Abstract
The Lyme disease spirochete Borrelia burgdorferi has bundles of periplasmic flagella subpolarly located at each cell end. These bundles rotate in opposite directions during translational motility. When not translating, they rotate in the same direction, and the cells flex. Here, we present evidence that asymmetrical rotation of the bundles during translation does not depend upon the chemotaxis signal transduction system. The histidine kinase CheA is known to be an essential component in the signaling pathway for bacterial chemotaxis. Mutants of cheA in flagellated bacteria continually rotate their flagella in one direction. B. burgdorferi has two copies of cheA designated cheA1 and cheA2. Both genes were found to be expressed in growing cells. We reasoned that if chemotaxis were essential for asymmetrical rotation of the flagellar bundles, and if the flagellar motors at both cell ends were identical, inactivation of the two cheA genes should result in cells that constantly flex. To test this hypothesis, the signaling pathway was completely blocked by constructing the double mutant cheA1∷kan cheA2∷ermC. This double mutant was deficient in chemotaxis. Rather than flexing, it failed to reverse, and it continually translated only in one direction. Video microscopy of mutant cells indicated that both bundles actively rotated. The results indicate that asymmetrical rotation of the flagellar bundles of spirochetes does not depend upon the chemotaxis system but rather upon differences between the two flagellar bundles. We propose that certain factors within the spirochete localize at the flagellar motors at one end of the cell to effect this asymmetry.
Keywords: spirochete|periplasmic flagella|Lyme disease|chemotaxis|motility
Spirochetes are a structurally unique group of bacteria. The organelles for motility, the periplasmic flagella (PFs), reside between the outer membrane sheath and cell cylinder. These PFs are subterminally attached at each end of the cell cylinder and propel the spirochetes by rotation (1–3). Several lines of evidence indicate that the PFs are not only involved in motility, but also influence the shape of that part of the cell in the domain where they reside. For example, in the Leptospiraceae, the relatively short PFs cause the ends of the cell to form hook or spiral shaped ends (4–6). In Treponema phagedenis, the PFs are associated with the bent-shaped ends of the cell. In fact, the length of the bent-shaped ends in T. phagedenis correlates precisely with that of the PFs (7). In each of these organisms, mutants lacking PFs still retain a helical cell body. In contrast, the entire shape of the Lyme disease spirochete Borrelia burgdorferi is markedly impacted by the PFs. These spirochetes have 7–11 PFs attached at each end of the cell that overlap at the cell center. Mutants lacking PFs of B. burgdorferi are no longer wave-like as are the wild type but are rod-shaped (8).
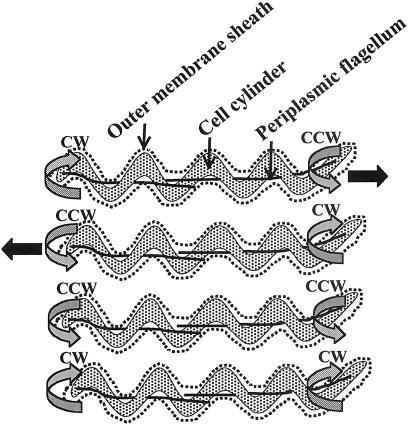
Unique to both spirochete and Spirillum spp. is asymmetrical rotation of the bipolarly attached flagellar bundles during translational motility (1, 5, 9, 10). Several lines of evidence indicate that in both Leptospiraceae and B. burgdorferi, the anterior PFs rotate counterclockwise (CCW) and the posterior PFs clockwise (CW) (Fig. 1; as a frame of reference, we view the PF filament from its distal end along its shaft; refs. 1, 3, 11–13). A given cell can run, pause, and run again either in the same or opposite direction; these intervals are the result in changes in rotational direction of the PFs. Depending on the species, pausing is often accompanied with a major change in shape, often with bending at the cell center, and is referred to as a flex (1, 3, 10). Flexing occurs when the PFs rotate in the same direction.
Figure 1.
Swimming cells of B. burgdorferi as a function of direction of rotation of the periplasmic flagella. Black arrows indicate direction of swimming. Dotted lines represent the outer membrane sheath. Gray arrows indicate direction of rotation of the periplasmic flagella. For simplification, only one periplasmic flagellum is shown attached at each end of the cell cylinder. In B. burgdorferi, there are between 7–11. The top two are translation forms, and the bottom two are nontranslational forms.
Escherichia coli and Salmonella enterica serovar Typhimurium serve as the model systems for the understanding of bacterial motility and chemotaxis (see refs. 14–16 for recent reviews). Flagellar rotation is driven by a proton gradient. These flagella act as propellers and allow an organism to swim in a given direction. During chemotaxis, cells swim by a biased random walk toward a favorable medium or away from one that is toxic. Initially, an effector molecule binds directly or indirectly via a periplasmic binding protein to a membrane-bound chemoreceptor protein. The chemoreceptor regulates the autophosphorylation of the sensor histidine kinase CheA. Activated CheA phosphorylates the response regulator CheY, which then interacts with the switch complex at the flagellar motor to increase the probability of the flagellar clockwise (CW) rotation and destabilize the counterclockwise (CCW) rotation. CCW flagellar rotation results in smooth swimming or runs, and CW rotation results in tumbling. Null mutants in cheY or cheA continuously rotate their flagella CCW and consequently fail to tumble (17). Cells showing a positive chemotactic response have longer runs and suppress the intervals spent tumbling.
