Abstract
Both paramutation and Mutator (Mu) transposon inactivation involve heritable changes in gene expression without concomitant changes in DNA sequence. The mechanisms by which these shifts in gene activity are achieved are unknown. Here we present evidence that these two phenomena are linked mechanistically. We show that mutation of a gene, modifier of paramutation 1 (mop1), which prevents paramutation at three different loci in maize, can reverse methylation of Mutator elements reliably. In mop1 mutant backgrounds, methylation of nonautonomous Mu elements can be reversed even in the absence of the regulatory MuDR element. Previously silenced MuDR elements are reactivated sporadically after multiple generations of exposure to mop1 mutations. MuDR methylation is separable from MuDR silencing, because removal of methylation does not cause immediate reactivation. The mop1 mutation does not alter the methylation of certain other transposable elements including those just upstream of a paramutable b1 gene. Our results suggest that the mop1 gene acts on a subset of epigenetically regulated sequences in the maize genome and paramutation and Mu element methylation require a common factor, which we hypothesize influences chromatin structure.
Although a great deal has been learned about the biochemistry and molecular biology of maize transposable elements, the means by which they are silenced epigenetically is only beginning to be understood. The most consistent molecular correlate with silencing in maize is cytosine methylation (1–4). The causal relationship between methylation of maize transposons and transposon activity, however, is not known. Work in maize and other systems such as Caenorhabditis elegans (5, 6) and Arabidopsis (7, 8) suggests that transposon regulation likely involves both transposon- (9, 10) and nontransposon-encoded gene products.
Methylation is a characteristic feature of inactivation of Mutator transposons (1, 3, 11, 12). The Mutator system includes a number of classes of elements; all classes share similar terminal inverted repeats (TIRs), but each carries unique internal sequences. MuDR is the regulatory element for the entire system, and the presence of active MuDR elements is required for transposition of the nonautonomous elements (11, 13). MuDR carries two genes, mudrA and mudrB. The mudrA gene is the presumptive transposase (14) and is absolutely required for all aspects of Mutator activity. Methylation of nonautonomous Mu elements is strictly correlated with the absence of functional MuDR elements and specifically with the absence of the mudrA gene product (11, 13, 15). Functional MuDR elements can be lost in three ways: segregation in genetic crosses, internal deletions, and epigenetic silencing correlated with methylation (16, 17). In Arabidopsis, mutations in DDM1 (homologous to SWI2/SNF2, a component of a chromatin-remodeling complex) are known to result in both reduced methylation and sporadic activation of silenced Mu-like (8) and Spm-like (18) elements.
Paramutation is another phenomenon that results in the heritable alteration of gene expression (19). In maize, paramutation has been observed at the r1, b1, pl1, and p1 loci, all of which are involved in flavonoid pigment production (reviewed in ref. 20). Paramutation occurs when a paramutagenic allele is heterozygous with a paramutable allele, resulting in a directed reduction in the expression of the paramutable allele. That change is heritable, and the paramutable allele can become paramutagenic.
Recently a mutation was isolated that prevents paramutation at b1, r1, and pl1 (21). When this mutation, mop1-1, is homozygous, the normally low-expressing paramutagenic allele of b1, B′, expresses at the level of the highly expressing paramutable allele, B-I, which results in the dark-purple plant color characteristic of the B-I allele as opposed to the lighter and sporadic purple color characteristic of the B′ allele. Further, when B′ is heterozygous with B-I in a mop1-1 homozygous background, paramutation is prevented, with both the B′ and B-I phenotypes transmitted after out-crossing. In the absence of the mop1-1 mutation only the B′ phenotype is transmitted from a B′/B-I heterozygote, because B-I is changed into B′ at a 100% frequency. The mop1-1 mutation also prevents the establishment of paramutation at the p1l and r1 loci, indicating that the mop1 gene product is generally required for paramutation (21).
Herein we show that mutations in mop1 can reverse Mu element methylation. We also show that somatic activity of previously silenced MuDR elements can be reactivated in a mop1 mutant background. Because mutations in mop1 affect multiple loci and phenomena, each of which requires different promoter sequences, we hypothesize that this gene operates on chromatin configuration rather than specific sequences.
Materials and Methods
Genetic Crosses.
Throughout the article, a single allele listing indicates homozygosity, whereas heterozygous individuals are indicated with alleles separated by a slash (/). Following maize genetic nomenclature, recessive alleles are indicated by lowercase and dominant alleles are indicated by capitalization of the first letter. The gene is indicated by lowercase, and the protein is indicated by uppercase and no italics. All stocks carry functional alleles of the c1 regulatory gene and all the anthocyanin biosynthetic genes except for a1, as indicated below. The alleles of the regulatory genes r1, b1, and pl1 in each stock are indicated for each cross.
Using MuKiller to Inactivate MuDR Elements.
