Abstract
We have analyzed a systematic flaw in the current system of gene identification: the oligo(dT) primer widely used for cDNA synthesis generates a high frequency of truncated cDNAs through internal poly(A) priming. Such truncated cDNAs may contribute to 12% of the expressed sequence tags in the current dbEST database. By using a synthetic transcript and real mRNA templates as models, we characterized the patterns of internal poly(A) priming by oligo(dT) primer. We further demonstrated that the internal poly(A) priming can be effectively diminished by replacing the oligo(dT) primer with a set of anchored oligo(dT) primers for reverse transcription. Our study indicates that cDNAs designed for genomewide gene identification should be synthesized by use of the anchored oligo(dT) primers, rather than the oligo(dT) primers, to diminish the generation of truncated cDNAs caused by internal poly(A) priming.
One of the major goals in any genome study is to identify the full set of genes in the targeted genome (1). Among various tools designed for gene identification, physical isolation of the genes through analyzing mRNA is the most reliable one (2). Expressed sequence tags (ESTs) are used most often for that purpose (3). Through single-pass sequencing of multiple individual clones in many cDNA libraries, ESTs with hundreds of bases long are generated to represent each clone. The performance of various EST projects in the past decade has resulted in the accumulation of nearly 4 million ESTs from the human genome and large numbers of ESTs from genomes of other species (http://www.ncbi.nlm.nih.gov/UniGene/).
Besides the collection of numerous expressed sequences for gene identification, the quality of the collected sequences is equally important. It has been observed that the quality of many expressed sequences in current databases is questionable, especially for the human EST data (4, 5). Among many factors influencing the quality of collected sequences, errors originating from experimental procedures are of special importance. Eliminating the “noise” from the very beginning of sequence collection will be an effective way of providing high-quality data.
In this study, we focused on the issue of the quality of cDNA generated by oligo(dT) priming. Since the beginning of DNA recombination technologies, oligo(dT) priming has been the method most widely used for conversion of mRNA into cDNA through reverse transcription (6–8). In reverse transcription, an oligo(dT) primer is first annealed to the poly(A) sequences universally present at the 3′ end of nearly every mRNA by T:A base-pairing. The reverse transcriptase then extends from the annealed oligo(dT) primer along the mRNA template, resulting in the copying of the mRNA sequence into the cDNA sequence. The single-strand cDNA is then converted into double-strand cDNA for construction of cDNA libraries for gene identification. Nearly all of the EST cDNA libraries have been constructed exclusively from cDNAs generated by oligo(dT) priming (http://www.ncbi.nlm.nih.gov/UniLib/).
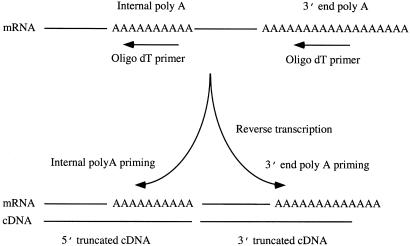
Besides being located universally at the 3′ end of mRNA, poly(A) sequences can also be located within mRNA sequences. We speculated that these internal poly(A) sequences could also be primed by oligo(dT) primers during reverse transcription, which would result in the generation of two kinds of truncated cDNA sequences: one starting from the internal poly(A), the other starting from the 3′ poly(A), but terminated before the internal poly(A) sequence (Fig. 1). The occurrence of 3′-truncated ESTs related to internal poly(A) was observed in earlier studies based on limited data, with estimated rate of 2.5–14% of the ESTs (5, 9). We performed a series of experiments and analyzed the current databases to investigate systematically the issue of internal poly(A) priming by oligo(dT) primer. Our studies reveal the patterns of internal poly(A) priming and the prevalence of the resulting truncated cDNAs in the public EST database. We also demonstrated that the problem of internal poly(A) priming could be minimized by simple replacement of the oligo(dT) primer with a set of anchored oligo(dT) primers for cDNA synthesis.
Figure 1.
Schematics of internal poly(A) priming by oligo(dT) primer. Besides priming the poly(A) sequence at the 3′ end of mRNA, the oligo(dT) primer can also prime the poly(A) sequences present internally within mRNA. The initiation of cDNA synthesis from the oligo(dT) primed at both locations results in the generation of two truncated cDNAs from a single mRNA template.