The understanding of spirochete chemotaxis is in its early stages (18). A membrane potential mediates the chemotactic response in Spirochaeta aurantia (19). Several compounds and media components have been shown to serve as attractants and repellents for S. aurantia (20) and to a lesser degree for Treponema denticola (21, 22) and B. burgdorferi (23). Disruption of the cheA and chemoreceptor genes dmcA and dmcB in T. denticola results in cells that fail to penetrate monolayers of eukaryotic cells (18, 24). Genomic analysis of B. burgdorferi, T. denticola, and Treponema pallidum indicate that these spirochetes contain flagellar and chemotaxis gene homologs common to other bacteria (refs. 25 and 26; http://www.hgsc.bcm.tmc.edu/microbial). B. burgdorferi differs from these other spirochete species in that it has multiple chemotaxis gene homologs such as two copies of cheA and three copies of cheY. The cheA genes are located on two different gene clusters that map far from one another on the linear B. burgdorferi chromosome (25).
The basis for the asymmetrical rotation of the PF bundles during translation is poorly understood. Two hypotheses are readily apparent with respect to how these spirochetes achieve asymmetry. One states that the chemotaxis system plays an essential role in this asymmetry. Thus, asymmetry depends on localized concentrations of CheY-P, and upon the fact that, during translation, one cell end has a higher concentration of CheY-P than the other. Because CheY-P synthesis depends on CheA, this hypothesis predicts that null mutants in cheA would result in cells that continuously rotate their flagella in the same direction as that found with other bacteria. For spirochetes, if all the periplasmic flagella rotate in the same direction, cells would continuously flex. The other hypothesis states that the motors at the two ends of the cells are different. This difference results in cells whereby the bundles rotate asymmetrically in the absence of CheY-P. Accordingly, this hypothesis predicts that null mutants in cheA would constantly run. In this communication, we present evidence in support of the latter hypothesis.
Materials and Methods
Bacterial Strains, Growth and Chemotaxis Assay Conditions, and Plasmids.
A single clone of avirulent B31, referred to as B31-A, was used for all gene transfer experiments and served as the reference wild-type strain (27). The nonmotile PF-deficient mutant MC-1, liquid BSK-II medium, agarose plates, growth conditions, and swarm agar plates have been described (8). Capillary tube chemotaxis assays were carried out similarly to the method described by Shi et al. for B. burgdorferi (23). Assays were done by using 75-μl capillary tubes incubated for 2 h at 34°C in a 3% CO2 atmosphere. E. coli strains were grown in LB broth (28). The plasmids for the construction of mutants and expression of recombinant proteins are listed in Table 1. Alignment and DNA analysis were done by using GENETOOL and PEPTOOL (Biotools, Edmonton, Canada) and GCG WISCONSIN package (Accelrys, Madison, WI).
Table 1.
Plasmids and oligonucleotides
| Plasmids |
| pGME-T: Ampr, PCR cloning vector |
| pGA1: pGME-T Easy with 1,690 bp cheA1 gene amplified by PCR |
| pGA2: pGEM-T Easy with 2,521 bp cheA2 gene amplified by PCR |
| pGA1kan: pGA1 with 1.3kb flgB-kan replacing 393 bp HindIII/HindIII fragment |
| pGA2kan: pGA2 with 1.3kb flgB-kan replacing 1,326 bp HindIII/HindIII fragment |
| pGA2ery: pGA2 with 1.1 kb ermC replacing 1,326 bp HindIII/HindIII fragment |
| pQE-31: Expression vector |
| pQE-CheA1: cheA1 cloned into pQE-31 BamHI/PstI sites |
| DNA Primers: |
| 5′-ATATTTAATCTCAATAGC-3′ (cheW2 reverse, primer extension) |
| 5′-AAGTAATTTCTGAGATCG-3′ (cheA1, 5′ +, inactivation) |
| 5′-AAAGTCTGATAACAGTTCGG-3′ (cheA1, 5′ +, inactivation) |
| 5′-TAGTTATTGCATCTATGTC-3′ (cheA1, 3′ −, inactivation) |
| 5′-ATGGAAATATTAGATTTGG-3′ (cheA2, 5′ +, inactivation) |
| 5′-ATTTGGAAAATGAAGAGC-3′ (cheA2, 5′ +, inactivation) |
| 5′-TTACCATTGCCAAGCGTAG-3′ (cheA2, 3′ −, inactivation) |
| 5′-AAGCTTTAATACCCGAGCTTCAAG-3′ (flgB-kan, 5′ +, kan cassette) |
| 5′-AAGCTTTCAAGTCAGCGTAATGCT-3′ (flgB-kan, 3′ −, kan cassette) |
| 5′-AAGCTTAACACACTAGACTTATTTAC-3′ (ermC, 5′ +, ermC cassette) |
| 5′-AAGCTTAAAAAATAGGCACACGAAAA-3′ (ermC, 3′ −, ermC cassette) |
| 5′-ACGGATCCGGATAGTAGTGATGTTATTG-3′ (cheA1, 5′ +, expression) |
| 5′-CTGCAGTATAAGTTTAGTTATTGCATC-3′ (cheA1, 3′ −, expression) |
Italics indicate engineered restriction cut sites.
