Abstract
A 3,673-bp murine cDNA predicted to encode a glycosylphosphatidylinositol-anchored protein of 1,088 amino acids was isolated during a study aimed at identifying transcripts specifically expressed in the inner ear. This inner ear-specific protein, otoancorin, shares weak homology with megakaryocyte potentiating factor/mesothelin precursor. Otoancorin is located at the interface between the apical surface of the inner ear sensory epithelia and their overlying acellular gels. In the cochlea, otoancorin is detected at two attachment zones of the tectorial membrane, a permanent one along the top of the spiral limbus and a transient one on the surface of the developing greater epithelial ridge. In the vestibule, otoancorin is present on the apical surface of nonsensory cells, where they contact the otoconial membranes and cupulae. The identification of the mutation (IVS12+2T>C) in the corresponding gene OTOA in one consanguineous Palestinian family affected by nonsyndromic recessive deafness DFNB22 assigns an essential function to otoancorin. We propose that otoancorin ensures the attachment of the inner ear acellular gels to the apical surface of the underlying nonsensory cells.
The mammalian inner ear consists of the cochlea, the organ of hearing, and the vestibule, which is responsible for balance. The vestibule is composed of five separate organs, the saccule, the utricle, and three cristae of the semicircular canals. The apical surface of each sensory organ is covered by an acellular gel. The tectorial membrane (TM) lies over the surface of the organ of Corti in the cochlea, an otoconial membrane covers the macula of the utricle and saccule, and a cupula sits on top of each crista in the ampullae of the semicircular canals. Relative motion generated between the apical surface of the sensory epithelium and the overlying acellular gel, in response to sound-induced basilar membrane motion in the cochlea or head motion in the vestibule, results in deflection of the hair cell's stereociliary bundle, thereby modulating the gating of mechanotransducer channels.
With the exception of prestin (1), all of the inner ear-specific proteins described so far in mammals, namely α- and β-tectorin (2), otogelin (3), and otoconin-95 (4), are components of these acellular gels. In the mouse inner ear, α- and β-tectorin are components of the tectorial and otoconial membranes but are not present in the cupulae (2). Otogelin is present in all of the acellular gels (3). Otoconin-95 is the major component of the otoconia, dense biominerals that load the otoconial membrane and provide inertial mass (4, 5), and is also present in the cupulae. The mammalian TM is a complex structure composed of collagen fibrils imbedded in a collagenase-insensitive striated-sheet matrix (6). Otogelin is associated mainly with the collagen fibril bundles (3, 7), whereas α- and β-tectorin are major components of the striated-sheet matrix (8, 9). Collagens are not prominent components of either the otoconial membranes or cupulae. Thus far, little is known about how these acellular gels of the inner ear are attached to the apical surface of the sensory epithelia.
Identifying genes specifically or preferentially expressed in the inner ear is a powerful approach for understanding its molecular physiology. It is also a useful way to identify genes involved in hereditary deafness. Several strategies based on cDNA subtraction have been developed to isolate such genes (1, 3, 4, 10–12). In this study, we describe the isolation of an inner ear-specific transcript derived from a subtracted cDNA library prepared from the vestibular sensory patches of the mouse inner ear (12). The encoded protein, which we called otoancorin, is specifically located on the apical surface of epithelial cells in the sensory organs of the inner ear at sites where they contact the overlying gels. Furthermore, we establish that otoancorin is defective in nonsyndromic deafness DFNB22.
Materials and Methods
Cloning of the Full-Length Otoa cDNA.
The 731-bp pB4D7 sequence was extended by rapid amplification of cDNA ends (RACE)–PCR. Oligo(dT)-primed double-stranded vestibular cDNA was prepared from postnatal day (P)0-P1 mice, ligated to adaptors, and amplified as described previously (3). The 5′ and 3′ RACE-PCR products, obtained with primer 5′-2432-AGGAAGCAACACCCCAACCATCT and primer 5′-2599-TGGAGGAGGACACTTTCATCAGG, respectively, were cloned into the pGEM-T Easy Vector (Promega) and sequenced. The cDNA was completed in the 5′ direction by using primer 5′-1137-GCATCTGTGGAGAAGGACT.
Reverse Transcription (RT)–PCR Analysis.