Materials and Methods
Generation of in Vitro Transcripts.
A 140-bp oligonucleotide was synthesized (Oligos Etc., Guilford, CT) containing the following components: 5′-T7 promoter-M13F-A20-Random nucleotides (20 bases)-A50-T3 promoters (5′-CGTAATACGACTCACTATAAGGGGTAAAACGACGGCCAGTACGN*B**A20N20B**A50CTTTAGTGAGGGTTAATTTC-3′; N* = A, G, C, T; B** = A, G, C). The oligo was converted into double-strand DNA by PCR by use of T7 and T3 primers. In vitro transcripts were generated by use of the double-strand DNA and T7 RNA polymerase (Life Technologies, Grand Island, NY). After the reaction, the double-strand DNA templates were destroyed by DNase I digestion. The absence of DNA templates in the in vitro transcripts was verified by direct PCR by use of T7 and T3 primers and RNase A-digested in vitro transcripts with negative amplification.
Reverse Transcription, Cloning, and Sequencing.
One microgram of in vitro transcript was used as the template for reverse transcription. The primers used for priming were either the regular oligo dT16 (5′-ACTATCTAGAGCGGCCGCTTT16–3′) (200 ng per reaction) or a mixture of five anchored oligo dT16 primers (5′-ACTATCTAGAGCGGCCGCTTT16-A, -G, -CA, -CG, and -CC-3′) (10) (200 ng per reaction). Reverse transcription was performed by use of a cDNA synthesis kit containing Moloney murine leukemia virus reverse transcriptase (Life Technologies). The resulting single-strand cDNA was converted into double-strand cDNA by PCR with M13F primer (5′-GTAAAACGACGGCCAGTACG-3′) and antisense primer (5′-ACTATCTAGAGCGGCCGCTT-3′) and cloned into TOPO TA vectors (Invitrogen). Clones were picked randomly and the insert in each clone was amplified with T7 primer located in the vector and T3 primer located in the insert, and used for sequencing reactions with T7 primer and a Big-Dye sequencing kit (Applied Biosystems). Sequences were collected with an ABI377 DNA sequencer (Applied Biosystems).
Selection of mRNAs of Known Genes with Internal Poly(A) Sequences for Reverse Transcription and Quantitative PCR.
A group of mRNA sequences, each containing a minimum of eight continuous internal As, were selected from the genes expressed in CD15+ myeloid progenitor cells (11). Four micrograms of total RNA from CD15+ myeloid progenitor cells was used for reverse transcription with either 200 ng oligo(dT) primer or 200 ng of the set of anchored oligo(dT) primers as described above. Quantitative PCR was used to determine the relative abundance of internally primed cDNA and 3′ primed cDNA extending beyond the internal poly(A) tract (12). One sense and two antisense (antisense 1 and antisense 2) primers were designed for each gene. The sense and antisense 1 primers were located upstream of the internal poly(A) sequence and were used to detect the cDNAs originating from the internal priming; antisense primer 2 was located downstream of internal poly(A), and the combination of sense and antisense 2 primers was used for detecting the cDNA originating from 3′ priming and passing the internal poly(A) sequence. A control template for quantitative PCR was generated for each gene by use of a control sense primer and an antisense primer 2. The control sense primer had 40 bases, including the sense primer joined to downstream sequences with a small deletion. The resulting control template is shorter than the wild-type template while retaining the sense, antisense 1, and antisense 2 primer sequences. Such control templates were successfully generated for eight mRNAs. Quantitative PCR was performed for each of these eight mRNAs by use of the corresponding sense primer, antisense primer 1, or antisense primer 2, cDNA templates generated by oligo(dT) or anchored oligo(dT) primers, and an optimized amount of the control templates. The PCR products were separated on a 2% agarose gel with ethidium bromide staining. The signals from the wild type and the control template for each gene were quantified with ID20 software (Kodak). The levels of cDNA originating from the internal poly(A) priming were determined by subtracting the signals of the sense/antisense 1 from the signals of the sense/antisense 2. All of the signals were normalized by using the internal control before being used for the subtraction. The sequences for all primers used for the analysis will be provided on request.
