Abstract
We have developed an alternative to transgenesis for producing antigen-specific T cells in vivo. In this system, clonal naive T cells with defined antigen specificity are generated by retrovirus-mediated expression of T cell antigen receptor cDNAs in RAG1-deficient murine hematopoietic precursor cells. These T cells can be stimulated to proliferate and produce cytokines by exposure to antigen in vitro, and they become activated and expand in vivo after immunization. IL-2-deficient T cells generated by this technique show decreased proliferation and cytokine production, both of which can be rescued by exogenous addition of this growth factor. Thus, retrovirus-mediated expression of T cell antigen receptor cDNAs in hematopoietic precursor cells permits the rapid and efficient analysis of the life history of antigen-specific T cells in different genetic backgrounds and may allow for the long-term production of antigen-specific T cells with different functional properties for prophylactic and therapeutic purposes.
T cells provide protection against infections and tumors by recognizing peptides presented on major histocompatibility molecules using a highly specific antigen receptor, the T cell receptor (TCR; refs. 1–4). The genetic template for this receptor is created during T cell development in the thymus by the V(D)J DNA rearrangement process, which imparts a unique antigen specificity on each TCR (5–7). The TCR plays an essential role in T cell function, development, and survival. Genetic lesions that interfere with the generation of antigen receptors block T cell development and result in immunodeficiencies (8–10). By defining the specificity of pathogenic T cells, TCRs also are involved centrally in the initiation of autoimmunity and allergies (11, 12).
Because TCRs play such a critical role in T cell development and function, much has been learned about the life history of T cells and the immune responses that they elicit by studying the composition and function of these antigen receptors. The ability to generate mice that express a specific TCR on most T cells by transgenesis has enabled the study of T cell development and function in vivo (13–20). It has also made possible the creation of antigen-specific animal models of many important immunological diseases (18, 21, 22). The first successful strategy to generate TCR transgenic mice involved pronuclear injection of large fragments of genomic DNA encoding the rearranged α and β chains of the TCR (20). In these mice, the expression of the TCR was driven by the homologous promoter and enhancer elements. Subsequent studies have demonstrated that it is possible also to generate TCR transgenic mice expressing constructs that use heterologous promoters that can be either T cell-specific, such as the CD2 promoter (23), or nonspecific, such as the MHC class I promoter, which is expressed in all nucleated cells (17). In TCR transgenic mice that use promoters with activity that is not restricted to T cells, TCRα and TCRβ chains are found in other cell types. However, these proteins are displayed only on the plasma membrane in T cells, because these are the only cells that express CD3, a protein complex required for surface expression of TCRs (24).
Recently a number of groups have shown that functional expression of a TCR can be obtained in mature T cells by using retroviral vectors to carry DNA into these cells (25–29). In these studies, TCRα and TCRβ genes were introduced and stably expressed in T cells that had been activated with a mitogen. By using this approach, T cells derived from nonspecific, heterogeneous populations were converted into antigen-specific T cells capable of responding to protein antigens and tumors (25–29). These results suggest that transgenic expression of TCRs might be used not only to study T cell function but also as a therapeutic strategy to generate antigen-specific T cells capable of targeting tumor antigens and infectious agents.
Although retrovirus-mediated expression of TCR genes shows promise as a strategy to produce antigen-specific T cells, the published approaches have certain limitations. The T cells that are engineered to express TCRs are activated mature cells that already express an endogenous TCR of unknown specificity. Their effector function may be restricted by the conditions under which they are activated in vitro, and it is unclear how long they persist in vivo (30, 31). To attempt to overcome these problems, we have tested whether it is possible to generate naive antigen-specific T cells by expressing TCRα and TCRβ genes in hematopoietic precursor cells derived from RAG1-deficient mice using retroviruses and then injecting these cells into irradiated mice to reconstitute their T cell population. We report here that this approach has been successful, and that T cells produced in this manner respond normally to antigen challenge both in vitro and in vivo. Furthermore, we demonstrate that this strategy can be used to generate T cells with different functional properties by expressing TCRα and TCRβ genes in hematopoietic cells from wild-type and IL-2 knockout mice. Our results demonstrate that retrovirus-mediated expression of TCR genes in hematopoietic precursor cells provides an efficient and rapid new approach for generating antigen-specific T cells that can be used for basic research and might be valuable in the clinic.
Materials and Methods
Mice.
C57BL/6 (B6) mice were purchased from Charles River Breeding Laboratories, and RAG1 and IL-2 knockout mice were purchased from The Jackson Laboratory. IL-2/RAG1 double-knockout mice were generated by breeding. All mice were housed in the California Institute of Technology animal facility according to institute regulations.
MIG-TCR Retroviruses Construction.
The generation of the MIG [murine stem cell virus/internal ribosome entry site/green fluorescent protein (GFP)] retroviral expression vector has been described before (32). OTII TCRα cDNA and OTII TCRβ cDNA were cloned separately into MIG. Retroviruses were generated by transfecting HEK293 T cells using a standard calcium phosphate precipitation technique (33). Viruses were harvested 48 and 72 h after transfection.
Generation of the THZ Hybridoma Cell Line and Infection with Retroviruses.