Total RNA from brain, eye, inner ear, heart, liver, lung, kidney, and small intestine of P2 mice and the testis and skeletal muscle of adult mice were prepared by the guanidium isothiocyanate procedure (13). RT was performed by using 1 μg of total RNA, an oligo(dT) primer, and SuperScript II Rnase H− Reverse Transcriptase (GIBCO/BRL). One-tenth of the reaction product was PCR-amplified in a total volume of 50 μl by using forward primer B4D7ProF 5′-33-GGGGACCGGAGTGAACAGACA and reverse primer B4D7ProR 5′-3488-GGACTTGGCCGTCAGAAGCAG. PCR for the ubiquitously expressed Hprt gene with forward primer 5′-576-GCTGGTGAAAAGGACCTCT and reverse primer 5′-824-CACAGGACTAGAACACCTGC provided a positive control. The reactions were carried out following a standard protocol, using 30 cycles with the Expand High Fidelity PCR system [Roche Biochemicals (Indianapolis, IN)] for amplication with the B4D7 primers and 35 cycles with Taq polymerase (Promega) for the Hprt primers.
Nucleotide and Protein Sequence Analysis.
Sequence analysis was performed by using gcg software (14) and the blast Network Service of the National Center for Biotechnology Information (15). The dgpi program was used to predict the glycosylphosphatidylinositol (GPI)-anchoring motif (http://129.194.186.123/GPI-anchor/index_en.html). The search for protein domains and internal repeated motifs was carried out by smart (http://smart.embl-heidelberg.de). Multiple alignment was performed by using multalin (http://www.toulouse.inra.fr/multalin.html).
Anti-otoancorin Sera.
Two rabbits were injected with a mixture of two nonoverlapping synthetic peptides derived from the sequence of the mature otoancorin protein, peptide 1: NH2–465-CDHKDLWQVLRSPLS-COOH, and peptide 2: NH2–730-CRLLEQWGPPENWTAE-COOH. Antisera were first tested by immunofluorescence and immunoblotting with transfected HeLa or Hek cells expressing otoancorin tagged with green fluorescent protein (GFP). Nontransfected cells were used as a control. Labeling of transfected cells with antisera from either rabbit revealed colocalization of GFP signals and otoancorin immunostaining. Serum J151L2 recognized a band of the expected size (≈153 kDa) on a Western blot and was subsequently affinity purified.
Immunofluorescence and Electron Microscopy of Inner Ear Sections.
Mouse inner ears were fixed by immersion in 4% paraformaldehyde (pH 7.4) for 2–5 h at 4°C. After three PBS rinses, they were immersed in 20% sucrose–PBS for at least 12 h at 4°C and then frozen in Tissue-Tek OCT embedding medium [Sakura Finetek (Zoeterwoude, The Netherlands)]. Cryostat sections (10–12 μm) were stored at −20°C until use. Immunolabeling using the otoancorin antiserum J151L2 (1/1,000) was performed as described (16). Sections were analyzed by conventional epifluorescent microscopy. The specificity of J151L2 was ascertained by immunocompetition with a mixture of the two immunizing peptides. The preimmune serum was used as negative control. For immunoelectron microscopy, cochleae from TectaΔENT/ΔENT mice were labeled with affinity-purified J151L2p antibodies or, as a control, nonimmune rabbit IgG, both at a concentration of 2.3 μg/ml, essentially as described previously (17).
Patients.
Informed consent was obtained from adult subjects and parents of under-aged patients. Pure tone audiometry was systematically performed for each individual over 5 years of age. In younger children, the auditory function was explored by recording the auditory brainstem response.
Genotyping and Linkage Analysis.
Genomic DNA was prepared from blood samples by using a standard phenol-chloroform extraction protocol. Fluorescent microsatellites markers (18) were PCR-amplified and analyzed as described (19). Logarithm of odds (LOD) scores were calculated by using the mapmaker/homoz program. The defect was assumed to be inherited in a recessive mode and fully penetrant. The allele frequencies of the polymorphic markers and the meiotic recombination frequencies for males and females were assumed to be equal.
Mutation Screening of DFNB22 Patients.
Twenty-one DNA fragments encompassing the 28 coding exons of OTOA were PCR-amplified from genomic DNA (100 ng) by using the primers listed in Table 1 (which is published as supporting information on the PNAS web site, www.pnas.org). PCR products were sequenced by using either the amplification primers or additional internal primers (Table 1). Once identified, the IVS12+2T>C mutation was screened by restriction digest analysis with Cac8I (recognition sequence: GCNNGC) of a 499-bp fragment covering exon 12. The majority of human DNA control samples was obtained from the National Laboratory for the Genetics of Israeli Populations at Tel Aviv University, Israel.
Results
Identification of a Mouse Inner Ear cDNA Encoding a GPI-Anchored Protein.