MuKiller (MuK), a dominant factor present in some lines but not in our minimal Mutator line, is competent to heritably silence MuDR elements (17). A plant carrying a single MuDR element at position 1 on chromosome 2L [MuDR(p1); ref. 11] with the genotype B′ pl-sr (allele of pl1 that confers sun-red anthers and husks) R-g (r1 allele that confers purple seed) a1-mum2 MuDR(p1) was crossed to a line carrying MuK and no active MuDR elements (B′ pl-sr R-g a1-mum2 MuK/+). The resulting plants were scored for the presence of newly silenced MuDR elements as indicated by no excision of the Mu1 element from a1-mum2 and methylation of Mu1 elements in the presence of intact MuDR elements. The a1-mum2 mutation contains a Mu1 insertion in the 5′ promoter region of the a1 anthocyanin biosynthesis gene (22). In the presence of active MuDR elements, excisions of Mu1 can be scored as somatic purple sectors on the kernels. Of nine individuals examined in this family, three carried at least one full-length MuDR element and several methylated Mu1 elements, which is consistent with the epigenetic silencing of MuDR elements by the segregating MuK activity.
The first ear of one such plant [MuK/+, silenced MuDR(p1), B′ pl-sr R-g a1-mum2] was crossed with a mop1-1 homozygous B′ Pl A1 r-g (an r1 allele that has no pigment and no paramutation activity) plant derived from an active Mutator stock and thus carried multiple MuDR and Mu1 elements. The second ear was test-crossed by a plant lacking MuDR elements, wild-type Mop1, and homozygous for the a1-mum2 reporter gene (11). None of the kernels from the second ear showed evidence of excisions of Mu1 from a1-mum2, confirming the silencing of MuDR and the presence of MuK in the parent. Progeny from the first ear were self-fertilized, yielding mop1-1, mop1-1/Mop1, and Mop1 (Fig. 1A). These progeny also were segregating a1-mum2 versus A1 and R-g versus r-g. Kernels with A1 or r-g are fully purple or colorless, respectively, preventing the scoring of Mu1 excision from a1-mum2. To generate a second generation of mop1-1 homozygous plants and heterozygous siblings, B′ mop1-1 plants resulting from the above cross were identified by their dark plant pigment and crossed to lightly pigmented B′ mop1-1/Mop1 siblings, and the resulting progeny were scored for the mop1-1 phenotype (Fig. 1B). To test for reversals of Mu element methylation, B′ Mop1/mop1-1 progeny, which carried methylated Mu elements, were self-fertilized (Fig. 1C).
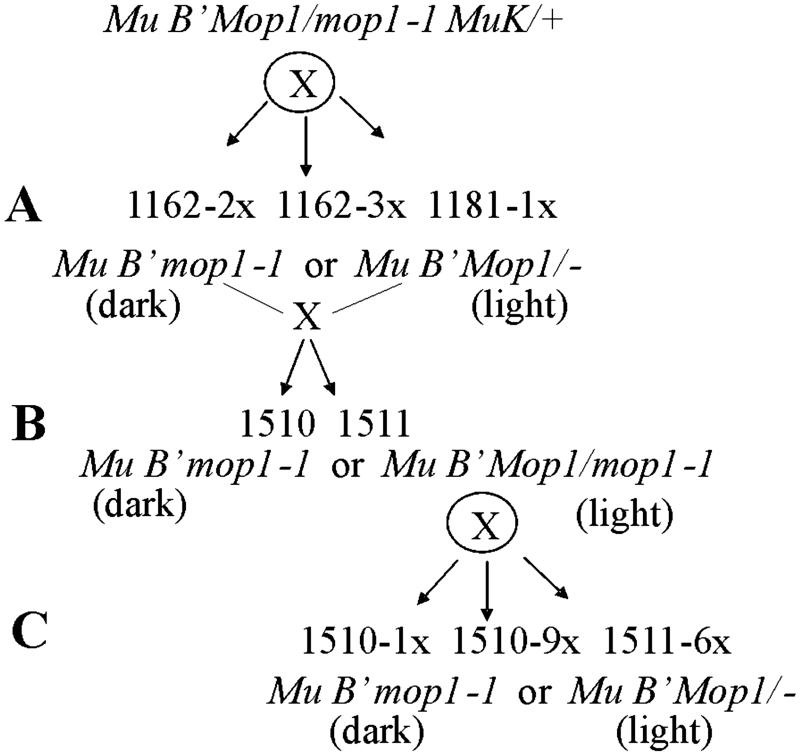
Figure 1.
Diagram of the crosses used to generate the families segregating mop1-1 and silenced Mu elements that were analyzed in Table 1 and Fig. 2. Individual plants with the genotype indicated at the top of the diagram were self-fertilized. The resulting progeny from each family were segregating mop1-1 (darkly pigmented plants) and wild type (lightly pigmented plants) as indicated in A. The symbol Mop1/− indicates that the other allele could have been Mop1 or mop1-1. Dark and light individuals were crossed to generate the families shown in B. Several plants with the wild-type phenotype then were self-fertilized, and the resulting progeny were as indicated in C.
Generation of Seeds with a Silenced MuDR Element and Homozygous for mop1-2.
Plants homozygous for a silenced MuDR element [B′ a1-mum2 MuDR(p1) R-g] were crossed to plants that were B′ A1 r-g mop1-2, an ethyl methanesulfonate-induced allele of mop1 (21, 23), from a non-Mutator line (lacking full-length MuDR elements). The progeny plants heterozygous for MuDR(p1) were self-fertilized, and the resulting families were screened for spotted kernels. If mop1-2 fully reactivated silenced MuDR(p1), then we would expect kernels homozygous for a1-mum2 (1/4), R-g/R-g or R-g/r-g (3/4), homozygous for mop1-2 (1/4) and carrying MuDR(p1) (3/4) to be spotted. Thus 100% reactivation would show 3.5% spotted kernels (1/4 × 3/4 × 1/4 × 3/4). In addition to self-fertilization, some plants were crossed to plants that were homozygous for mop1-2, a1-mum2, and R-g, with the expected frequency for 100% reactivation to be 12.5% spotted kernels (1/2 a1-mum2 × 1/2 MuDR(p1) × 1/2 mop1-2) for 100% reactivation.