Activated mouse CD4+ T cells were fused with the BWZ cell line, which contains a reporter gene (lacZ) that is expressed under the control of the nuclear factor of activated T cells (NFAT) elements of the human IL-2 promoter by standard methodology (34). The resulting T cell hybridomas were cloned by limiting dilution. One specific clone, named THZ, lacked TCR expression but still maintained CD3 and CD4 expression. It was sorted by flow cytometry three times to stabilize the TCR-CD3+CD4+ phenotype. THZ has allowed us to test surface expression and specificity of TCRs by assaying antigen responses using a lacZ assay.
THZ cells expressing the OTII TCR were generated by spin infection with both MIG/OTIIα and MIG/OTIIβ retroviruses in the presence of 10 μg/ml polybrene for 90 min at 2,500 rpm at 30°C using a Beckman Allegra 6R centrifuge. The efficiency of infection was determined by assaying GFP fluorescence by flow cytometry. The response of THZ OTII cells to residues 323–339 of chicken ovalbumin (OVAp) was assayed by stimulating these cells with OVAp and B6 spleen cells overnight. The next day, TCR signaling was measured by lacZ assay.
Bulk LacZ Assay.
THZ OTII cells were cultured in 96-well plates, washed once with PBS, lysed, and exposed to the colorogenic β-galactosidase substrate, chlorophenol red β-galactoside (Calbiochem), in 100 μl of Z buffer (100 mM 2-mercaptoethanol/9 mM MgCl2/0.125% Nonidet P-40 in PBS) and incubated at 37°C overnight. The development of the colored lacZ product was assayed by using a plate reader with a 570-nm filter and a 630-nm filter for reference.
Primary CD4+ T Cell Infection and Stimulation.
To obtain CD4+ T cells, splenocytes were harvested from B6 mice. CD8+ T cells were depleted by using Dynabeads against mouse CD8 (Dynal, Oslo) according to manufacturer instructions. CD4+ T cells and spleen antigen-presenting cells (APCs) then were cultured at 2 × 106 per ml in the presence of 0.5 μg/ml anti-CD3 (PharMingen) for 3 days. On days 1 and 2, the cells were spin-infected with both MIG/OTIIα and MIG/OTIIβ retroviruses in the presence of 5 μg/ml polybrene for 90 min at 2,500 rpm at 30°C. After each infection, the retroviral supernatant was replaced. On day 3, the cells were collected, and some were used to assay the expression of OTII TCR by flow cytometry. The rest were cultured O/N with 40 units/ml IL-2 (BioSource International, Camarillo, CA) at 1 × 106 cells per ml. The next day, the rested cells were tested for their responsiveness to OVAp by culturing them at 2 × 105 cells per well in a 96-well plate with 2 × 105 cells per well of irradiated B6 spleen cells as APCs in T cell culture medium containing OVAp at 0–10 μg/ml. Three days later, [3H]thymidine was added to the wells at 0.01 mCi/ml (1 Ci = 37 GBq) followed by O/N culture. The proliferation of the cells was analyzed with a Wallac (Gaithersburg, MD) 3H counter.
Bone Marrow (BM) Precursor Cell Isolation, Infection, and Transfer.
RAG1 knockout mice in a wild-type or IL-2 knockout background were treated with 250 μg/g of body weight of 5-flurouracil (Sigma) dissolved in PBS. BM cells were harvested 5 days later from the tibia and femur and cultured for 5 days at 2 × 106 cells per ml with 20 ng/ml recombinant mouse IL-3, 50 ng/ml recombinant mouse IL-6, and 50 ng/ml recombinant mouse stem cell factor (SCF) (all from Biosource International) in DMEM containing 10% FCS. After 48–72 h, the BM cells were spin-infected with a mixture of MIG/OTIIα and MIG/OTIIβ retroviruses supplemented with 8 μg/ml polybrene for 90 min at 2,500 rpm at 30°C. After infections, the retroviral supernatant was replaced. Recipient RAG1−/− mice of desired genetic background received a total of 480 rads of whole-body radiation and then were injected intravenously with 1–2 × 106 infected BM cells. BM-recipient mice were maintained on the mixed antibiotic sulfamethoxazole and trimethoprim oral suspension (Hi-Tech Pharmacal, Amityville, NY) in a sterile environment for 6–11 weeks until analysis.
Immunization of BM-Recipient Mice.
Ten weeks after irradiation and receiving BM, mice were immunized by intraperitoneal injection of 200 μg of OVAp in PBS and then left for 6 days until analysis.
In Vitro T Cell Stimulation and Proliferation Assays.
Spleen cells from BM-recipient mice were harvested and cultured at 2 × 105 cells per well in a 96-well plate with 2 × 105 cells per well irradiated B6 spleen cells as APCs in T cell culture medium containing OVAp at 0–10 μg/ml. Three days later, culture supernatants were collected and assayed for IL-2 and IFN-γ levels by ELISA. Proliferation was assessed by [3H]thymidine incorporation.
IL-2 and IFN-γ ELISA.