DNA Manipulation, PCR Conditions, and Primers.
Restriction mapping, enzyme modification, and transformation were carried out by standard procedures (28). Amplification of target genes, primer sequences, and plasmids are listed in Table 1. Amplified products were purified by using Qiagen PCR purification kits or gel removing kits (Qiagen, Chatsworth, CA). The resultant products were cloned into the respective plasmids for further manipulation.
Construction of Plasmids, pGA1kan, pGA2kan, pGA2ery, and Targeted Mutagenesis.
cheA1 and cheA2 were amplified by PCR from chromosomal DNA, and the resultant products were cloned into pGEM-T Easy vector (Promega). HindIII was used to generate an internal deletion in each cheA gene. The 393-bp deletion in cheA1 (nucleotides 1,016–1,409) and the 1,326-bp deletion in cheA2 (nucleotides 773–2,099) were each replaced with a 1.3-kb flgB-kanamycin resistance cassette (27). The plasmids obtained, pGA1kan and pGA2kan, were used as the source of DNA for targeted mutagenesis. To construct the plasmid pGA2ery, the HindIII-generated deletion in cheA2 was replaced by the amplified erythromycin resistance cassette ermC (29). Restriction digest mapping indicated that the direction of transcription of kan or ermC was the same as that of cheA1 or cheA2. Preparation of competent B. burgdorferi, electroporation, and plating of transformants were done as described (8, 27). Amplified input DNA (≈2 μg) were used for electroporation. Growth media were supplemented with 350 μg/ml kanamycin/0.05 μg/ml erythromycin, or both.
RNA Preparation, Reverse Transcription (RT)-PCR, Primer Extension, and Construction of Recombinant Proteins.
Total RNA was prepared for both RT-PCR and primer extension analysis as described (30). RT-PCR was carried out by using the One-Step RT-PCR kit (Qiagen). Primer extension was done by using the AMV reverse transcriptase Primer Extension System (Promega; ref. 31). B. burgdorferi CheA1 was over-expressed as a His-tagged fusion protein. The complete gene was amplified and cloned into pQE31 vector (Qiagen) at BamHI/PstI sites, resulting in pQE-CheA1. This construct was transformed into host cell M15(pREP4) (Qiagen). Cells readily overproduced CheA1 after induction with isopropyl β-d-thiogalactoside. The overproduced protein was purified with the ProBond Purification System (Invitrogen) according to the manufacturer's instructions. Approximately 350 μg of recombinant CheA1 protein was used to immunize rats to produce a specific antiserum. An antiserum to recombinant CheY3 was made in a similar manner and is described elsewhere (M.A.M., R.G.B., C.L., N.W.C., unpublished work).
Gel Electrophoresis and Western Blotting.
SDS/PAGE and Western blotting with the enhanced chemiluminescence detection method (Amersham Pharmacia) were carried out as reported (30, 32). Rabbit antiserum to E. coli CheA was generously provided by P. Matsumura (University of Illinois, Chicago, IL). Monoclonal antibodies to FlaA, FlaB, and DnaK were kindly provided by B. Johnson (Centers for Disease Control, Fort Collins, CO), A. Barbour (University of California, Irvine, CA), and G. Benach (State University of New York, Stony Brook, N.Y.), respectively.
Electron and Light Microscopy, and Computer-Assisted Motion Analysis.
Standard methods for electron microscopy and negative staining were used to view spirochetes with attached periplasmic flagella (4). Live cells were observed by dark-field or phase microscopy by using Zeiss optics with cells held at 34°C with a Physitemp temperature controlled stage (8). Video recording of images was carried out as described (3). The Hobson BacTracker was used to track the motion of individual cells to determine reversal frequency and velocity. For tracking experiments, cells were first gently centrifuged at room temperature and then suspended in motility buffer that was supplemented with 5% BSK-II/1% methylcellulose. Because of the large size of B. burgdorferi relative to its slow velocity, certain modifications were made for data analysis. Cells were videotaped with dark-field illumination at 200× for at least 1 min. We used the XY module of the tracking system whereby the position of the center of a cell was determined every 1/60th sec. To obtain specific data on velocity and reversing, we averaged every 12 data points (0.2 sec). The results were displayed as a two-dimensional track and a bar chart of distance vs. time. In addition, the video was digitized to allow for frame-by-frame verification of run and reversal intervals. At least three cells were tracked for a given strain, and the results are expressed as means ± SD. To analyze tethered cells, cells in BSK-II (without methylcellulose) that adhered to the glass in the central part of the cell were video-recorded by phase microscopy at 2,500× (3).
Results
Expression of CheA1 and CheA2.