DNA Preparation and Genomic Blotting.
DNA preparation and genomic blotting were performed as described (21). A plasmid containing Mu1 was as described (24). To generate an internal probe for Mu1, the plasmid was digested with AvaI and BstEII, and the internal fragment was gel-isolated. An internal fragment of MuDR bounded by EcoRI and BamHI was as described (11). The b1 upstream SB (SalI-BglII fragment) probe used to generate data shown in Figs. 2F and 5 was described previously (25, 26). Its location in b1 is shown in Fig. 5C. As a control for partial digestion of DNA, blots with HinfI-digested DNA samples were reprobed with a KpnI fragment of the a1 gene spanning a region of the coding sequence adjacent to (but not including) the Mu1 insertion in this gene (22). As a control for blots with SacI digests, the blots were probed with a single copy of PstI fragment flanking but not including the MuDR insertion on chromosome 2L (13).
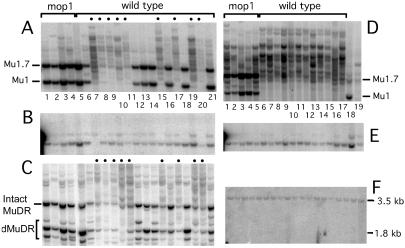
Figure 2.
(A–C) Methylation status of Mu1 and MuDR elements in family 1162-3x, segregating for MuK and mop1-1. (A) A HinfI digest of DNA from this family probed with a Mu1 internal fragment. The lanes marked “mop1” are samples from plants that were homozygous for mop1-1. The lanes marked “wild type” are samples from plants that were either mop1-1/Mop1 or Mop1/Mop1. The fragments expected for unmethylated Mu1 and slightly larger Mu1.7 elements are indicated. Individuals were scored as methylated (indicated by ●) if most of the Mu1 and Mu1.7 elements generated HinfI fragments larger than the 1.4- and 1.7-kb fragment expected from complete digestion. The individual in lane 20 was retested, and the presence of methylated Mu1 elements was confirmed (data not shown). (B) To control for complete digestion by the restriction enzyme, the blot shown in A was stripped and rehybridized with a fragment of the a1 gene. The two fragments represent complete digestion by HinfI. (C) A SacI digest of the same DNA samples shown in A probed with an internal MuDR fragment. The 4.8-kb MuDR fragment diagnostic for the presence of unmethylated, intact MuDR elements is indicated. Smaller deleted versions of MuDR are indicated as dMuDR. The larger fragments observed in lanes 1–4 are sequences related to MuDR that are found in all maize lines (13), which serve as controls for loading differences and estimating the number of unmethylated, intact MuDR elements. (D–F) Reversal of Mu1 element methylation. (D) A blot of HinfI-digested DNA samples from family 1511, segregating mop1-1 homozygotes (mop1) and mop1-1/Mop1-1 heterozygotes (wild type) was probed with a Mu1 fragment. As controls, the last two lanes contain DNA of plants with Mu1 and carrying (lane 18) or lacking (lane 19) MuDR. (E) To control for complete digestion by the restriction enzyme, the blot shown in D was stripped and rehybridized with a fragment of the a1 gene. (F) The same blot was stripped and rehybridized with the SB probe from a region upstream of the b1 gene (see Fig. 5C for location). The 1.8-kb fragment is the size expected for no methylation, and the larger fragment represents methylation of a particular site upstream of the b1 gene.
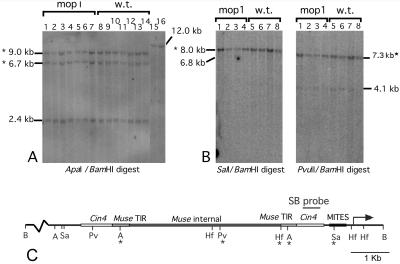
Figure 5.
DNA blots and summary map of digests of DNA from mutant (mop1) and wild-type (w.t.) individuals using several methyl-sensitive enzymes. Hybridization was with the SB probe upstream of the b1 gene (shown in C). (A) ApaI/BamHI digestions. Lanes 15 and 16 are mop1-1 homozygous and wild-type individuals, respectively, digested with only BamHI. (B) SalI/BamHI and PvuII/BamHI digestions. The fragments marked with asterisks represent cutting at the BamHI site in the b1 coding region and not cutting at one or more of the indicated upstream sites. (C) Restriction map of the region upstream of the start of transcription of B′. Sites marked with asterisks are methylated partially or completely in all genotypes tested (B′ Mop1 or Mop1/mop1-1 or B′ mop1-1). The other sites are unmethylated in all genotypes tested. A, ApaI; B, BamHI; Pv, PvuII; Hf, HinfI; Sa, SalI.
Results
Reduced Methylation of Mu Elements Correlates with the mop1-1 Mutation.