To isolate genes specifically expressed in the inner ear, we searched the subtracted cDNA library prepared from the sensory epithelia of the mouse vestibular apparatus (12) for clones lacking homology to known expressed sequences or only sharing homology with embryonic expressed sequence tags. Among the 231 different sequences of the library, 27 fulfilled these criteria. Clone pB4D7 (for partial B4D7), with a 731-bp ORF, was selected for further study on the basis of its inner ear-specific RT-PCR expression profile (see below). Search for sequence similarity revealed a 81.4% nucleotide identity with a human BAC, clone (CIT987SK-A-270G1), derived from a region of chromosome 16p12. A 3,673-bp poly(A)+ cDNA was reconstituted by using rapid amplification of cDNA ends–PCR. The translation initiation site was identified in a Kozak consensus site (AGGatgT) (20) at position 78 and is preceded by a stop codon at position 63. The ORF of 3,411 bp is followed by a 3′ untranslated region of 185 bp. We found no evidence for alternative splicing. The expression of B4D7 was investigated in several mouse tissues by using RT-PCR (Fig. 1). The B4D7 transcript was exclusively detected in the inner ear.
Figure 1.
RT-PCR analysis of Otoa expression in mouse tissues. (A) Amplification products obtained with B4D7 primers located in the 5′UTR (B4d7ProF) and the last exon (B4d7ProR). A 3,336-bp product is observed only in the inner ear. (B) Products obtained with Hprt primers (positive control). ± indicates presence (+) or absence (−) of reverse transcriptase in the cDNA synthesis reaction. M, 1-kb DNA ladder.
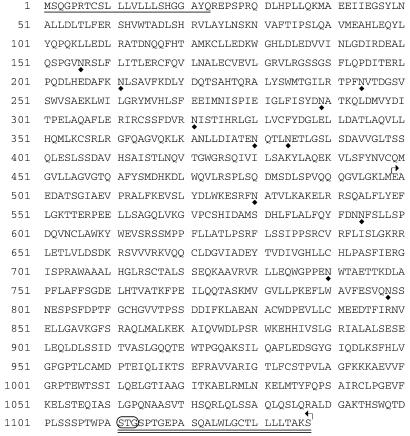
The ORF encodes a polypeptide of 1,137 amino acids with a calculated molecular mass of 126.4 kDa. The predicted protein starts with a 23-amino acid signal peptide (Fig. 2). The deduced protein terminates with a sequence of predominantly hydrophobic residues, characteristic of proteins that are membrane-bound via a GPI anchor (21) (Fig. 2). Eleven potential N-glycosylation sites were detected (Fig. 2). The corresponding protein was named otoancorin in reference to its proposed function (see below). The proteins predicted from the murine and human (deduced from the genomic sequence) otoancorin nucleotide sequences share 77.3% identity. Comparison of the predicted amino acid sequences with the current databases revealed weak similarity between the C-terminal 639-amino acid region of otoancorin and the entire megakaryocyte potentiating factor (MPF)/mesothelin precursor sequence from mouse, rat, and human (≈21–22% identity) (see Fig. 2 and Fig. 8, which is published as supporting information on the PNAS web site, www.pnas.org). Mouse and rat MPF/mesothelin proteins share 58 and 56.1% identity with human MPF/mesothelin, respectively. The MPF/mesothelin precursor, initially attached to the membrane via a GPI anchor, gives rise to MPF, a soluble factor, and to mesothelin, a GPI-anchored cell surface antigen, as a result of a cleavage by a furin protease (22, 23). The function(s) of MPF/mesothelin has not yet been elucidated. However, preliminary data suggest a role of mesothelin in cell to substrate adhesion (23).
Figure 2.
Amino acid sequence of mouse otoancorin. The putative signal peptide is underlined. The characteristic C-terminal signal sequence for addition of a GPI anchor is doubly underlined, with the predicted cleavage site circled. Potential N-glycosylation sites are indicated below the sequence by a diamond-shaped symbol. The region of homology with MPF/mesothelin precursors is delineated by broken arrows.
Expression Pattern of Otoancorin in the Inner Ear.