Before targeting cheA1 and cheA2 by allelic exchange mutagenesis, we first determined whether these genes were expressed in growing cells. Previous analysis has shown that cheA2 resides within a motility and chemotaxis gene cluster with the following gene order: flaA, cheA2, cheW3, cheX, cheY3. Transcriptional analysis has shown that this gene cluster consists of an operon initiated by a σ70 promoter (30, 33, 34). On the other hand, the expression of cheA1 has not been characterized. cheA1 maps within a cluster consisting of cheW2, orf566, cheA1, cheB2, orf569, cheY2 (25). By using RT-PCR analysis, all six genes were found to be cotranscribed as a polycistronic mRNA. Primer extension analysis revealed a σ70 recognition sequence directly upstream of cheW2 (TTGATA—N20—TAAAT—N6-A). These results indicate that both cheA1 and cheA2 are transcribed in growing cells as part of σ70 initiated operons. Western blot analysis, as described below, indicate that both CheA1 and CheA2 are synthesized in growing cells.
Construction and Analysis of LC-A1 and LC-A2 Mutants.
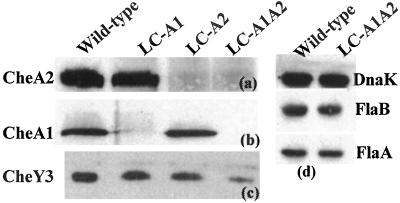
To examine the functions of the two cheA genes, we targeted each by allelic-exchange mutagenesis by using a kan cassette with accompanying deletion formation. PCR analysis indicated that the resultant cheA1∷kan (LC-A1) and the cheA2∷kan (LC-A2) mutants each contained the kan insert as expected. These inserts were transcribed in the same direction as cheA1 or cheA2. Western blot analysis was used to test for synthesis of both CheA1 and CheA2 in the wild-type and mutant cells. We used two different sources of antibodies for the analysis. First, an antiserum directed to E. coli CheA was found to react with a band corresponding to CheA2 in wild-type cells (Fig. 2a). The reacting protein was clearly CheA2, as it migrated at approximately 98 kDa on SDS/PAGE. In addition, a similar reaction was detected in lysates of LC-A1 but not in LC-A2. These results indicate that CheA2 is expressed in wild-type cells, and that LC-A2 suffered a mutation in the cheA2 gene encoding that protein. To test for CheA1 expression, we raised an antiserum to recombinant CheA1. By using this antiserum, we found strong reactivity in both wild-type and LC-A2 cell lysates at a band corresponding to CheA1 (79 kDa; Fig. 2b). No reactivity was detected in LC-A1 lysates. These results indicate that both CheA1 and CheA2 were expressed in wild-type cells, and that mutants in the respective cheA genes were deficient in the proteins that these genes encode.
Figure 2.
Western blot analysis using different antisera reacted against whole-cell lysates of wild-type and cheA mutants. Anti-DnaK was used as an internal control. FlaB is the major and FlaA is the minor PF filament proteins (30).
The insertion of the kan cassette within cheA1 or cheA2 could conceivably alter gene expression of downstream genes and thus complicate subsequent interpretations. However, several lines of evidence indicate that kan insertion into cheA1 or cheA2 did not have a polar effect on downstream gene expression. First, RT-PCR analysis indicated that all of the motility and chemotaxis genes downstream in both the cheA1∷kan and cheA2∷kan mutations were still transcribed (not shown). Second, although insertion of kan in cheA1 had no obvious phenotype (see below), mutations in the downstream gene orf569 resulted in cells that fail to reverse (C.L. and N.W.C., unpublished work). Thus, the mutation in cheA1 did not negatively impact the expression of orf569. Third, because cheY3 is downstream of cheA2, we directly tested for CheY3 expression in mutant LC-A2 by Western analysis (Fig. 2c). By using an antiserum directed to the recombinant protein, CheY3 was found to be expressed at approximately the same level in mutant LC-A2 as in the wild type. These results suggest that there was no inhibition of CheY3 synthesis by the kan insert. Finally, mutations in cheX, which is immediately downstream of cheA2 and is of unknown function, result in cells that constantly flex (M.A.M. and N.W.C., unpublished work). Because the cheA2∷kan mutant had such a markedly different phenotype than cheX∷kan (see below), these results suggest that cheX is still expressed in the cheA∷kan mutant.
Behavior of Single cheA Mutants.