For determining the dosage effects of the oligo(dT) primer on internal priming, mRNA of Proteoglycan 1(X17042), which contains an internal A10 between base 909 and 918, was used as the template. Different amounts of oligo(dT) primers were used for reverse transcription. The steps and primers used for quantitative PCR were the same as above.
Estimation of Database Prevalence of ESTs Originating from Internal Poly(A) Priming.
The databases used in this study include the EST database (dbEST human, release 10/04/2001, http://www.ncbi.nlm.nih.gov/dbEST/index.html) and the Reference Sequence database (RefSeq human, release 8/14/2001, http://www.ncbi.nlm.nih.gov/LocusLink/refseq.html; ref. 13). Four sets of analysis were performed:
Identification of RefSeq sequences containing internal poly(A) tracts.
An A tract was defined as at least eight bases in length with at most two non-A bases, and with A at the first and last positions.
Matching of 3′ labeled ESTs to RefSeq sequences.
ESTs labeled as 3′ and longer than 100 bases were extracted from dbEST. A sequence-similarity search of these ESTs was conducted in RefSeq by using BLAST (14). The following conditions were used in determining a match between EST query sequences and RefSeq: (i) Only correctly oriented (±) alignments between the EST and RefSeq sequences were accepted. (ii) The EST was matched along at least 90% of its length. (iii) Identity in the matched region was greater than 90%.
Determination of 3′ end-matched ESTs and internal-matched ESTs in the 3′ ESTs.
Because many sequences in the RefSeq database have short nongenomic tails after their 3′ end poly(A), which most likely were derived from the cloning vector, the following criteria were used for defining the ESTs that matched the 3′ poly(A): (i) For the reference sequences without tail, an EST should be mapped within 20 bases from the 3′ end of a reference sequence. (ii) For the reference sequences with the tail, an EST should be mapped within the 100 bases of the 3′ end of a reference sequence that contains more than 50% As.
Estimates of ESTs originating from internal poly(A) priming.
3′-Labeled ESTs that matched to a region within 30 bases upstream of an internal poly(A) tract in a RefSeq sequence were classified as internally primed sequences.
Results
Oligo(dT) Primer Generates High Frequency of Truncated cDNAs through Internal Priming.
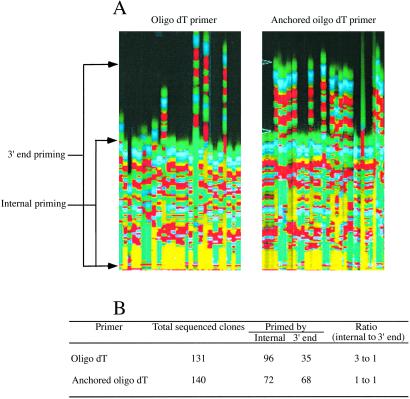
We used a synthetic transcript as the model to investigate whether internal poly(A) priming by oligo(dT) primer occurs. This transcript contains a 20-base internal poly(A) and a 50-base 3′ poly(A) to mimic the real mRNA with internal poly(A). The use of the 20-base internal poly(A) serves two purposes: (i) to provide various lengths of poly(A) sequence for possible internal priming and (ii) to determine the minimal A residuals required for internal priming. This transcript was used for reverse transcription with an oligo(dT) primer. The results show that the internal poly(A) priming not only occurs, but at a high frequency; the ratio between the internal poly(A) priming and the 3′ poly(A) priming was 3:1 (Fig. 2). Sequence analyses of the resulting 96 clones indicate that the minimal length of internal poly(A) for the priming was eight As, and the priming efficiency increased significantly when the internal A residues increased. By using the anchored oligo(dT) primers as a control for the same reaction, we observed that the rate between the internal poly(A) priming and 3′ poly(A) priming was reduced to 1:1 (Fig. 2). This result also normalized the potential biased PCR amplification of shorter internally primed templates.
Figure 2.