ELISA plates (96-well) were coated with 50 μl per well of 1 μg/ml anti-mIL-2 or anti-mIFN-γ antibody (PharMingen) in carbonate buffer for 2 h at 37°C. The plates then were washed twice with PBS, blocked by adding 100 μl per well of dilution buffer [2% borate buffered saline (BBS)/0.002% sodium azide], and incubated for 30 min at 37°C. After being washed four times with PBS, sample supernatants were added to the plates at a final volume of 50 μl per well and incubated for 3 h at 37°C. The plates were then washed four times followed by the addition of 50 μl per well of 1 μg/ml of the detecting biotinylated antibody (PharMingen) in dilution buffer and incubated for 45 min at room temperature. The plates were then washed six times with PBS, followed by addition of 50 μl/well of the Avidin-Alkaline Phosphotase (PharMingen) at a dilution of 1:400 in dilution buffer. The plates were then incubated for 30 min at room temperature. Finally, the plates were washed six times with PBS. Developing solution Sigma 104 phosphatase substrate (Sigma) was made up at 1 mg/ml in DEA buffer right before use and then added at 50 μl per well. Absorbance was assayed by using a plate reader with a 405-nm filter.
Results
Functional Expression of the OTII TCR in T Cell Lines and Primary CD4+ T Cells Using Retroviruses.
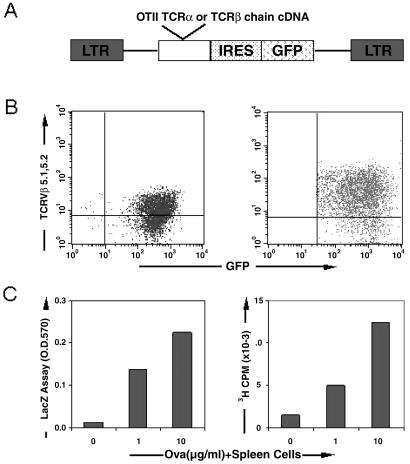
We previously generated a bicistronic retroviral expression vector MIG that allows efficient expression of genes in primary cells of hematopoietic origin (32). This retroviral vector expresses both GFP, to mark infected cells, as well as a test gene. To examine whether we could use retroviruses to functionally express TCRs in T cell populations, we cloned the OTII TCRα or TCRβ chain cDNAs into MIG (Fig. 1A). The OTII TCR is a well defined class II MHC-restricted TCR that responds to residues 323–339 of chicken ovalbumin (35). MIG retroviruses expressing the cDNA for the OTII TCRα or OTII TCRβ chains were used to coinfect the THZ hybridoma line. This cell line expresses CD3, and thus it can express TCRs on its surface. It also contains a reporter gene (lacZ) that is expressed under the control of the NFAT element of the human IL-2 promoter and can be used to assay TCR signaling (30). The infected THZ cells (marked by GFP fluorescence) expressed the β chain of the OTII TCR at the cell surface (Fig. 1B Left) and expressed the lacZ reporter gene when stimulated with OVAp presented by APCs (Fig. 1C Left), indicating that we had achieved functional expression of TCR in this hybridoma line.
Figure 1.
Functional expression of the OTII TCR in a T cell hybridoma line and primary CD4+ T cells using retroviruses. (A) Schematic representation of the MIG retrovirus construct expressing the cDNA for the OTII TCRα or TCRβ chain. LTR, long terminal repeat; IRES, internal ribosomal entry site. (B) Surface expression of the OTII TCRβ chain in infected (GFP+) THZ cells (Left) and primary CD4+ T cells (Right). Cells were coinfected with MIG retroviruses expressing the cDNA for the OTII TCRα or OTII TCRβ chain and then stained with an antibody against TCR Vβ5, which is the Vβ element used by the OTII TCRβ chain. (C) Functional expression of the OTII TCR in THZ cells (Left) and primary CD4+ T cells (Right). Cells were coinfected with MIG retroviruses expressing OTII TCRα chain or OTII TCRβ chain and restimulated for 48 h with OVAp in the presence of B6 spleen cells as APCs. Antigen responses were evaluated by assaying the induction of β-galactosidase expression for THZ cells and by [3H]thymidine incorporation for primary CD4+ T cells.
We next tested whether we could express the OTII TCR in primary T cells by using retroviruses. In these experiments, purified CD4+ T cells from wild-type C57BL/6 mice were activated with an antibody to CD3ɛ and infected with MIG OTIIα and MIG OTIIβ viruses. The infected T cells (marked by GFP fluorescence) expressed the β chain of the OTII TCR at the cell surface (Fig. 1B Right) and proliferated when cultured with OVAp presented by APCs (Fig. 1C Right), indicating that we had achieved functional expression of TCR in primary CD4+ T cells.
Generation of OTII TCR Monospecific T Cells in Vivo After Retrovirus-Mediated Gene Delivery into BM Precursor Cells.
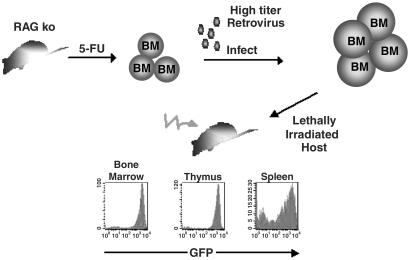
We previously developed a strategy to infect BM precursor cells with the MIG retrovirus (36). To test whether it is possible to generate functional T cells by expressing TCRs in these cells, we harvested BM from wild-type and IL-2 knockout RAG1-deficient mice and infected them with retroviruses expressing OTIIα and OTIIβ. These cells were injected into lethally irradiated RAG1-deficient recipient mice and allowed to reconstitute their immune system, thus producing OTII-monospecific mice (Fig. 2).
Figure 2.