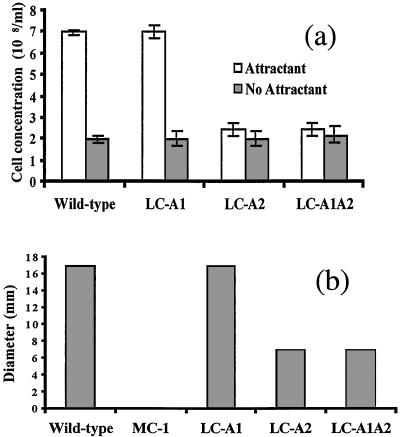
We determined whether the mutants obtained were deficient in chemotaxis. Two different assays were used to test for chemotaxis in the wild-type and cheA mutants (23, 35). Rabbit serum has been previously shown to be an attractant for B. burgdorferi (23). Using the capillary tube assay, wild type and LC-A1 had a strong chemotactic response to 0.5% rabbit serum (Fig. 3a). In contrast, LC-A2 failed to have a response with this assay. We also used the swarm plate assay to test for chemotaxis. Bacteria are known to swarm out in rings when undergoing chemotaxis in soft-agar plates (35). Wild-type and LC-A1 cells swarmed on soft-agar plates in a ring-formation with BSK-II medium diluted 1:5 (Fig. 3b). Because swarms on undiluted medium were considerably smaller (not shown), evidently a gradient of attractant in the diluted medium was generated during incubation. No swarming occurred when testing the nonmotile PF-deficient mutant MC-1 in the diluted medium. These results indicate swarm formation is motility-dependent, and that the cells are chemotactic to components such as rabbit serum present in the BSK-II medium. We found that the size of the ring of mutant LC-A2 was considerably smaller than that of the wild type. Thus, both the capillary tube and swarm-plate assays indicate that mutations in cheA2 but not cheA1 result in an inhibition of chemotaxis.
Figure 3.
Chemotaxis assays of the wild type and mutants. (a) Capillary tube assay using 0.5% rabbit serum. (b) Swarm agar plates assay using BSK-II medium diluted 1:5. Approximately 4 μl containing 1 × 106 cells was deposited on the center of agar plates and incubated 3 days. The results are expressed as ring diameter of a given strain minus that of the nonmotile MC-1 mutant. MC-1 formed had a ring diameter of 15 mm, which was equivalent to the size of the initial drop of cells on the agar.
We analyzed the behavior of LC-A1 and LC-A2 by tracking individual cells in 1% methylcellulose for at least 1 min. Cell velocity and reversal frequency were determined for the individual mutants and compared with the wild type. The run velocity of the mutants was approximately the same as that of the wild type (Table 2). Thus, the mutations in cheA1 or cheA2 did not alter cell velocity. On the other hand, wild-type and mutant LC-A1 cells were found to reverse quite frequently (18–21 reversals per min), but mutant LC-A2 swam in only one direction and failed to reverse. This mutant swam continuously with no stopping or flexing when tracked for as long as 5 min. These results indicate that inactivation of cheA2 but not cheA1 markedly altered the flagellar reversal frequency.
Table 2.
Reversal frequency and velocity of cheA mutants
| Strains | Reversal frequency, reversals per min | Velocity, μm/sec |
|---|---|---|
| Wild type | 18.89 ± 5.08 | 3.02 ± 0.13 |
| LC-A1 | 21.00 ± 2.75 | 2.48 ± 0.71 |
| LC-A2 | 0 | 3.01 ± 0.61 |
| LC-A1A2 | 0 | 3.10 ± 0.32 |
Construction and Analysis of cheA1cheA2 Double Mutant.
The above results indicate that both CheA1 and CheA2 were expressed in growing cells. To ensure that no CheA was synthesized in the cells, we constructed a double mutant cheA1∷kan cheA2∷ermC (LC-A1A2). Essentially the 1.3-kb kan cassette in pGA2kan was replaced by a 1.1-kb ermC cassette (29). After electroporation of LC-A1 and selection with erythromycin, the resultant double mutant was characterized. PCR analysis confirmed that ermC was inserted into cheA2 as expected. Western blot analysis verified that both CheA1 and CheA2 synthesis were inhibited in the LC-A1A2 mutant (Fig. 2 a and b). We tested for downstream effects of the erythromycin cassette in LC-A1A2. CheY3 was still synthesized, but it was ≈1/2 that of the wild type and LC-A2 (Fig. 2c).
The swimming behavior of LC-A1A2 was compared with that of the wild-type and the single cheA mutants. This double mutant had a similar velocity to that of the wild-type and single mutants, and it was identical to LC-A2 in its inability to flex and to reverse direction (Table 2). In addition, it was deficient in chemotaxis using both the capillary tube and swarm plate assays (Fig. 3 a and b). These results indicate that LC-A1A2 resembles LC-A2 with respect to chemotaxis and cell-reversal frequency. It also suggests that cells completely deficient in CheA swim in only one direction. Taken together, the results indicate that asymmetry of PF bundle rotation is based on differences of the flagellar bundles rather than a localized effect of chemotactic signal transducers.
LC-A1A2 Periplasmic Flagella Analysis.
We were concerned that the mutations in LC-A1A2 had unexpected effects on flagellar synthesis. One possible explanation for its constantly running phenotype is that the mutations resulted in the inhibition of synthesis of one bundle of PFs with the other bundle constantly rotating in one direction. To test for this possibility, we analyzed cells by electron microscopy and by Western blot analysis. Electron microscopy of LC-A1A2 revealed bundles of PFs at both cell ends (not shown). In addition, Western blot analysis indicated that the amount of the PF filament proteins FlaA and FlaB in LC-A1A2 were the same as the wild type (Fig. 2d). These results indicate that the phenotype in LC-A1A2 is not caused by the inhibition of PF synthesis at one cell end.