Comparison of internal poly(A) priming and 3′ poly(A) priming between oligo(dT) primer and anchored oligo(dT) primers. An in vitro transcript containing an internal A20 and 3′ A50 was used for reverse transcription with either oligo(dT) primer or anchored oligo(dT) primers. (A) Sequencing image of cDNA clones primed by oligo(dT) primer and anchored oligo(dT) primers. (B) Summary of the results of the analyses.
Internal Poly(A) Priming Prevents the Extension of 3′ End-Primed cDNA along the mRNA Template.
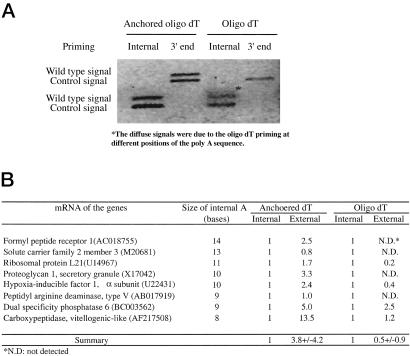
In a given mRNA template, if the internal poly(A) sequence is occupied by an oligo(dT) primer, the extension reaction from the 3′ end priming would be blocked by the internal primed oligo(dT) at the internal poly(A) sequences. This blocking could result in the generation of two truncated cDNAs from both internal priming and 3′ end priming. We investigated such a possibility. mRNAs containing 8–14 bases of internal poly(A) sequence were selected from well-characterized genes. Whereas the cDNAs originating from internal poly(A) priming by oligo(dT) primer predominated in seven of eight mRNAs, very low levels of cDNAs primed from 3′ poly(A), and passed the internal poly(A) sequence were detected (Fig. 3). In contrast, when the anchored oligo(dT) primers were used for the same reaction, the cDNAs primed from the 3′ end poly(A) predominated for all eight mRNAs, whereas low-level cDNAs originating from the internal poly(A) sequences were detected (Fig. 3). Although the 3′ primed cDNA with the oligo(dT) primer was higher than the internal primed one in mRNA from dual specificity phosphatase 6 (1–2.5), the level increased when the anchored oligo(dT) primers were used (1–5.0).
Figure 3.
Internal poly(A) priming prevents the extension of 3′ end-primed cDNA along the mRNA template. A group of mRNAs with internal poly(A) sequences was primed with oligo(dT) or anchored oligo(dT) primers for reverse transcription. Quantitative PCR was used to determine cDNA levels originating from internal poly(A) priming, and 3′ end poly(A) priming extending beyond the internal poly(A) tract. (A) Demonstration of the levels of cDNA from internal poly(A) priming and 3′ end poly(A) priming in proteoglycan mRNA. It shows the diminishing of cDNA originating from 3′ end priming when oligo(dT) primer was used, whereas the level was high between internal and 3′ end-primed cDNA when anchored oligo(dT) primers were used. (B) Summary of the results from the analyses. The value from internal priming is set as 1 for comparing the relative levels between the cDNA from internal priming and cDNA from external priming.
Increasing the Dose of Oligo(dT) Primers for Reverse Transcription Results in an Increase in Truncated cDNA by Internal Poly(A) Priming.
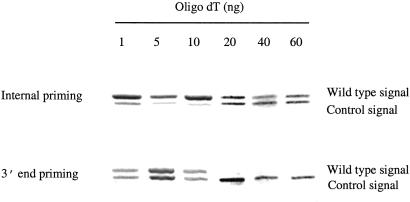
Because of the presence of long 3′ poly(A) tails in mRNAs, cDNAs converted from mRNA by oligo(dT) priming will include varying lengths of poly(dA/dT) sequences at their 3′ end. These poly(dA/dT) sequences generate many problems, such as poor quality of 3′ sequencing (15), and the loss of cDNA templates during subtraction hybridization because of the formation of nonspecific poly(dA)/poly(dT) hybrids between unrelated cDNA templates (10). Increasing the concentration of oligo(dT) primer has been used in cDNA synthesis in the hope of preventing the incorporation of such longer poly(dA/dT) sequences (16, 17). As the 3′ poly(A) sequence becomes increasingly annealed by oligo(dT) primers, extension occurs only from the most 5′ annealed primer, and the resulting cDNAs contain shorter poly(dA/dT) sequences. We tested the dosage effect of the oligo(dT) primer on internal poly(A) priming. We observed an unexpected result from this experiment: increasing the amount of oligo(dT) primer from 1 to 60 ng resulted in a dramatic decrease of cDNAs that were initiated from 3′ end priming and passed the internal poly(A) sequence, whereas increasing the primer had little effect on the level of cDNAs initiated from internal poly(A) priming (Fig. 4). Such different effects of high-dose oligo(dT) primer on the 3′ end and internal poly(A) priming result in the generation of predominantly truncated cDNAs from internal priming.