The mop1-1 mutation was isolated originally in a Mutator active line (21) with multiple copies of both MuDR and Mu1, all of which were unmethylated (data not shown). To examine the effect of the mop1-1 mutation on the process of Mutator silencing, a mop1-1 homozygote that carried multiple active MuDR elements was crossed to a plant carrying MuK, a factor that dominantly inactivates the Mutator system (17). Fig. 1 shows a schematic of the crosses performed with this stock, and details are in Materials and Methods. In all cases, progeny were scored visually for mop1-1 (dark-purple plants) versus wild-type (light-purple plants) phenotypes. In addition to their dark pigment, the mop1-1 plants often were significantly shorter than their wild-type siblings, and some did not produce ears, consistent with previous observations of pleiotropic effects of this mutation (21). The methylation status of both the autonomous MuDR elements and the nonautonomous Mu1 and related Mu1.7 elements (hereafter referred to collectively as Mu1) was determined by using DNA blots. Table 1 summarizes the genotype and methylation status of Mu1 elements for all the progeny.
Table 1.
Mu1 methylation in families (see Fig. 1) segregating for the mop1-1 mutation
| Families | Progeny phenotypes*
|
||
|---|---|---|---|
| mop1-1 | Wild type | ||
| A | 1162-2x† | 3 (0) | 7 (7) |
| 1162-3x‡ | 4 (0) | 17 (9) | |
| 1181-1x‡ | 3 (0) | 7 (4) | |
| B | 1510 | 14 (0) | 15 (15) |
| 1511 | 5 (0) | 12 (12) | |
| C | 1510-1x† | 1 (0) | 13 (13) |
| 1510-9x† | 3 (0) | 7 (7) | |
| 1511-6x† | 3 (0) | 11 (11) | |
| Total plants | 36 (0) | 89 (78) | |
Progeny were phenotyped based on pigment (dark for mop1-1 and light for wild type). DNA from leaves was examined for the methylation status of Mu1 as described in the Fig. 2 legend. The number methylated is indicated in parentheses.
These families were derived from a parent that had all methylated Mu elements.
These families were derived from a parent that had predominantly unmethylated Mu1 elements.
Several plants resulting from the cross between the mop1-1 mutant and the plant carrying MuK were self-fertilized (1162-2x, 1162-3x, and 1181-1x). An example of the methylation data for family 1162-3x is shown in Fig. 2 A–C. To determine the effect of mop1-1 on Mu1 methylation, DNA from the 21 plants in this family was digested with HinfI and probed with an internal Mu1 fragment (Fig. 2A). The Mu1 and Mu1.7 elements contain unique HinfI sites near the ends of their TIRs (1). Complete digestion (no methylation) results in 1.4- and 1.7-kb fragments for Mu1 and Mu1.7, respectively, whereas larger fragments are indicative of methylation in the TIRs. Mul methylation as assayed by HinfI digestion has been a reliable indicator of transposon activity in previous studies (1, 13, 16). To control for complete digestion, the blot was probed with a fragment of the a1 gene flanked by HinfI sites that normally are not methylated (Fig. 2B). To assay for changes in methylation of MuDR termini, the same samples were digested with SacI and probed with an internal MuDR fragment (Fig. 2C). SacI cuts once near the ends of each of the MuDR TIRs, generating a 4.8-kb fragment if both sites are not methylated. As documented (13, 16), a plant is scored as having methylated MuDR elements if the expected 4.8-kb band is missing or reduced in intensity relative to the fragments seen in all maize lines and concomitantly larger fragments appear (Fig. 2C). Control hybridization with a single-copy clone from chromosome 2L revealed complete digestion by SacI (data not shown).
In the four mop1-1 homozygotes in family 1162-3x, the majority of the Mu1 and MuDR elements were unmethylated (Fig. 2 A and C, lanes 1–4). In contrast, of the wild-type (mop1-1/Mop1 or Mop1/Mop1) siblings, half (9/17) had mostly methylated Mu1 elements, and half (8/17) had mostly unmethylated Mu1 elements (Fig. 2A, lanes 5–21). The MuDR elements were not methylated in mop1-1 plants, nor were they methylated in wild-type individuals with mostly unmethylated Mu1 elements (Fig. 2C). The wild-type plants with methylated Mu1 elements did have the fragment diagnostic for the unmethylated full-length MuDR element, but it was reduced in intensity, and that reduction was accompanied by the appearance of additional, larger fragments (Fig. 2C). These results are consistent with the methylation of sites within the MuDR and Mu1 TIRs seen previously in plants undergoing epigenetic silencing (1, 16, 17) and presumably are caused by the activity of MuK in this family.
A total of 41 progeny from self-fertilizations of mop1-1/Mop1 heterozygotes from this first generation were examined including 10 mop1-1 homozygotes and 31 of their wild-type siblings (Table 1). The Mu1 elements in all 10 of the mop1-1 homozygotes from the three families were mostly unmethylated (1181-1x, 1162-3x, and 1162-2x). Among wild-type siblings the frequency of Mu1 methylation in Mop1/Mop1 and Mop1/mop1-1 individuals depended on the methylation status of the parent, which varies because it sometimes takes more than one generation for MuK to silence the Mutator system fully (D.L., unpublished data). In the two families generated from the self-fertilization of plants that had unmethylated Mu1 elements (1162-3x and 1181-1x), 13 of the 24 plants with the wild-type phenotype (Mop1/Mop1 or mop1-1/Mop1) had methylated Mu1 elements. In the family generated from a parent with methylated Mu1 elements (1162-2x), all seven wild-type progeny had methylated Mu1 elements. If mop1-1 were not affecting methylation, we would have expected six of the mop1-1 mutants to have had methylated Mu1 elements based on the frequency of methylation in the wild-type sibs (20/31). The absence of any such plants is significant (P = 0.05 from χ2 test).