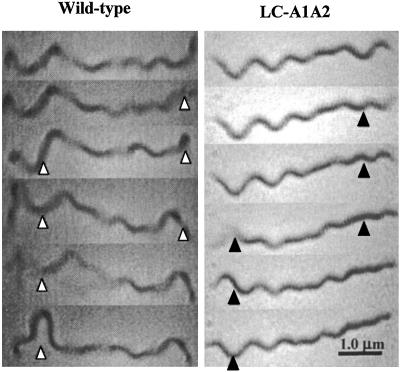
Another hypothesis for the constantly running phenotype of LC-A1A2 is that although both bundles are present in the mutants, only one bundle is capable of rotation, and it does so only in one direction. Accordingly, we monitored the motion of cells tethered to a glass surface. We know from previous studies that PFs influence the shape of the cell in the domain where they reside (4–8), and, in some species, they generate independent gyrational motion (3, 5, 7, 12, 13). We analyzed those occasional tethered cells that gyrated their cell ends with little or no motion in the central part of the cell. Wild-type cells were found to change their gyrational direction (e.g., CW, then CCW) of one end relative to the other. Occasionally, the ends would alternate gyration, with one end stopping, and the other starting (Fig. 4a). Mutant LC-A1A2 was similar, as cells would occasionally alternate the gyration of their cell ends (Fig. 4b). This mutant did not change the direction of gyration of the ends as seen in the wild type, which is consistent with observations of free-swimming cells. These results indicate that both bundles of PFs are capable of rotation in LC-A1A2.
Figure 4.
Sequential video frames of wild-type and LC-A1A2 mutant taken every 0.33 s. Arrow points to the cell end having a change in position relative to the above frame.
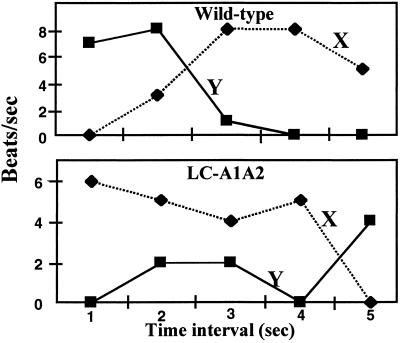
Although the above experiments indicate that both bundles are capable of rotation in LC-A1A2, they did not determine if both were simultaneously active. We tested for simultaneous gyration of both cell ends by analyzing tethered cells as described above. For most wild-type and LC-A1A2 mutant cells, the beat frequencies at both cell ends were identical. Thus, it was difficult to distinguish whether PFs at both cell ends generated rotational motion, or one passively rotated with the other driving the rotation. However, cells of the wild type and LC-A1A2 were observed occasionally with the beat frequency at one end varying with that of the other end (Fig. 5). These results indicate that both bundles of PFs in the wild type and LC-A1A2 can simultaneously rotate and generate gyrational motion.
Figure 5.
Beat frequencies (gyrations or beats per s) of ends arbitrarily designated X and Y of wild-type and LC-A1A2 mutant. The mean beat frequency was determined for each 1-s interval.
Discussion
The two cheA genes inactivated in this study resemble cheA genes of other bacteria, but their phylogenetic origins may be different. cheA1 and cheA2 map far from one another on the linear B. burgdorferi chromosome (25). CheA from several bacterial species has been shown to consist of five functional domains joined by linker regions (16, 17). We found that both CheA1 and CheA2 had extensive homology in each of these five domains to CheA in other bacteria, with less conservation to the cheY binding region known as P2 (not shown). Alignment analysis indicated that B. burgdorferi CheA2 is most similar to CheA proteins of the spirochetes T. pallidum (40% identity) and T. denticola (42% identity). Only one cheA gene is present in these latter species. In contrast, B. burgdorferi CheA1 had its highest homology to that of Rhodobacter sphaeroides CheA2 (35% identity) and Vibrio cholerae CheA1 (35% identity). These results suggest that CheA1 is likely to be a recent acquisition from the Proteobacteria whereas CheA2 is well conserved among the spirochetes. A similar conclusion was reached in analyzing cheY2 which maps within the cheA1 operon and cheY3 which is in the cheA2 operon (I. Zhulin, personal communication). B. burgdorferi resides in both the tick and the mammal, whereas T. pallidum and T. denticola dwell only in the mammal. Perhaps the operon containing cheA1 primarily functions within the tick, whereas the operon with cheA2 is most active within the mammalian host.
Transcription of the operon containing cheA1 was found to be initiated by a σ70 promoter. All five motility operons analyzed to date are initiated by σ70 promoters (1, 31, 34, 36), and no motility gene-specific promoters or σ factors have been identified (25). Several spirochete species have been shown to have σ28-specific motility promoters (1, 37). To our knowledge, B. burgdorferi is the only bacterial species that lacks transcriptional cascade control of motility gene expression by alternative σ factors. Our working hypothesis is that motility and chemotaxis are so vital for the survival of B. burgdorferi in both the tick and mammal that many of the genes are constitutively expressed (1, 31). In support of this hypothesis, B. burgdorferi expresses flaB message and produces PFs in both these hosts (38, 39). Perhaps B. burgdorferi also relies on a translational control system to regulate motility and chemotaxis gene expression (40–43). Preliminary results with insertion mutants in flaA, flaB, flgE, and fliF support this possibility (M. Sal, M.A.M., and N.W.C., unpublished work).