Figure 4.
Effects of high-dose oligo(dT) primers on the internal poly(A) priming and 3′ end poly(A) priming. mRNA from proteoglycan with 10 internal As was used in this analysis. Increased doses of oligo(dT) primer were used for the first-strand cDNA synthesis. Quantitative PCR was used to determine cDNA levels originating from internal poly(A) priming and 3′ end poly(A) priming extending beyond the internal poly(A) tract. The same amount of control templates was used in the quantitative PCR for each set of reactions.
Many ESTs in dbEST Are Derived from Truncated cDNAs Originating from Internal Poly(A) Priming.
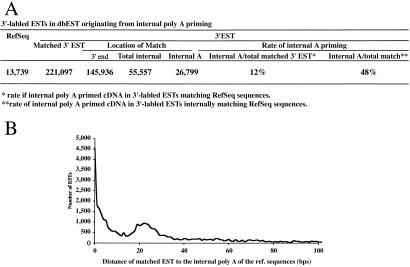
Our experimental results raise a serious question about the levels of truncated sequences originated from internal poly(A) priming in current expression databases. To understand this issue, we compared the sequences between the dbEST and RefSeq databases. The dbEST database contains most of the expressed sequences collected so far. The RefSeq database contains the well-annotated genes with full-length cDNA sequences. By matching 3′-labeled ESTs in dbEST to known sequences in RefSeq, we estimated the number of ESTs originated from internal poly(A) priming. By setting the standard of the internal poly(A) sequence with eight poly(A)s including up to two mismatches, we first identified that 86% (11,806) of the 13,739 sequences in the RefSeq database are internal poly(A)+ with a total of 78,114 internal poly(A) sequences. A total of 221,097 ESTs were matched to the sequences in RefSeq, 27% of these ESTs (58,770) were matched to internal regions in the RefSeq sequences, and 12% of these ESTs (26,799) were matched to a region within 30 bases of an internal poly(A) tract (Fig. 5). Of the 72% (145,936) of 3′ end-matched sequences, up to the equal rate (12%) would be expected to be the truncated sequences that terminated before the internal poly(A) sequences, although the precise number of such ESTs cannot be determined unless the corresponding 5′ sequences are used for the analysis.
Figure 5.
Estimated prevalence of ESTs in dbEST originating from internal poly(A) priming. (A) Summary of the analyses. See the text for detailed description. (B) Distance between the sequence matched by EST to the internal poly(A) sequence in the matched reference sequence.
Discussion
With the human genome as a typical example, efforts at genomewide gene identification in many species are currently under way. For many of these studies, gene identification through analysis of mRNA rather than gene identification through complete genomic sequencing and computational prediction will be the main method used for accomplishing the goals, largely because of the high complexity of some of the targeted genomes and the issue of cost effectiveness (18–20). An analysis of the limitations of the current status of gene identification through mRNA and suggestions for improvements will benefit future studies.
Our present studies build on our earlier analysis which showed that the cDNAs generated by oligo(dT) priming contain long 3′ poly(dA/dT) sequences that were largely responsible for the low efficiency of identification of the genes expressed at lower levels (10). We now report that the oligo(dT) primer also generates truncated cDNA through internal poly(A) priming. Our analysis shows that 12% of 3′-labeled ESTs in the current dbEST database are likely to be originated from internal poly(A) priming. We speculate that a similar rate of such truncated sequences may exist for the entire set of 4 million human ESTs. These results also suggest that a similar phenomenon may be present in data generated by SAGE (serial analysis of gene expression), most of which also uses oligo(dT) primer for cDNA synthesis (21). Considering the common occurrence of internal poly(A) tracts in most mRNAs, and the high rate of internal poly(A) priming by oligo(dT) demonstrated here, the consequence of internal poly(A) priming is clearly of great significance.