The mop1-1 Mutation Reverses the Effects of Previous Methylation of the Mu Elements.
To test whether mop1-1 reverses Mu1 methylation, mop1-1 homozygotes from the 1162-3x family were crossed to three different wild-type siblings with inactive methylated Mu elements. One ear produced no mop1-1 plants, suggesting that the inactive parent was Mop1-homozygous. Progeny from the two other ears (1510 and 1511) segregated plants with the mop1-1 phenotype. A methylation analysis of family 1511 is shown in Fig. 2 D–F. This family was derived from a cross between the two siblings, the DNA of which was analyzed in Fig. 2A, lanes 4 and 15. In both families, all the resulting mop1-1 plants carried predominantly unmethylated Mu1 elements (Table 1 and Fig. 2D, lanes 1–5). In contrast, none of the wild-type Mop1/mop1-1 plants carried any unmethylated Mu1 elements (Fig. 2D, lanes 6–17). These results suggest that the mop1-1 mutation reverses previously established Mu1 element methylation. Additional plants heterozygous for mop1-1 and carrying methylated Mu1 elements were self-fertilized (Fig. 1C), and scored for both the mop1-1 phenotype (dark-purple pigment) and methylation of Mu1 elements (families 1510-1x, 1510-9x, and 1511-6x, Table 1). All seven of the mop1-1 homozygotes had predominantly unmethylated Mu1 elements relative to their wild-type siblings, all of which carried only methylated Mu1 elements. Thus, the mop1-1 mutation reliably reverses previously established methylation of Mu1 elements.
The mop1-1 Mutation Reverses Mu1 Methylation in the Absence of Full-Length MuDR Elements.
Although mop1-1 was derived from a Mutator line, in the process of propagating the stocks we identified several lineages that lacked intact, functional MuDR elements by using SacI or XbaI or with an EcoRI/HindIII double-digest (data not shown). These stocks allowed us to test whether an intact MuDR element is required for mop1-1-mediated decreases in Mu1 element methylation. We examined progeny from two families derived from the self-fertilization of plants heterozygous for mop1-1 and carrying no intact MuDR elements. Eight mop1-1 mutant and eight wild-type siblings were examined. The Mu1 elements of all the mop1-1 plants were unmethylated relative to the Mu1 elements of their wild-type siblings (one family in Fig. 3A). Thus, reversal of Mu1 element methylation occurs efficiently in mop1-1 plants independent of intact MuDR elements (Fig. 3B). Normally, there is a very tight correlation between reduced Mu1 element methylation and the presence of intact, active MuDR elements, suggesting that the MuDR transposase is required to prevent a default methylation pathway that targets Mu elements for inactivation (27). The mop1-1 homozygous plants lacking functional MuDR elements provide the first exception to this rule.
Figure 3.
Reversal of Mu1 element methylation in the absence of MuDR. DNA was isolated from a family segregating for mop1-1/mop1-1 (mop1, lanes 1–3) and mop1-1/Mop1 or Mop1/Mop1 (wild type, lanes 4–6). In A and B, lane 7 contains DNA from a MuDR-active individual. The diagnostic fragments for a full-length unmethylated MuDR element and unmethylated Mu1 and Mu1.7 elements are indicated. (A) A HinfI digest of DNA was blotted and hybridized with the Mu1 internal probe. The 1.7-kb fragment visible in lanes 1–6 represents an endogenous Mu1.7-homologous HinfI fragment present in many maize lines (32). (B) DNA from the same family digested with SacI, blotted, and probed with an internal fragment of MuDR. Digestions with additional methylation-insensitive enzymes confirmed that no intact MuDR elements were present (data not shown).
A Second Allele of mop1 Also Reduces Mu1 Methylation.
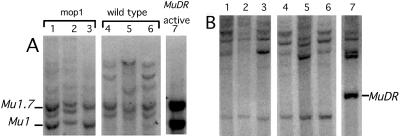
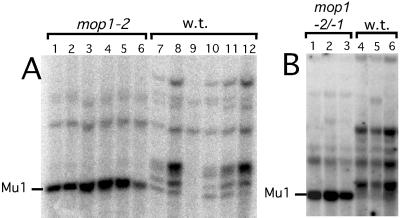
To eliminate the possibility that there was a factor modifying Mu1 methylation that was only coincidentally linked to mop1-1, we examined a family segregating for a different recessive allele of mop1, mop1-2. This family was derived from a non-Mutator stock that had been ethyl methanesulfonate-mutagenized (21, 23). Non-Mutator stocks typically contain one to a few methylated Mu1 elements but no MuDR elements (28). DNA from six mop1-2 mutants and six wild-type siblings, Mop1/mop1-2 and Mop1/Mop1, were digested with HinfI and probed with Mu1 (Fig. 4A). We also examined plants heterozygous for mop1-1 and mop1-2 (Fig. 4B). As with mop1-1, the Mu1 elements in the mop1-2 and mop1-2/mop1-1 plants were mostly unmethylated relative to their wild-type siblings (Fig. 4 A and B).
Figure 4.