Our results support the hypothesis that the asymmetrical rotation of the PFs during translation is independent of the CheA signal transduction system. LC-A1A2 was deficient in chemotaxis using both the capillary tube and swarm plate assays. This mutant swam only in one direction, and an analysis of tethered cells indicated that both bundles of the PFs simultaneously participate in translational motility; thus, the failure of these mutants to reverse is not the result of only one polar bundle of PFs being active. LC-A1A2 is similar to cheA mutants found in other bacteria, in that they constantly run and do not reverse or tumble. However, in these other species, cheA mutants constantly rotate their flagella in one direction (14, 16, 17). In contrast, the bundles of PFs in B. burgdorferi would necessarily rotate in opposite directions for translation to occur (Fig. 1). Thus, in the absence of CheA, i.e., the default state, one bundle of PFs rotates CCW, and the other bundle rotates CW. These results imply asymmetry and, thus, structural differences with respect to the flagellar motors at the opposite ends of the cell. Such flagellar asymmetry has not been noted in other bacterial species. Similar results have been noted in cheA mutants of T. denticola (R. Lux and W. Shi, personal communication); consequently, it is likely that other spirochetes have asymmetrical rotation of the PFs at opposite ends of the cells in the absence of CheA.
Asymmetrical localization of specific proteins and structures has been described for several bacterial species (44, 45). For example, the stalk structure localizes at one end of the cell in Caulobacter crescentus at a site previously occupied by the flagellum. The ActA protein of Listeria monocytogenes, and the IcsA protein of Shigella flexneri localize at one of the cell poles in each of these species. Localization of these proteins target the old cell pole, but their mechanisms of localization are different. In S. flexneri, localization of IcsA depends on direct targeting to that specific pole, whereas ActA seems to be excluded from the newly synthesized cell pole. Perhaps in B. burgdorferi, there is association with an unknown factor or factors with the flagellar switch complexes at one cell pole. This association could result in the PFs at that end rotating CW rather than CCW in the default state.
Several candidate effectors could be involved in flagellar asymmetrical rotation. One such possible effector is FliG. Specifically, the genomes of B. burgdorferi, T. pallidum, and T. denticola each have two homologs encoding FliG, fliG-1 and fliG-2 (1, 25,26 http://www.hgsc.bcm.tmc.edu/microbial). In B. burgdorferi, fliG-1 and fliG-2 are the only known duplicate homologs that encode proteins for the flagellar apparatus. FliG is part of the switch complex that determines the direction of flagellar rotation (15–17). Perhaps there is differential localization of one of the FliG proteins with motors at one of the cell ends relative to the other. Alternatively, as suggested by W. Shi, CheX also could be such an effector (W. Shi, personal communication). Our results with B. burgdorferi (M.A.M. and N.W.C., unpublished work), and those with T. denticola (L. Lux and W. Shi, unpublished work), suggest that cheX mutants constantly flex or rapidly reverse. Perhaps CheX associates with one cell end to result in flagellar asymmetrical rotation. Now that the tools are available for genetic analysis of spirochetes, the major factors for this asymmetrical rotation in spirochetes can begin to be deciphered.
Acknowledgments
We thank S. Goldstein, M. Sal, and D. Yelton for helpful discussions, and A. Barbour, J. Benach, B. Johnson, and P. Matsumura for antibodies. We also thank R. Lux, W. Shi, and I. Zhulin for stimulating discussions and communicating unpublished material. This research was supported by U.S. Public Health Service Grants AI29743 (to N.W.C.) and AI 43063 (to F.C.C.).
Abbreviations
- PFs
periplasmic flagella
- CW
clockwise
- CCW
counter-clockwise
- RT-PCR
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Li C, Motaleb A, Sal M, Goldstein S F, Charon N W. J Mol Microbiol Biotechnol. 2000;2:345–354. [PubMed] [Google Scholar]
- 2.Charon N W, Goldstein S F, Block S M, Curci K, Ruby J D, Kreiling J A, Limberger R J. J Bacteriol. 1992;174:832–840. doi: 10.1128/jb.174.3.832-840.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein S F, Charon N W, Kreiling J A. Proc Natl Acad Sci USA. 1994;91:3433–3437. doi: 10.1073/pnas.91.8.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bromley D B, Charon N W. J Bacteriol. 1979;137:1406–1412. doi: 10.1128/jb.137.3.1406-1412.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg H C, Bromley D B, Charon N W. Symp Soc Gen Microbiol. 1978;28:285–294. [Google Scholar]
- 6.Picardeau M, Brenot A, Saint Girons I. Mol Microbiol. 2001;40:189–199. doi: 10.1046/j.1365-2958.2001.02374.x. [DOI] [PubMed] [Google Scholar]