The truncated sequences caused by internal poly(A) priming can lead to many problems. First, they make the annotation of genes in the genome difficult. Although ESTs have been used extensively for annotation of the human genome, the presence of multiple ESTs from the same gene because of internal poly(A) priming can make the assembly of the gene rather difficult (22). Second, they confuse the analysis of alternative splicing. Different mRNAs can originate from the same gene through alternative splicing. However, the presence of truncated sequences makes the analysis of alternative splicing very complicated. Third, they interrupt the generation of full-length cDNA. One of the major challenges in gene identification is to generate a full-length cDNA for each gene (14, 23, 24), as shown by the fact that only about 11,000 human cDNAs with an ORF have been deposited in public databases, and most of the clones for these cDNA sequences are not available (24). The generation of truncated cDNA by internal poly(A) priming makes these efforts much more difficult. Fourth, they complicate the quantitative analysis of gene expression. By dispersing the signal from a single transcript among multiple detected sequences, truncated sequences contribute “noise” to EST-based expression analysis and other cDNA-driven methods, such as serial analysis of gene expression. Finally, the generation of truncated cDNA from internal priming wastes substantial resources used for gene identification.
Besides the physical blocking of the 3′ extension by the oligo(dT) primer annealed to the internal poly(A) sequence, another explanation for the differences of internal poly(A) priming and 3′ poly(A) priming may be related to the “overhang” partial annealing of the oligo(dT) primer in the 3′ poly(A) sequence. Because the lengths of most internal poly(A) sequences in mRNA templates are short, very limited space is available for primer annealing to form stable hybrids. In contrast, the longer poly(A) sequence at the 3′ end of mRNA provides much more space for the formation of stable hybrids. The far greater excess of molecules of oligo(dT) primers than mRNA templates in reverse transcription can result in the saturated binding of oligo(dT) primers along the poly(A) sequences, including the “overhang” partial annealing of a primer to the 5′ end of the poly(A) sequences which blocks the extension of reverse transcription. The more oligo(dT) primer is used, the worse the overhang effect will be. This phenomenon does not seem to be so serious for the internal poly(A). The situation becomes different when the anchored oligo(dT) primers are used for cDNA synthesis. The extension requires both the annealing of the oligo(dT) part and the anchored nucleotide to the 5′ end of the 3′ poly(A) sequences. Although the “overhang” effect also exists at the 3′ poly(A) sequence, the rate of extension from the internal primed site also decreases, which balances the rate of extension from both the internal primed and 3′ end-primed anchored oligo(dT)s. The conventional anchored oligo(dT) primers with two-base-anchored nucleotides are not suitable for the purpose of cDNA synthesis discussed here (25), because only the one with correct anchored pairing among the 12 primers annealed to the poly(A) sequence can be extended, which results in a very low yield of cDNA. The set of anchored oligo(dT) primers we used consists of a minimum of five primers. The yield of cDNA synthesis with these primers is not affected significantly (26).
The results from our analyses indicate that any cDNA used for genome-scale gene identification should be synthesized with the set of anchored oligo(dT) primers, rather than the oligo(dT) primer. Such a simple switch will diminish the generation of truncated cDNA caused by internal poly(A) priming and provide high-quality cDNAs derived from the real 3′ end of mRNA.
Acknowledgments
This work was supported by National Institutes of Health Grants CA84405 (to J.D.R.) and CA78862–01 (to J.D.R. and S.M.W.) and by the G. Harold and Lelia Y. Mathers Charitable Foundation (to S.M.W.).