Reversal of Mu1 element methylation in mop1-2 plants. (A) DNA was isolated from a family segregating mop1-2 and wild type (mop1-2/Mop1 and Mop1, w.t.). (B) DNA was isolated from a family that was segregating mop1-2/mop1-1 and wild type (mop1-2/Mop1). A HinfI digest of DNA was blotted and hybridized with the Mu1 interval probe. The diagnostic fragment for an unmethylated Mu1 element is indicated.
The mop1-1 Mutation Does Not Reverse Methylation of Sites Immediately Upstream of the b1 Locus.
Given that the mop1-1 mutation correlates with reduced Mu1 methylation, we wanted to determine whether methylation of sequences elsewhere in the genome were affected. Characterization of rDNA and centromere repeats had shown no methylation differences in these repeats between mop1-1 and wild-type siblings (21). Previous studies identified a number of sites upstream of the B′ transcription unit (GenBank accession nos. X70790 and S48060) that were methylated (25, 26). We tested whether these sites were undermethylated in mop1-1. In our experiments both B′ mop1-1 and B′ Mop1/mop1-1 plants were methylated equivalently at several sites as documented previously for B′ Mop1 (25, 26). These sites include the HinfI site 1.8 kb upstream of the transcriptional start site (Fig. 2F) and the ApaI, SalI, and PvuII sites (Fig. 5). Interestingly, these sites are all within sequences related to transposable elements, suggesting that methylation of transposon-related sequences in the promoter proximal region of B′ is not reversed by mop1-1. These elements include a MITE element immediately upstream of the start of transcription and the Muse element (a distant relative of MuDR; V.L.C., D. Selinger, and M. Stam, personal observation). This region is not involved in B′ paramutation (20, 25). Once key paramutation sequences are identified, it will be important to determine whether mop1 mutations influence their methylation.
Progeny of Plants Homozygous for the mop1-1 or mop1-2 Mutations Do Not Show Restoration of Mu Element Transposition or Somatic Excision.
To test for the effects of the mop1-1 mutation on reactivation of Mu element transposition, the progeny of two mop1-1 mutant plants and two of their wild-type siblings were assayed for the appearance of new Mu1 fragments, which is consistent with new transposition events. A total of 30 progeny of mop1-1 mutant plants were examined. These plants carried previously silenced MuDR elements, as well as multiple segregating Mu1 elements, most of which were unmethylated in mop1-1 homozygotes. Given the presence of an average of 12 Mu1 elements in the parents of these families and the average transposition frequency of Mu1 elements observed in the presence of MuDR(p1) (10% per element per generation; ref. 13), if MuDR elements were reactivated we would have expected at least 36 new restriction fragments representing transposition events (12 progenitor Mu1 elements × 10% transposition frequency × 30 mop1-1 individuals). No new fragments were observed in any of the progeny.
To explore this issue further, we used a more sensitive assay, reactivation of somatic excision of a reporter Mu1 element. Non-Mutator plants that lacked full-length MuDR elements and were homozygous for mop1-2 were crossed to plants homozygous for a single MuDR element [MuDR(p1)] that had been silenced by using MuKiller. The resulting plants were either self-fertilized or crossed to plants homozygous for a1-mum2 and mop1-2. The resulting ears were screened for the appearance of spotted kernels. If mop1-2 fully activated the silenced MuDR element, 3.5% of the progeny kernels from the self-fertilizations or 12.5% of the test-cross progeny kernels should have excisions (Materials and Methods). Twenty-two families consisting of 6,324 progeny kernels resulting from self-fertilization and three families with a total of 710 kernels generated from out-crosses to a1-mum2 testers were examined. The expected number of spotted kernels from these crosses was 221 and 89, respectively. No spotted kernels were observed in any of these families, indicating no reactivation of the silenced MuDR element.
MuDR Can Be Reactivated in Progeny of Plants Homozygous for the mop1-2 Mutation for Multiple Generations.
One concern with the above experiment is that MuK was potentially segregating, possibly preventing reactivation by mop1-2. It also was possible that the presence of MuK in previous generations may have set up a chromatin state that was more difficult to activate, and multiple generations in the presence of mop1-2 might be required for activation. To determine whether MuDR could be reactivated after multiple generations of exposure to mop1 mutants in a non-MuK background, additional crosses with mop1-2 were done. In all cases, plants homozygous for the mop1-2 mutation and heterozygous for the MuDR(p1) element were either self-fertilized or out-crossed with mop1-2/Mop1, a1-mum2 (no MuDR) plants. Progeny were subjected to DNA gel-blot analysis to determine which carried MuDR(p1) and unmethylated Mu1 elements, and their pigment phenotype was scored for mop1-2 versus wild type. Kernels from all ears were scored for spots indicative of excision of Mu1 from a1-mum2.
Progeny from one plant in which silenced MuDR(p1) had been in a mop1-2 homozygous background for two consecutive generations were examined. No spotted kernels were observed in the 288 progeny kernels from the self-fertilization or the 241 kernels from the out-cross. To examine a third consecutive generation, seeds from the out-cross were planted, and all plants were crossed with mop1-2 a1-mum2 testers, resulting in 33 ears. Of the six ears from plants homozygous for mop1-2 and carrying MuDR(p1), four had spotted kernels (29 of the 965 kernels were spotted). The excision frequency in these kernels was variable, ranging from only a few excisions per kernel to a frequency typical for a single active MuDR element. In contrast, in 13 ears generated from plants heterozygous for mop1-2 and that carried MuDR(p1), 0 of 3,309 kernels were spotted. Similarly, in 9 ears generated from plants homozygous for mop1-2 but that lacked MuDR(p1), 0 of 1,900 kernels were spotted, and in 5 ears generated from plants heterozygous for mop1-2 but that lacked MuDR(p1), 0 of 989 kernels were spotted. The presence of spotted kernels only in the plants homozygous for mop1-2 and carrying MuDR(p1) strongly suggests that the mutation is responsible for the sporadic reactivation of MuDR in these plants.