Strategy to generate OTII monospecific T cells in vivo using retroviruses. Hematopoietic precursor cells are obtained from wild-type and IL-2-deficient RAG knockout mice that have been treated with 5-fluorouracil. These cells then are cultured in the presence of cytokines and coinfected with MIG retroviruses expressing the cDNA for the OTII TCRα or OTII TCRβ chain. The infected hematopoietic precursor cells then are transferred into a lethally irradiated host mouse and allowed to reconstitute the immune system. Retrovirally transduced cells are identified by their expression of the GFP.
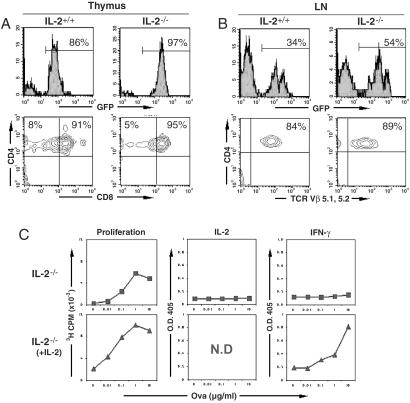
To determine whether retrovirus-mediated expression of the OTII TCR could drive T cell development, we harvested and stained thymocytes 11 weeks after the mice received BM cells. The first indication that T cell development had been rescued was that the cellularity of the thymi derived from mice expressing OTIIα and OTIIβ chains was increased greatly compared with that from control mice that received BM precursor cells infected with the empty MIG vector (data not shown). As shown in Fig. 3A, the majority (>80%) of cells in the thymi of mice receiving OTII-expressing cells were GFP-positive. These thymocytes showed the expected distribution of CD4 and CD8 markers for a developing class II-restricted T cell. Consistent with the absence of endogenous TCRs, all mature T cells in the thymus were CD4 single-positive. The development of OTII T cells was identical in wild-type and IL-2-deficient backgrounds, supporting the notion that IL-2 is not required for CD4+ T cell development (37–39).
Figure 3.
Development of functional wild-type and IL-2-deficient OTII monospecific T cells derived from retrovirally transduced hematopoietic precursor cells. (A) Normal thymic development of OTII monospecific CD4+ T cells in lethally irradiated mice receiving retrovirally transduced hematopoietic precursor cells. Thymocytes obtained from lethally irradiated host mice 11 weeks after injection of retrovirally transduced hematopoietic precursor cells were stained with antibodies to CD4 and CD8 and analyzed by flow cytometry. The distribution of CD4 and CD8 expression on GFP+ thymocytes is shown. (B) Presence of mature OTII monospecific CD4+ T cells in the peripheral lymphoid organs of mice receiving retrovirally transduced hematopoietic precursor cells. Lymph node (LN) and spleen (data not shown) cells obtained from lethally irradiated host mice 11 weeks after injection of retrovirally transduced hematopoietic precursor cells were stained with antibodies to CD4 and TCR Vβ5 and analyzed by flow cytometry. The distribution of CD4 and TCR Vβ5 expression on GFP+ lymph node cells is shown. (C) Normal functional responses of OTII monospecific CD4+ T cells obtained from the peripheral lymphoid organs of mice receiving retrovirally transduced hematopoietic precursor cells. Spleen cells obtained from lethally irradiated host mice 11 weeks after injection of retrovirally transduced hematopoietic precursor cells derived from IL-2-deficient mice were supplemented with B6 spleen cells as APCs and stimulated in vitro with OVAp in the presence or absence of exogenous IL-2. Proliferation was assayed after 72 h by [3H]thymidine incorporation and cytokine production by ELISA. The data were normalized for the number of GFP+CD4+TCR Vβ5+ cells present in the starting spleen cell populations. Proliferation and cytokine production were seen with wild-type OTII T cells both in the presence and absence of IL-2 (data not shown). N.D, not determined.
We also tested whether there were mature CD4+ T cells in the peripheral lymphoid organs of the OTII-monospecific mice. From 30 to 60% of the cells in the lymph nodes and spleens (data not shown) of these mice were GFP-positive (Fig. 3B). More than 80% of these cells were mature CD4+ T cells that expressed the OTII Vβ element, Vβ5. Similar results were observed in both wild-type and IL-2-deficient backgrounds. These observations demonstrated that retrovirus-mediated expression of TCR cDNAs in BM precursor cells could drive normal T cell development.
To determine the kinetics with which monospecific T cells populated recipient mice after BM transfer, we performed a time-course experiment. The earliest time point at which we observed mature (CD4 single-positive) T cells in the thymus of recipient mice was 6 weeks after reconstitution (data not shown). However, no T cells were detectable in the peripheral lymphoid organs of these mice at this time. At 8 weeks we started to observe small numbers of monospecific CD4 single-positive T cells in the lymph nodes and spleen. These cells were able to respond to OVAp, indicating that they were mature. By 10–11 weeks the reconstitution of the T cell compartment was complete, and substantial numbers of functional OTII T cells were detectable in all lymphoid organs analyzed.
Normal in Vitro and in Vivo Antigen Responses of OTII TCR Monospecific T Cells.