Abbreviation
- EST
expressed sequence tag
References
- 1.International Human Genome Sequencing Consortium. Nature (London) 2001;409:860–921. [Google Scholar]
- 2.Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, et al. Nature (London) 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 3.Adams M D, Kerlavage A R, Fields C, Venter J C. Nat Genet. 1993;4:256–267. doi: 10.1038/ng0793-256. [DOI] [PubMed] [Google Scholar]
- 4.Hillier L D, Lennon G, Becker M, Bonaldo M F, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- 5.Aaronson J S, Eckman B, Blevins R A, Borkowski J A, Myerson J, Imran S, Elliston K O. Genome Res. 1996;6:829–845. doi: 10.1101/gr.6.9.829. [DOI] [PubMed] [Google Scholar]
- 6.Weiss G B, Wilson G N, Steggles A W, Anderson W F. J Biol Chem. 1976;251:3425–3431. [PubMed] [Google Scholar]
- 7.Verma I M. J Virol. 1978;26:615–629. doi: 10.1128/jvi.26.3.615-629.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagenbuchle O, Krikeles M S, Sprague K U. J Biol Chem. 1979;254:7157–7162. [PubMed] [Google Scholar]
- 9.Gautheret D, Poirot O, Lopez F, Audic S, Claverie J. Genome Res. 1998;8:524–530. doi: 10.1101/gr.8.5.524. [DOI] [PubMed] [Google Scholar]
- 10.Wang S M, Fears S C, Zhang L, Chen J-J, Rowley J D. Proc Natl Acad Sci USA. 2000;97:4162–4167. doi: 10.1073/pnas.97.8.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, Zhou G, Clark T, Chen J, Rowley J D, Wang S M. Proc Natl Acad Sci USA. 2001;98:3340–3345. doi: 10.1073/pnas.051013798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S M, Rowley J D. Proc Natl Acad Sci USA. 1998;95:11909–11914. doi: 10.1073/pnas.95.20.11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pruitt K D, Katz K S, Sicotte H, Maglott D R. Trends Genet. 2000;16:44–47. doi: 10.1016/s0168-9525(99)01882-x. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan A S, Wilcox A S, Hopkins J A, Sikela J M. Nucleic Acids Res. 1991;19:1715. doi: 10.1093/nar/19.7.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonaldo M F, Lennon G, Soares M B. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- 17.Jiang H, Bivens N J, Ries J E, Whitworth K M, Green J A, Forrester L J, Springer G K, Didion B A, Mathialagan N, Prather R S, et al. BioTechniques. 2001;31:38–42. doi: 10.2144/01311bm05. [DOI] [PubMed] [Google Scholar]
- 18.Hacia J G. Trends Genet. 2001;17:637–645. doi: 10.1016/s0168-9525(01)02494-5. [DOI] [PubMed] [Google Scholar]
- 19.McConkey E H, Varki A. Science. 2000;289:1295–1296. doi: 10.1126/science.289.5483.1295b. [DOI] [PubMed] [Google Scholar]
- 20.Dunwell J M, Moya-Leon M A, Herrera R. Biol Res. 2001;34:153–164. doi: 10.4067/s0716-97602001000300003. [DOI] [PubMed] [Google Scholar]
- 21.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 22.Zhuo D, Zhao W D, Wright F A, Yang H Y, Wang J P, Sears R, Baer T, Kwon D H, Gordon D, Gibbs S, et al. Genome Res. 2001;11:904–918. doi: 10.1101/gr.164501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagase T, Seki N, Ishikawa K, Ohira M, Kawarabayasi Y, Ohara O, Tanaka A, Kotani H, Miyajima N, Nomura N. DNA Res. 1996;3:321–329. doi: 10.1093/dnares/3.5.321. [DOI] [PubMed] [Google Scholar]
- 24.Wiemann S, Weil B, Wellenreuther R, Gassenhuber J, Glassl S, Ansorge W, Bocher M, Blocker H, Bauersachs S, Blum H, et al. Genome Res. 2001;11:422–435. doi: 10.1101/gr.154701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang P, Pardee A B. Science. 1992;257:967–970. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 26.Zhou G, Chen J, Lee S, Clark T, Rowley J D, Wang S M. Proc Natl Acad Sci USA. 2001;98:13966–13971. doi: 10.1073/pnas.241526198. [DOI] [PMC free article] [PubMed] [Google Scholar]