Discussion
Our results demonstrate that in mop1 mutants, in which paramutation is inhibited at three genes, cytosine methylation of Mu elements is reduced. One explanation, based on the activation of transposons by “genomic shock” (29), is that the demethylation of Mu elements is an indirect effect of genomic stress caused by the defect in mop1. We think this explanation is unlikely for several reasons. First, although many mop1 mutants show pleiotropic developmental effects and look stressed, some mop1 mutants look quite normal (21), yet without exception all mop1 mutants show reduced methylation of Mu elements. For example, one mop1-2 homozygote was runty and five were healthy, yet all six had reduced Mu1 methylation (Fig. 4A). Second, we have isolated two paramutation mutants, rmr1-1 and rmr2-1, that have no obvious pleiotropic developmental phenotypes (23), but Mu1 elements in these mutants show reduced methylation (D.L., unpublished data). Third, we have examined Mu element methylation in plants that clearly were under stress, yet we have never seen reduced methylation of Mu elements in lines that were silenced previously. Thus, we favor an alternative explanation, which is that the product of the mop1 gene is involved in maintaining silent states, potentially by mediating chromatin changes that influence DNA methylation of certain sequences.
Our observation that mop1 mutants affect methylation of both SacI (lacks CG or CXG sequences) and HinfI (has CG sequence) sites suggests that the wild-type mop1 product may interact with both de novo and maintenance methylation pathways. The change in methylation did not lead to an immediate reactivation of transposition. However, after several generations of exposure of an epigenetically silenced MuDR element to a mop1 mutant background, MuDR activity was restored.
Because both Mu elements and paramutagenic alleles are affected by mutations in mop1, it is probable that these two genetic systems share targets for epigenetic modification. Clearly, the targets are not something as broad as all methylated sequences, because not everything that is methylated is altered by mutations in mop1. The targets for silencing are unlikely to be specific sequences or specific chromosomal locations, because there is little sequence similarity between the genetic elements affected by mop1 mutations, and the affected sequences are distributed throughout the genome. It is also unlikely that mop1 mutations are simply reversing silenced sequences within condensed heterochromatin, because Mu elements typically insert within or nearby single-copy sequences (30). However, not all Mu1 elements are unmethylated in mop1-1 homozygotes (e.g., see Figs. 2D and 3A). Thus, there may be variation in the capacity of the mop1-1 mutation to reverse Mu1 methylation as a function of the element's chromosomal position or chromatin context.
Although there is an excellent and immediate correlation between reduced Mu element methylation in plants homozygous for mop1 mutations, reactivation of Mu element somatic excision was delayed and sporadic. The lack of immediate reactivation of silenced MuDR elements in mop1 mutant backgrounds suggests that methylation is only part of the process of Mu element silencing. Previous studies with Mutator lines demonstrated that methylation follows rather than causes inactivation (13). In these experiments, methylation of the nonautonomous elements is restored rapidly after the loss of a single MuDR element because of deletions within the MuDR element during somatic development or loss of the element after genetic segregation, suggesting MuDR activity prevents Mu1 methylation. Our data suggest that MuDR methylation is separable from MuDR silencing, because removal of methylation does not cause immediate reactivation. Experiments with MOM mutants in Arabidopsis (31), which showed reactivation of silenced transgenes in the presence of continued methylation, also indicate that methylation and silencing are separable.
Our experiments demonstrate that MOP1 is required for Mu element methylation, but the absence of MOP1 only gradually results in the reversal of MuDR silencing, which is similar to the activation of transposons seen with ddm1 mutations in Arabidopsis (8, 18). Multiple mutations have been isolated that affect paramutation (23), and mutations in three genes, rmr1, rmr2 and mop3, also reduce Mu element methylation (D.L. and V.L.C., unpublished data). It will be interesting to examine whether silenced MuDR elements would be reactivated more rapidly in double- or triple-mutant backgrounds. The gradual, sporadic increase in MuDR activation by mop1 mutants contrasts with the immediate increase in the transcription of the paramutant B′ and Pl′ alleles in all homozygous mop1 mutant plants (21). The two phenomena are distinct at the transcriptional level as the transposons are silenced completely in wild-type maize and Arabidopsis plants, whereas transcription is reduced but detectable from the paramutant B′ and Pl′ alleles. The slower response to mop1 mutations by transposons versus paramutant alleles may reflect distinct chromatin states.
Until we know the identity of mop1, we can only speculate on its specific mode of action. One possibility is that there is a general chromatin code for directing both methylation and transcriptional repression. We hypothesize that the two genes, mop1 in maize and DDM1 in Arabidopsis, function early to interpret the code, because mutations in these genes affect both methylation and transcriptional repression. In contrast, the MOM gene may act downstream, translating the chromatin code into transcriptional repression but not methylation. Although the phenotypes of mop1 and DDM1 are very similar, there is an interesting difference in that DDM1 mutants have a global effect on DNA methylation, whereas the targets of mop1 appear more limited. Further studies should reveal whether this difference is because these two genes encode distinct functions or because of differences between chromatin regulation in maize and Arabidopsis.