To test whether the mature T cells present in OTII-monospecific mice were functional, we stimulated cells from lymph nodes and spleens derived from wild-type and IL-2 knockout mice with increasing concentrations of OVAp. OTII T cells derived from wild-type mice proliferated and secreted IL-2 when stimulated with OVAp (data not shown). IL-2 knockout OTII T cells did not secrete IL-2 or the effector cytokine IFN-γ after antigen stimulation and showed a weak proliferative response to antigen (Fig. 3C). The addition of exogenous IL-2 rescued proliferation and cytokine production (Fig. 3C). These results demonstrate that the mature T cells found in OTII-monospecific mice are functionally normal, and T cells with defined genetic lesions exhibit the expected effects of this mutation.
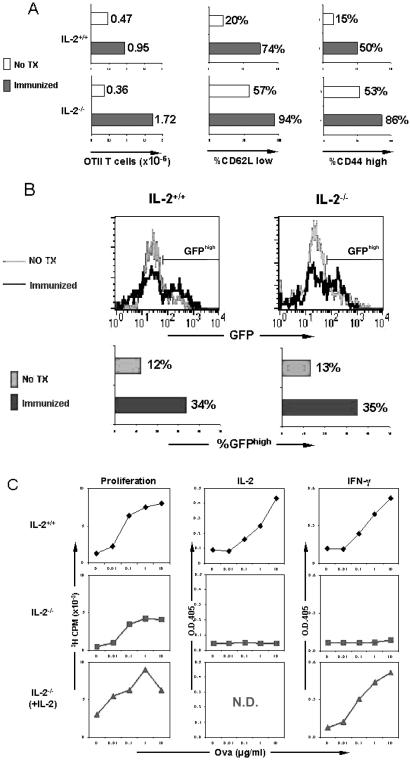
In the final set of experiments, we analyzed the response of OTII-monospecific T cells to antigen stimulation in vivo by immunizing mice with OVAp. Cells from the lymph nodes and spleen were harvested 6 days later and counted, stained with antibodies to activation markers, and restimulated in vitro. In both the wild-type and IL-2-deficient backgrounds, there was a significant increase in T cell number after immunization (Fig. 4A). The accumulating cells expressed surface markers typical of activated T cells (CD62Llow and CD44high; Fig. 4A; refs. 40 and 41). Interestingly, we noticed that immunization led to a preferential expansion of GFPhigh cells (Fig. 4B). Because the expression of GFP correlates with the expression of TCR in our vector system (32), this indicates that antigen exposure selects for T cells that express higher levels of TCR in the mice. After restimulation with OVAp in vitro, wild-type OTII T cells from immunized mice proliferated vigorously and secreted the effector cytokine, IFN-γ (Fig. 4C). As expected, immunized IL-2-deficient T cells showed reduced proliferation and IFN-γ production unless exogenous IL-2 was added to the cultures (Fig. 4C). These results demonstrated that retrovirus-mediated expression of TCR cDNAs in BM precursor cells can give rise to functionally mature T cells with different genetic backgrounds that respond normally to antigen exposure in vivo.
Figure 4.
In vivo responses of wild-type and IL-2-deficient OTII monospecific T cells after immunization with cognate antigen. (A) T cell expansion and expression of activation markers after immunization. Lethally irradiated host mice were immunized via an intraperitoneal injection of 200 μg of OVAp (Immunized) or left untreated (No TX) 10 weeks after receiving retrovirally transduced hematopoietic precursor cells. Spleen and lymph node cells were harvested and counted 6 days later. An aliquot of these cells was stained with antibodies to CD4 and TCR Vβ5, CD62L, or CD44 and analyzed by flow cytometry. The number of OTII monospecific T cells present in the spleen and lymph nodes of immunized and control mice was determined by multiplying the percentage of GFP+CD4+TCR Vβ5+ cells by the total number of cells present in these organs. The frequency of activated T cells was determined by gating on GFP+ CD4+TCR Vβ5+ and CD62Llow or CD44high cells. (B) Preferential expansion of GFPhigh OTII monospecific CD4+ T cells after immunization. Mice receiving retrovirally transduced hematopoietic precursor cells were immunized as described for A. Spleen and lymph node cells were collected and stained with antibodies to CD4 and TCR Vβ5 and analyzed by flow cytometry. The expression of GFP in Vβ125+CD4+OT.II T cells and the frequency of GFPhigh OTII T cells are shown. (C) Effector function of OTII monoclonal CD4+ T cells after immunization. Mice receiving retrovirally transduced hematopoietic precursor cells were immunized as described for A. Spleen cells were harvested and stimulated in vitro with OVAp in the presence of B6 spleen cells as APCs. Proliferation was assayed by [3H]thymidine incorporation, and cytokine production was assayed by ELISA. The data were normalized for the number of GFP+CD4+TCR Vβ5+ cells present in the starting populations. N.D., not determined.
Discussion
We report here a strategy to generate functional antigen-specific T cells by expressing TCRα and TCRβ chain cDNAs in RAG1-deficient hematopoietic precursor cells using a retrovirus-based vector and then injecting the cells into lethally irradiated host mice. The T cells that develop in these mice respond normally to peptide antigen when stimulated in vitro and can be expanded and activated in vivo by immunization. Our approach allows the rapid and efficient generation of antigen-specific T cells in different genetic backgrounds and provides a tool for studying T cell function. It may be possible to adapt this strategy to generate T cells with prophylactic and therapeutic value.
