y, et al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat Genet. 2017;49:765–72. [DOI] [PubMed] [Google Scholar]
- 8.Liang ZC, Duan SC, Sheng J, Zhu SS, Ni XM, Shao JH, Liu CH, Nick P, Du F, Fan PG, et al. Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nat Commun. 2019;10:1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang MY, Xue C, Hu HJ, Li JM, Xue YS, Wang RZ, Fan J, Zou C, Tao ST, Qin MF, et al. Genome-wide association studies provide insights into the genetic determination of fruit traits of pear. Nat Commun. 2021;12:1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie DR, Ma XS, Rahman MZ, Yang MC, Huang XM, Li JG, Wang HC. Thermo-sensitive sterility and self-sterility underlie the partial seed abortion phenotype of Litchi chinensis. Sci Hortic-Amsterdam. 2019;247:156–64. [Google Scholar]
- 11.Zhang CY, Wu JF, Fu DW, Wang LM, Chen JZ, Cai CH, Ou LX. Soaking, temperature, and seed placement affect seed germination and seedling emergence of Litchi chinensis. HortScience. 2015;50:628–32. [Google Scholar]
- 12.Li JM, Zhu RX, Zhang MY, Cao BB, Li XL, Song BB, Liu ZC, Wu J. Natural variations in the PbCPK28 promoter regulate sugar content through interaction with PbTST4 and PbVHA-A1 in pear. Plant J. 2023;114:124–41. [DOI] [PubMed] [Google Scholar]
- 13.Li MJ, Feng FJ, Cheng LL. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS One. 2012;7:e33055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang HP, Wu JY, Qin GH, Yao GF, Qi KJ, Wang LF, Zhang SL. The role of sucrose-metabolizing enzymes in pear fruit that differ in sucrose accumulation. Acta Physiol Plant. 2014;36:71–7. [Google Scholar]
- 15.Komatsu A, Takanokura Y, Moriguchi T, Omura M, Akihama T. Differential expression of three sucrose-phosphate synthase isoforms during sucrose accumulation in citrus fruits (Citrus unshiu Marc.). Plant Sci. 1999;140:169–78. [Google Scholar]
- 16.Malek JA, Mathew S, Mathew LS, Younuskunju S, Mohamoud YA, Suhre K. Deletion of beta-fructofuranosidase (invertase) genes is associated with sucrose content in date palm fruit. Plant Direct. 2020;4:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang ZY, Wei XY, Yang JJ, Li HX, Ma BQ, Zhang KK, Zhang YF, Cheng LL, Ma FW, Li MJ. Heterologous expression of the apple hexose transporter MdHT2.2 altered sugar concentration with increasing cell wall invertase activity in tomato fruit. Plant Biotechnol J. 2020;18:540–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Zhang Z, Sun X, Liu Z, Ma M, Fan J, Luo W, Wang L, Zhang S. Identification and characterization of invertase family genes reveal their roles in vacuolar sucrose metabolism during Pyrus bretschneideri Rehd. fruit development. Genomics. 2021;113:1087–97. [DOI] [PubMed] [Google Scholar]
- 19.Fan S, Wang D, Xie H, Wang H, Qin Y, Hu G, Zhao J. Sugar transport, metabolism and signaling in fruit development of Litchi chinensis Sonn: a review. Int J Mol Sci. 2021;22:11231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang JQ, Wu ZC, Hu FC, Liu L, Huang XM, Zhao JT, Wang HC. Aberrant seed development in Litchi chinensis is associated with the impaired expression of cell wall invertase genes. Hortic Res. 2018;5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, Zhao J, Qin Y, Qin Y, Hu G. Molecular cloning, characterization and expression profile of the sucrose synthase gene family in Litchi chinensis. Hortic Plant J. 2021;7:520–8. [Google Scholar]
- 22.Wang TD, Zhang HF, Wu ZC, Li JG, Huang XM, Wang HC. Sugar uptake in the aril of litchi fruit depends on the apoplasmic post-phloem transport and the activity of proton pumps and the putative transporter LcSUT4. Plant Cell Physiol. 2015;56:377–87. [DOI] [PubMed] [Google Scholar]
- 23.Wen YJ, Ou LX, Shi FC, Yan Q, Cai CH, Jiang YH, Liu HL, Chen JZ. Conservation status and innovative utilization of litchi resources in the national litchi and banana germplasm repository (Guangzhou). J Plant Genet Resour. 2023;24:1205–14. [Google Scholar]
- 24.Chen GB, Lee SH, Zhu ZX, Benyamin B, Robinson MR. EigenGWAS: finding loci under selection through genome-wide association studies of eigenvectors in structured populations. Heredity. 2016;117:51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan Q, Feng JT, Chen JZ, Wen YJ, Jiang YH, Mai YX, Huang K, Liu HL, Liu HS, Shi FC, Hao YW, Cai CH, Yu CY, Ou LX, Xia R: The genetic basis of fruit quality dissected through comprehensive genomic analyses and phenotypic profiling in litchi (Litchi chinensis Sonn.). Datasets, BioProject. 2024. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1107979/.