Acknowledgments
We thank David Braun for helpful advice on the manuscript. This work was supported by National Science Foundation Grants MCB-9603638 and MCB-9982447 (to V.L.C.) and a Syngenta Agricultural Discovery Institute University of California-Berkeley Strategic Alliance grant (to D.L.). The Molecular Dynamics Storm 860 system used in this work was purchased with funds from Department of Army Research Grant No. DAAG559710102.
Abbreviation
- TIR
terminal inverted repeat
References
- 1.Chandler V L, Walbot V. Proc Natl Acad Sci USA. 1986;83:1767–1771. doi: 10.1073/pnas.83.6.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chomet P S, Wessler S, Dellaporta S L. EMBO J. 1987;6:295–302. doi: 10.1002/j.1460-2075.1987.tb04753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennetzen J L. Mol Gen Genet. 1987;208:45–51. [Google Scholar]
- 4.Fedoroff N. Genetics. 1989;121:591–608. doi: 10.1093/genetics/121.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ketting R F, Haverkamp T H, van Luenen H G, Plasterk R H. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 6.Tabara H, Sarkissian M, Kelly W G, Fleenor J, Grishok A, Timmons L, Fire A, Mello C C. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 7.Hirochika H, Okamoto H, Kakutani T. Plant Cell. 2000;12:357–369. doi: 10.1105/tpc.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singer T, Yordan C, Martienssen R. Genes Dev. 2001;15:591–602. doi: 10.1101/gad.193701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlappi M, Smith D, Fedoroff N. Genetics. 1993;133:1009–1021. doi: 10.1093/genetics/133.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlappi M, Raina R, Fedoroff N. Cell. 1994;77:427–437. doi: 10.1016/0092-8674(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 11.Chomet P, Lisch D, Hardeman K J, Chandler V L, Freeling M. Genetics. 1991;129:261–270. doi: 10.1093/genetics/129.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hershberger R J, Warren C A, Walbot V. Proc Natl Acad Sci USA. 1991;88:10198–10202. doi: 10.1073/pnas.88.22.10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lisch D, Chomet P, Freeling M. Genetics. 1995;139:1777–1796. doi: 10.1093/genetics/139.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen J A, Benito M I, Walbot V. Nucleic Acids Res. 1994;22:2634–2636. doi: 10.1093/nar/22.13.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lisch D, Girard L, Donlin M, Freeling M. Genetics. 1999;151:331–341. doi: 10.1093/genetics/151.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martienssen R, Baron A. Genetics. 1994;136:1157–1170. doi: 10.1093/genetics/136.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lisch D, Freeling M. Maydica. 1994;39:289–300. [Google Scholar]
- 18.Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Nature (London) 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- 19.Brink R A. Annu Rev Genet. 1973;7:129–152. doi: 10.1146/annurev.ge.07.120173.001021. [DOI] [PubMed] [Google Scholar]
- 20.Chandler V L, Eggleston W B, Dorweiler J E. Plant Mol Biol. 2000;43:121–145. doi: 10.1023/a:1006499808317. [DOI] [PubMed] [Google Scholar]
- 21.Dorweiler J E, Carey C C, Kubo K M, Hollick J B, Kermicle J L, Chandler V L. Plant Cell. 2000;12:2101–2118. doi: 10.1105/tpc.12.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Reilly C, Shepherd N S, Pereira A, Schwarz-Sommer Z, Bertram I, Robertson D S, Peterson P A, Saedler H. EMBO J. 1985;4:591–597. doi: 10.1002/j.1460-2075.1985.tb03713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollick J B, Chandler V L. Genetics. 2001;157:369–378. doi: 10.1093/genetics/157.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talbert L E, Patterson G I, Chandler V L. J Mol Evol. 1989;29:28–39. doi: 10.1007/BF02106179. [DOI] [PubMed] [Google Scholar]
- 25.Patterson G I, Thorpe C J, Chandler V L. Genetics. 1993;135:881–894. doi: 10.1093/genetics/135.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson G I, Kubo K M, Shroyer T, Chandler V L. Genetics. 1995;140:1389–1406. doi: 10.1093/genetics/140.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandler V L, Hardeman K J. Adv Genet. 1992;30:77–122. doi: 10.1016/s0065-2660(08)60319-3. [DOI] [PubMed] [Google Scholar]
- 28.Chandler V, Rivin C, Walbot V. Genetics. 1986;114:1007–1021. doi: 10.1093/genetics/114.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClintock B. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 30.Hanley S, Edwards D, Stevenson D, Haines S, Hegarty M, Schuch W, Edwards K J. Plant J. 2000;23:557–566. doi: 10.1046/j.1365-313x.2000.00830.x. [DOI] [PubMed] [Google Scholar]
- 31.Amedeo P, Habu Y, Afsar K, Scheid O M, Paszkowski J. Nature (London) 2000;405:203–206. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- 32.Talbert L E, Chandler V L. Mol Biol Evol. 1988;5:519–529. doi: 10.1093/oxfordjournals.molbev.a040510. [DOI] [PubMed] [Google Scholar]