TCR-Expressing Retroviruses Drive T Cell Development.
The ability to impart on T cells a defined antigen specificity by transgenic expression of TCR genes has proven extremely valuable as an approach for studying the immune system. Traditionally, this feat has been accomplished by injecting linear DNA fragments that express TCRα and TCRβ genes under the control of homologous or heterologous promoters into the nuclei of embryos. Recent studies have shown that it is possible also to functionally express TCRs directly in mature T cells by using retroviruses. This finding suggests that TCR transgenesis might be used for therapeutic as well as scientific applications.
Our current findings demonstrate that it is possible to obtain functional antigen-specific T cells in vivo by retrovirus-mediated expression of TCRα and TCRβ cDNAs in hematopoietic precursor cells. We accomplished this by using a well-studied retroviral vector, based on the murine stem cell virus (42). The advantage of the murine stem cell virus is that it maintains long-term and stable expression in hematopoietic precursor cells and their differentiated progeny (43). Importantly, the levels of TCRα and TCRβ cDNAs obtained from this vector in the T cell compartment are sufficiently high to drive T cell development and function. We have detected large numbers of mature antigen-specific T cells up to 3 months after injection of infected hematopoietic precursor cells. Because these cells are derived from long-lived progenitor cells, it is likely that they will be generated continually and persist for many months, possibly even for the lifetime of the mouse (36). Previous attempts to generate monospecific T cell populations by using other oncoretrovirus-based vectors have failed to show detectable or functional expression of these receptors, presumably because the vectors were silenced during hematopoietic development (40).
The only significant drawback of the retroviral vectors that we have used in this study is that they can only introduce DNA into cells that are proliferating. Hematopoietic stem cells usually are quiescent. To induce them to proliferate, they have to be harvested from mice that have been treated with the cytotoxic agent 5-fluorouracil and then cultured with cytokines in vitro. In contrast, lentivirus-based vectors are able to express genes in noncycling cells (44). We recently developed protocols to express TCR genes in unmanipulated hematopoietic precursor cells by using well defined HIV-derived vectors (data not shown).
Use of Retroviruses to Study T Cell Development and Function.
Traditional TCR transgenic mice are designed to express a complex genomic fragment of the TCRα and TCRβ locus, not the cDNA for these genes (45). In fact, transgenic mice have been created with the TCRα and TCRβ cDNAs for the OTII TCR that we use here, but they did not show surface expression of this TCR or produce any OTII T cells (35). It is thought that genomic fragments produce better-regulated expression of TCRs, leading to more efficient generation of transgenic T cells. The fact that we have been successful with cDNAs may reflect the circumstance that in each experiment we generate a large number of hematopoietic precursor cells that express TCR genes at different levels. Any individual retrovirus transgenic mouse receives ≈1 million of these cells. It is likely that during thymic development only those progenitor cells that express an appropriate amount of TCR proteins give rise to mature T cells. Supporting the idea that populations of T cells can be selected on the basis of the level of TCR that they express in our monospecific T cell mice, we observed that immunization led to an increase in GFPhigh (TCRhigh) cells. It is likely that the limited amount of cognate peptide antigen present in immunized mice led to a preferential expansion of T cells expressing high amounts of the OTII TCR.
The available data suggest that neither transgenic nor retrovirus-mediated expression of TCRs faithfully reproduce the patterns of endogenous TCR expression. How this impacts the development of T cells remains unclear. However, a growing body of anecdotal and direct evidence suggests that the timing and level of TCR expression plays an important role in determining the fate of a T cell (46–50). New retroviruses that drive expression of genes under the control of tissue-specific and regulated promoters should provide a novel and efficient tool to address this important question in T cell biology further (44).
An additional advantage of using retroviruses to study T cell development is that they can be used to introduce TCR genes into hematopoietic stem cells derived from mice of different genetic backgrounds or with defined genetic lesions, which has enabled us to generate antigen-specific T cells that are deficient for IL-2, and should allow the rapid and efficient evaluation of genetic effects more generally on the development and function of antigen-specific T cells.
Therapeutic and Prophylactic Uses of Retrovirus-Generated T Cells.
As well as providing a tool for studying T cell biology and immune responses, our approach to generate antigen-specific T cells with defined functional properties might be applied in a clinical setting. Most importantly, our approach allows the production of large numbers of naive T cells for prolonged periods of time. These cells can be activated into effector T cells both in vitro and in vivo by using a known peptide antigen. By expressing TCRs with the appropriate antigen specificity, these T cells might provide protection against infectious diseases such as HIV/AIDS and cancers. Alternatively, they could be used to regulate the development of allergies and autoimmune diseases.
The TCR monospecific T cells generated in this study were derived from hematopoietic precursor cells deficient in the RAG1 gene. These cells were used because they produce lymphoid cells that fail to express endogenous TCR chains. Such endogenous proteins might compete with the retrovirally expressed TCR for surface expression and reduce the efficiency with which antigen-specific T cells were generated. Although our study shows that transgenic T cells can be generated readily from RAG1-deficient progenitor cells, it would be useful, as a model for potential human therapeutics, to be able to generate these cells with wild-type cells as well. We already have been able to generate T cells expressing a defined TCRβ chain by retrovirus-mediated gene expression in wild-type hematopoietic precursor cells (data not shown). It will also be informative to test whether it is possible to generate transgenic T cells by injecting infected progenitor cells into untreated mice or mice that receive myeloablative treatments that are less severe than whole body irradiation. These treatments are less likely to produce long-lasting damage to the host and more closely resemble protocols that might be used in humans. Similar approaches have been developed by using autologous hematopoietic stem cells to produce long-term microchimerism that permits the transplantation of immunologically incompatible organs (51).