- 26.Wang Y, Yang L, Tang YM, Tang RJ, Jing YP, Zhang C, Zhang B, Li XJ, Cui YN, Zhang CH, et al. Arabidopsis choline transporter-like 1 (CTL1) regulates secretory trafficking of auxin transporters to control seedling growth. Plos Biol. 2017;15:e200431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phoeurk C, Somana J, Sornwatana T, Udompaisarn S, Traewachiwiphak S, Sirichaiyakul P, Phongsak T, Arthan D. Three novel mutations in α-galactosidase gene involving in galactomannan degradation in endosperm of curd coconut. Phytochemistry. 2018;156:33–42. [DOI] [PubMed] [Google Scholar]
- 28.Liu ZJ, Zheng LM, Pu L, Ma XF, Wang X, Wu Y, Ming HN, Wang Q, Zhang GF. ENO2 affects the seed size and weight by adjusting cytokinin content and forming ENO2-bZIP75 complex in Arabidopsis thaliana. Front Plant Sci. 2020;11:574316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang N, Huang HJ, Ren ST, Li JJ, Sun Y, Sun DY, Zhang SQ. The rice wall-associated receptor-like kinase gene OsDEES1 plays a role in female gametophyte development. Plant Physiol. 2012;160:696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao YW, Han ZG, He L, Huang CJ, Chen JW, Dai F, Xuan LS, Yan SY, Si ZF, Zhang TZ. UDP-glucosyltransferase 71C4 regulates seed development by redistributing phenylpropanoid metabolism in cotton. bioRxiv. 2023;11:566436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Durme M, Carrillo YO, Pfeiffer ML, Doll NM, De Winter F, Lin ZC, Nowack MK. Fertility loss in senescing Arabidopsis ovules is controlled by the maternal sporophyte via a NAC transcription factor triad. Proc Natl Acad Sci. 2023;120:e2219868120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang ZY, Wang TD, Wang HC, Huang XM, Qin YH, Hu GB. Patterns of enzyme activities and gene expressions in sucrose metabolism in relation to sugar accumulation and composition in the aril of Litchi chinensis Sonn. J Plant Physiol. 2013;170:731–40. [DOI] [PubMed] [Google Scholar]
- 33.Roitsch T, González MC. Function and regulation of plant invertases: sweet sensations. Trends Plant Sci. 2004;9:606–13. [DOI] [PubMed] [Google Scholar]
- 34.Chang CC, Chow CC, Tellier L, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muranty H, Denancé C, Feugey L, Crépin JL, Barbier Y, Tartarini S, Ordidge M, Troggio M, Lateur M, Nybom H, et al. Using whole-genome SNP data to reconstruct a large multi-generation pedigree in apple germplasm. BMC Plant Biol. 2020;20:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myles S, Boyko AR, Owens CL, Brown PJ, Grassi F, Aradhya MK, Prins B, Reynolds A, Chia JM, Ware D, et al. Genetic structure and domestication history of the grape. Proc Natl Acad Sci USA. 2011;108:3530–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menzel C. Lychee, its origin, distribution and production around the world. West Aust Nut Tree Crop Assoc Yearbook. 1995;19:41–8. [Google Scholar]
- 39.Zhang SL, Yao J, Wang L, Wu N, van Nocker S, Li Z, Gao M, Wang XP. Role of grapevine SEPALLATA-related MADS-box gene VvMADS39 in flower and ovule development. Plant J. 2022;111:1565–79. [DOI] [PubMed] [Google Scholar]
- 40.Mejía N, Soto B, Guerrero M, Casanueva X, Houel C, Miccono MD, Ramos R, Le Cunff L, Boursiquot JM, Hinrichsen P, Adam-Blondon AF. Molecular, genetic and transcriptional evidence for a role of VvAGL11 in stenospermocarpic seedlessness in grapevine. BMC Plant Biol. 2011;11:11–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen JW, Yan Q, Li JW, Feng L, Zhang Y, Xu J, Xia R, Zeng ZH, Liu YL. The GRAS gene family and its roles in seed development in litchi (Litchi chinensis Sonn). BMC Plant Biol. 2021;21:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kader AA. Flavor quality of fruits and vegetables. J Sci Food Agr. 2008;88:1863–8. [Google Scholar]
- 43.Sievenpiper JL, de Souza RJ, Cozma AI, Chiavaroli L, Ha V, Mirrahimi A. Fructose vs. glucose and metabolism: do the metabolic differences matter? Curr Opin Lipidol. 2014;25:8–19. [DOI] [PubMed] [Google Scholar]
- 44.Mayuoni-Kirshinbaum L, Porat R. The flavor of pomegranate fruit: a review. J Sci Food Agr. 2014;94:21–7. [DOI] [PubMed] [Google Scholar]
- 45.Gomez M, Lajolo F, Cordenunsi B. Evolution of soluble sugars during ripening of papaya fruit and its relation to sweet taste. J Food Sci. 2002;67:442–7. [Google Scholar]
- 46.Wang HC, Huang HB, Huang XM. Sugar accumulation and related enzyme activities in the litchi fruit of ‘Nuomici’ and ‘Feizixiao.’ Acta Horticulturae Sinica. 2003;30:1. [Google Scholar]
- 47.Wang L, Ruan YL. Regulation of cell division and expansion by sugar and auxin signaling. Front Plant Sci. 2013;4:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sosso D, Luo DP, Li QB, Sasse J, Yang JL, Gendrot G, Suzuki M, Koch KE, McCarty DR, Chourey PS, et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet. 2015;47:1489–93. [DOI] [PubMed] [Google Scholar]
- 49.OU LX, Chen JZ. Description and data standard for litchi (Litchi chinensis Sonn.), China Agriculture Press. 2006; Chapter4:44–71.