- 7.Charon N W, Goldstein S F, Curci K, Limberger R J. J Bacteriol. 1991;173:4820–4826. doi: 10.1128/jb.173.15.4820-4826.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motaleb M A, Corum L, Bono J L, Elias A F, Rosa P, Samuels D S, Charon N W. Proc Natl Acad Sci USA. 2000;97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg H C. Annu Rev Biophys Bioeng. 1975;4:119–136. doi: 10.1146/annurev.bb.04.060175.001003. [DOI] [PubMed] [Google Scholar]
- 10.Fosnaugh K, Greenberg E P. J Bacteriol. 1989;170:1678–1774. [Google Scholar]
- 11.Goldstein S F, Buttle K F, Charon N W. J Bacteriol. 1996;178:6539–6545. doi: 10.1128/jb.178.22.6539-6545.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstein S F, Charon N W. Proc Natl Acad Sci USA. 1990;87:4895–4899. doi: 10.1073/pnas.87.13.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charon N W, Daughtry G R, McCluskey R S, Franz G N. J Bacteriol. 1984;160:1067–1073. doi: 10.1128/jb.160.3.1067-1073.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bren A, Eisenbach M. J Bacteriol. 2000;182:6865–6873. doi: 10.1128/jb.182.24.6865-6873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armitage J P. Adv Microb Physiol. 1999;41:229–289. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- 16.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkinson J S. Annu Rev Genet. 1977;11:397–414. doi: 10.1146/annurev.ge.11.120177.002145. [DOI] [PubMed] [Google Scholar]
- 18.Lux R, Moter A, Shi W. J Mol Microbiol Biotechnol. 2000;2:355–364. [PubMed] [Google Scholar]
- 19.Goulbourne E A, Jr, Greenberg E P. J Bacteriol. 1981;148:837–844. doi: 10.1128/jb.148.3.837-844.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg E P, Canale-Parola E. J Bacteriol. 1977;130:485–494. doi: 10.1128/jb.130.1.485-494.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kataoka M, Li H, Arakawa S, Kuramitsu H. Infect Immun. 1997;65:4011–4016. doi: 10.1128/iai.65.10.4011-4016.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Arakawa S, Deng Q D, Kuramitsu H. Infect Immun. 1999;67:694–699. doi: 10.1128/iai.67.2.694-699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi W, Yang Z, Geng Y, Wolinsky L E, Lovett M A. J Bacteriol. 1998;180:321–235. doi: 10.1128/jb.180.2.231-235.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lux R, Miller J N, Park N H, Shi W. Infect Immun. 2001;69:6276–6283. doi: 10.1128/IAI.69.10.6276-6283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, et al. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 26.Fraser C M, Norris S J, Weinstock C M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, et al. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 27.Bono J L, Elias A F, Kupko J J, III, Stevenson B, Tilly K, Rosa P. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniatias T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. [Google Scholar]
- 29.Sartakova M, Dobrikova E, Cabello F C. Proc Natl Acad Sci USA. 2000;97:4850–4855. doi: 10.1073/pnas.080068797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge Y, Li C, Corum L, Slaughter C A, Charon N W. J Bacteriol. 1998;180:2418–2425. doi: 10.1128/jb.180.9.2418-2425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ge Y, Old I G, Saint Girons I, Charon N W. J Bacteriol. 1997;179:2289–2299. doi: 10.1128/jb.179.7.2289-2299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, Corum L, Morgan D, Rosey E L, Stanton T B, Charon N W. J Bacteriol. 2000;182:6698–6706. doi: 10.1128/jb.182.23.6698-6706.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge Y, Charon N W. FEMS Microbiol Lett. 1997;153:425–431. doi: 10.1111/j.1574-6968.1997.tb12606.x. [DOI] [PubMed] [Google Scholar]
- 34.Ge Y, Charon N W. J Bacteriol. 1997;179:552–556. doi: 10.1128/jb.179.2.552-556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adler J. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 36.Ge Y, Old I, Saint Girons I, Charon N W. Microbiol. 1997;143:1681–1690. [Google Scholar]
- 37.Charon N W, Greenberg E P, Koopman M B, Limberger R J. Res Microbiol. 1992;143:597–603. doi: 10.1016/0923-2508(92)90117-7. [DOI] [PubMed] [Google Scholar]
- 38.Gilmore R D, Jr, Mbow M L, Stevenson B. Microbes Infect. 2001;3:799–808. doi: 10.1016/s1286-4579(01)01435-6. [DOI] [PubMed] [Google Scholar]
- 39.Das S, Barthold S W, Giles S S, Montgomery R R, Telford S R, III, Fikrig E. J Clin Invest. 1997;99:987–995. doi: 10.1172/JCI119264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlinsey J E, Lonner J, Brown K L, Hughes K T. Cell. 2000;102:487–497. doi: 10.1016/s0092-8674(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 41.Bonifield H R, Yamaguchi S, Hughes K T. J Bacteriol. 2000;182:4044–4050. doi: 10.1128/jb.182.14.4044-4050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aldridge P, Hughes K T. Trends Microbiol. 2001;9:209–214. doi: 10.1016/s0966-842x(01)02014-5. [DOI] [PubMed] [Google Scholar]
- 43.Chilcott G S, Hughes K T. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lybarger S R, Maddock J R. J Bacteriol. 2001;183:3261–3267. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shapiro L, Losick R. Cell. 2000;100:89–98. doi: 10.1016/s0092-8674(00)81686-4. [DOI] [PubMed] [Google Scholar]