Acknowledgments
We thank Francis Carbone and William Heath for providing us with the OTII TCRα-12 and TCRβ-12 cDNAs and Nilabh Shastri for providing us with the BWZ cell line. We are grateful to Susan Kovats for her assistance in cell-proliferation assays. X.-F.Q. is the recipient of Damon Runyon–Walter Winchell Fellowship DRG 1568. This work was supported by National Institutes of Health Grants R01 GM39458 (to D.B.) and R21 AI 494897 (to L.V.P.).
Abbreviations
- TCR
T cell antigen receptor
- MIG
murine stem cell virus/internal ribosome entry site/green fluorescent protein
- GFP
green fluorescent protein
- OVAp
chicken ovalbumin peptide, residues 323–339
- APC
antigen-presenting cell
- BM
bone marrow
References
- 1.Hedrick S M, Cohen D I, Nielsen E A, Davis M M. Nature (London) 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- 2.Yanagi Y, Yoshikai Y, Leggett K, Clark S P, Aleksander I, Mak T W. Nature (London) 1984;308:145–149. doi: 10.1038/308145a0. [DOI] [PubMed] [Google Scholar]
- 3.Fremont D H, Rees W A, Kozono H. Curr Opin Immunol. 1996;8:93–100. doi: 10.1016/s0952-7915(96)80111-7. [DOI] [PubMed] [Google Scholar]
- 4.Garcia K C, Teyton L, Wilson I A. Annu Rev Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 5.Schatz D G, Oettinger M A, Baltimore D. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 6.Fugmann S D, Lee A I, Shockett P E, Villey I J, Schatz D G. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 7.Hesslein D G, Schatz D G. Adv Immunol. 2001;78:169–232. doi: 10.1016/s0065-2776(01)78004-2. [DOI] [PubMed] [Google Scholar]
- 8.Shinkai Y, Rathbun G, Lam K P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, et al. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 9.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 10.Villa A, Sobacchi C, Notarangelo L D, Bozzi F, Abinun M, Abrahamsen T G, Arkwright P D, Baniyash M, Brooks E G, Conley M E, et al. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 11.Ji H, Korganow A S, Mangialaio S, Hoglund P, Andre I, Luhder F, Gonzalez A, Poirot L, Benoist C, Mathis D. Immunol Rev. 1999;169:139–146. doi: 10.1111/j.1600-065x.1999.tb01312.x. [DOI] [PubMed] [Google Scholar]
- 12.Kreuwel H T, Sherman L A. Curr Opin Immunol. 2001;13:639–643. doi: 10.1016/s0952-7915(01)00272-2. [DOI] [PubMed] [Google Scholar]
- 13.Teh H S, Kisielow P, Scott B, Kishi H, Uematsu Y, Bluthmann H, von Boehmer H. Nature (London) 1988;335:229–233. doi: 10.1038/335229a0. [DOI] [PubMed] [Google Scholar]
- 14.Sha W C, Nelson C A, Newberry R D, Kranz D M, Russell J H, Loh D Y. Nature (London) 1988;336:73–76. doi: 10.1038/336073a0. [DOI] [PubMed] [Google Scholar]
- 15.Kisielow P, Bluthmann H, Staerz U D, Steinmetz M, von Boehmer H. Nature (London) 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 16.Berg L J, Pullen A M, Fazekas de St Groth B, Mathis D, Benoist C, Davis M M. Cell. 1989;58:1035–1046. doi: 10.1016/0092-8674(89)90502-3. [DOI] [PubMed] [Google Scholar]
- 17.Pircher H, Burki K, Lang R, Hengartner H, Zinkernagel R M. Nature (London) 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 18.Ohashi P S, Oehen S, Buerki K, Pircher H, Ohashi C T, Odermatt B, Malissen B, Zinkernagel R M, Hengartner H. Cell. 1991;65:305–317. doi: 10.1016/0092-8674(91)90164-t. [DOI] [PubMed] [Google Scholar]
- 19.Schonrich G, Kalinke U, Momburg F, Malissen M, Schmitt-Verhulst A M, Malissen B, Hammerling G J, Arnold B. Cell. 1991;65:293–304. doi: 10.1016/0092-8674(91)90163-s. [DOI] [PubMed] [Google Scholar]
- 20.Bluthmann H, Kisielow P, Uematsu Y, Malissen M, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. Nature (London) 1988;334:156–159. doi: 10.1038/334156a0. [DOI] [PubMed] [Google Scholar]
- 21.Katz J D, Wang B, Haskins K, Benoist C, Mathis D. Cell. 1993;74:1089–1100. doi: 10.1016/0092-8674(93)90730-e. [DOI] [PubMed] [Google Scholar]
- 22.Goverman J, Woods A, Larson L, Weiner L P, Hood L, Zaller D M. Cell. 1993;72:551–560. doi: 10.1016/0092-8674(93)90074-z. [DOI] [PubMed] [Google Scholar]