- 50.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He ZL, Zhang HK, Gao SH, Lercher MJ, Chen WH, Hu SN. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016;44:236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Earl DA, Vonholdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–61. [Google Scholar]
- 55.Zhang C, Dong SS, Xu JY, He WM, Yang TL. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35:1786–8. [DOI] [PubMed] [Google Scholar]
- 56.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–5. [DOI] [PubMed] [Google Scholar]
- 58.Huang M, Liu XL, Zhou Y, Summers RM, Zhang ZW. BLINK: a package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience. 2019;8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu XL, Huang M, Fan B, Buckler ES, Zhang ZW. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. Plos Genet. 2016;12:1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang JB, Zhang ZW. GAPIT version 3: boosting power and accuracy for genomic association and prediction. Genom Proteom Bioinf. 2021;19:629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D, Landmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357-U121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnol. 2015;33:290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen CJ, Wu Y, Li JW, Wang X, Zeng ZH, Xu J, Liu YL, Feng JT, Chen H, He YH, Xia R. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining. Mol Plant. 2023;16:1733–42. [DOI] [PubMed] [Google Scholar]
- 65.Zhong HY, Chen JW, Li CQ, Chen L, Wu JY, Chen JY, Lu WJ, Li JG. Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep. 2011;30:641–53. [DOI] [PubMed] [Google Scholar]
- 66.Wu CY, Yang Y, Su DD, Yu CY, Xian ZQ, Pan ZL, Guan HL, Hu GJ, Chen D, Li ZG, et al. The SlHB8 acts as a negative regulator in tapetum development and pollen wall formation in tomato. Hortic Res. 2022;9:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figures S1–S17. This file contains all supplementary figures. Fig. S1 PCA scatter plots of the 277 accessions. Fig. S2 Phenotypic data distribution for 21 fruit traits. Fig. S3 Differences of fruit trait phenotypic data in subpopulations. Figs. S4 and S5 Pearson correlation between 19 traits and different PCAs in 2 years. Fig. S6 Venn diagram of the overlapped genes between the PC2 and PC4 groups. Fig. S7 EigenGWAS of the PC1 and PC3 traits. Fig. S8 The expression level of the 35 seed-related genes assessed by RNA-seq. Figs. S9 and S10 Genome-wide association studies of seed traits by Blink and FarmCPU models in 2 years. Fig. S11 The expression of GWAS associated seed-related genes by MLM model. Fig. S12 Expression levels of candidate seed-related genes validated by qRT-PCR. Fig. S13 Genome-wide association studies of glucose and sucrose contents. Fig. S14 Phylogenetic tree of LcSAI in 11 plant species. Fig. S15 The sugar contents of varieties with different genotypes at the same SNP site. Fig. S16 The function of LcSAI validated in T0 transgenic tomato fruits. Fig. S17 Numbers of accessions related to maturity in four subpopulations
Additional file 2: Tables S1–S11. This file contains all supplementary tables. Table S1 Litchi samples and their origins and sequencing results. Table S2 Statistical traits and phenotypes of the 276 accessions. Tables S3 and S4 Genes in the regions under PC2 and PC4 by EigenGWAS. Table S5 Differential expression genes of the two-type seed development. Table S6 The overlap genes between PC2, PC4, and DEGs. Table S7 Genome-wide association signals of seed traits. Table S8 Sugar composition and contents under HPLC method. Table S9 Genotype and sugar accumulation type of different accessions under PCR amplification. Table S10 Linkage disequilibrium values between 7:22496380 G:A and their nearby sites. Table S11 The primers of qRT-PCRs and markers
Data Availability Statement
The raw sequence data have been deposited in the NCBI Sequence Read Archive (SRA) under accession PRJNA1107979 [25] and PRJNA747875 [2]. The litchi reference genome used in this study is available from the ‘Feizixiao’ of our previous study [2]. All other relevant data are contained within the paper and its Additional files. All other data generated or analyzed during this study are included in the article or its additional files. Specifically, Additional file 1 provides all supplementary figures, which present supporting data for the population structure analysis, GWAS results, and functional gene verification; Additional file 2 contains the comprehensive phenotypic data for 21 fruit traits, lists of candidate genes from GWAS and transcriptome analyses, significant SNP loci, sugar content measurements, and all primer sequences used.