- 23.Mamalaki C, Elliott J, Norton T, Yannoutsos N, Townsend A R, Chandler P, Simpson E, Kioussis D. Dev Immunol. 1993;3:159–174. doi: 10.1155/1993/98015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oettgen H C, Pettey C L, Maloy W L, Terhorst C. Nature (London) 1986;320:272–275. doi: 10.1038/320272a0. [DOI] [PubMed] [Google Scholar]
- 25.Clay T M, Custer M C, Sachs J, Hwu P, Rosenberg S A, Nishimura M I. J Immunol. 1999;163:507–513. [PubMed] [Google Scholar]
- 26.Cooper L J, Kalos M, Lewinsohn D A, Riddell S R, Greenberg P D. J Virol. 2000;74:8207–8212. doi: 10.1128/jvi.74.17.8207-8212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujio K, Misaki Y, Setoguchi K, Morita S, Kawahata K, Kato I, Nosaka T, Yamamoto K, Kitamura T. J Immunol. 2000;165:528–532. doi: 10.4049/jimmunol.165.1.528. [DOI] [PubMed] [Google Scholar]
- 28.Kessels H W, Wolkers M C, van den Boom M D, van der Valk M A, Schumacher T N. Nat Immunol. 2001;2:957–961. doi: 10.1038/ni1001-957. [DOI] [PubMed] [Google Scholar]
- 29.Stanislawski T, Voss R H, Lotz C, Sadovnikova E, Willemsen R A, Kuball J, Ruppert T, Bolhuis R L, Melief C J, Huber C, Stauss H J, Theobald M. Nat Immunol. 2001;2:962–970. doi: 10.1038/ni1001-962. [DOI] [PubMed] [Google Scholar]
- 30.Jamieson B D, Ahmed R. J Exp Med. 1989;169:1993–2005. doi: 10.1084/jem.169.6.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Opferman J T, Ober B T, Ashton-Rickardt P G. Science. 1999;283:1745–1748. doi: 10.1126/science.283.5408.1745. [DOI] [PubMed] [Google Scholar]
- 32.Van Parijs L, Refaeli Y, Lord J D, Nelson B H, Abbas A K, Baltimore D. Immunity. 1999;11:281–288. doi: 10.1016/s1074-7613(00)80103-x. [DOI] [PubMed] [Google Scholar]
- 33.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanderson S, Shastri N. Int Immunol. 1994;6:369–376. doi: 10.1093/intimm/6.3.369. [DOI] [PubMed] [Google Scholar]
- 35.Barnden M J, Allison J, Heath W R, Carbone F R. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 36.Van Parijs L, Refaeli Y, Abbas A K, Baltimore D. Immunity. 1999;11:763–770. doi: 10.1016/s1074-7613(00)80150-8. [DOI] [PubMed] [Google Scholar]
- 37.Sadlack B, Merz H, Schorle H, Schimpl A, Feller A C, Horak I. Cell. 1993;75:253–261. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki H, Kundig T M, Furlonger C, Wakeham A, Timms E, Matsuyama T, Schmits R, Simard J J, Ohashi P S, Griesser H, et al. Science. 1995;268:1472–1476. doi: 10.1126/science.7770771. [DOI] [PubMed] [Google Scholar]
- 39.Willerford D M, Chen J, Ferry J A, Davidson L, Ma A, Alt F W. Immunity. 1995;3:521–530. doi: 10.1016/1074-7613(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 40.Huet S, Groux H, Caillou B, Valentin H, Prieur A M, Bernard A. J Immunol. 1989;143:798–801. [PubMed] [Google Scholar]
- 41.Mobley J L, Rigby S M, Dailey M O. J Immunol. 1994;153:5443–5452. [PubMed] [Google Scholar]
- 42.Hawley R G, Lieu F H, Fong A Z, Hawley T S. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 43.Cherry S R, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R. Mol Cell Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lois C, Refaeli Y, Qin X F, Van Parijs L. Curr Opin Immunol. 2001;13:496–504. doi: 10.1016/s0952-7915(00)00247-8. [DOI] [PubMed] [Google Scholar]
- 45.Kouskoff V, Signorelli K, Benoist C, Mathis D. J Immunol Methods. 1995;180:273–280. doi: 10.1016/0022-1759(95)00002-r. [DOI] [PubMed] [Google Scholar]
- 46.von Boehmer H. Annu Rev Immunol. 1990;8:531–556. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- 47.von Boehmer H, Kirberg J, Rocha B. J Exp Med. 1991;174:1001–1008. doi: 10.1084/jem.174.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu C P, Kappler J W, Marrack P. J Exp Med. 1996;184:1619–1630. doi: 10.1084/jem.184.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruno L, Fehling H J, von Boehmer H. Immunity. 1996;5:343–352. doi: 10.1016/s1074-7613(00)80260-5. [DOI] [PubMed] [Google Scholar]
- 50.Lacorazza H D, Tucek-Szabo C, Vasovic L V, Remus K, Nikolich-Zugich J. J Immunol. 2001;166:3184–3193. doi: 10.4049/jimmunol.166.5.3184. [DOI] [PubMed] [Google Scholar]
- 51.Sykes M. Immunity. 2001;14:417–424. doi: 10.1016/s1074-7613(01)00122-4. [DOI] [PubMed] [Google Scholar]