The distribution of otoancorin in the mouse inner ear was studied by immunofluorescence. Otoancorin was first detected in the cochlea at embryonic day 16.5 (Fig. 3A). At this stage, three regions can be distinguished within the dorsal wall of the cochlear duct. These are (i) the immature spiral limbus (SL), which will give rise to the interdental cells; (ii) the greater epithelial ridge (GER), a zone of tall columnar cells, that will progressively recede to form the inner sulcus; and (iii) the lesser epithelial ridge (LER) (Fig. 3A). The inner hair cells (IHCs) develop from the lateral margin of the GER, and the outer hair cells (OHCs) from the LER. Otoancorin staining, as it first appeared, was restricted to the surface of the SL (Fig. 3A). Otoancorin labeling was shown to persist at the surface of the SL at later stages of development (Fig. 3 B and C) and in adults (not shown). The protein was also detected within the flask-shaped interdental cells at P3 (Fig. 3B). From this stage to P10, an additional patch of otoancorin staining was observed on the luminal surface of the GER, toward its lateral boundary (Fig. 3C). To precisely localize this site of transient otoancorin labeling, immunofluorescence was performed on transgenic mice (line M3H11-R) expressing GFP under control of a Myo7A promoter (24). In these mice, GFP is expressed by IHCs throughout the length of the cochlea and by OHCs in the apical coil. Otoancorin was observed along the surface of cells lying immediately adjacent and medial to the IHCs (see Fig. 3C Inset).
Figure 3.
Distribution of otoancorin in the mouse cochlea revealed by immunofluorescence microscopy. (A) A profile of the cochlear duct at embryonic day 16.5. (B) High magnification of a cross section of the spiral limbus (SL) at P3, showing detail of interdental cells (ic). (C) Section of the organ of Corti at P6. (Inset) A merged image showing a detailed view of the otoancorin-cya3 staining (red) relative to GFP-labeled hair cells (green) in an M3H11-R mouse at P6. GER, greater epithelial ridge; ic, interdental cells; ihc, inner hair cell; IS, inner sulcus; LER, lesser epithelial ridge; ohc, outer hair cells; RM, Reissner's membrane; SL, spiral limbus; TM, tectorial membrane. (Bar = A, B, 30 μm; C, 60 μm.)
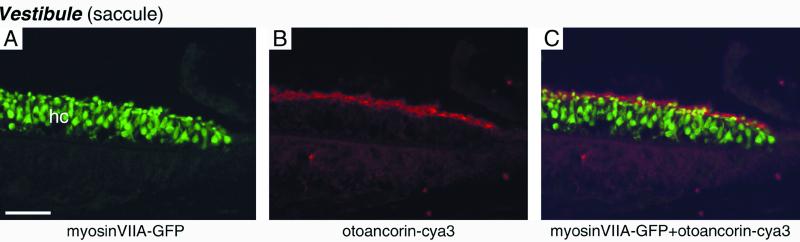
The otoancorin expression pattern was also analyzed in the vestibule of M3H11-R mice, where all hair cells express GFP (Fig. 4A). Otoancorin was observed at the apical surface of the supporting cells surrounding the hair cells in the sensory epithelia of the saccule (Fig. 4 B and C), the utricle, and the cristae ampullaris (not shown). Otoancorin was also found on the apical surface of epithelial cells surrounding the sensory areas; these peripheral regions were always covered by extensions of the acellular gels (not shown).
Figure 4.
Distribution of otoancorin in the saccule of a M3H11-R mouse at P8 revealed by immunofluorescence microscopy. (A–C) Section of the saccular macula. (A) Myosin VIIA-GFP labeling. The sensory hair cells (hc) are labeled in green. (B) Otoancorin-cya3 staining (red). (C) Merged images from A and B. (Bar = 45 μm.)
Although these observations and sequence data strongly suggested that otoancorin was present at the surface of sensory epithelia, it could not be formally excluded that otoancorin was an intrinsic component of the overlying acellular gel. To address this issue, we used α-tectorin mutant mice (TectaΔENT/ΔENT mice), in which the TM is completely detached from the underlying neuroepithelium (25). At P8 and adult stages (Figs. 5 A and B), otoancorin distribution was indistinguishable in α-tectorin mutants from that seen in wild-type control mice. Furthermore, immunoelectron microscopy performed on 7-day-old TectaΔENT/ΔENT mice demonstrated labeling for otoancorin on the apical membrane of the interdental cells (Fig. 5C) and of the epithelial cells lying medial to the IHCs (Fig. 5D).
Figure 5.
Distribution of otoancorin in the cochlea of α-tectorin mutant mice (TectaΔENT/ΔENT mice). Immunohistofluorescence at P8 (A) and adult stage (B). Immunoelectron microscopy at P7, showing the ultrastuctural distribution of otoancorin at the apical surface of the spiral limbus (C) and at the apical surface of border epithelial cells (arrowhead in A) lying medial to IHCs (D). Some gold particles are indicated by arrows. (Bars = A and B, 50 μm; C and D, 200 nm.)
Mutation Analysis in DFNB22 Patients.
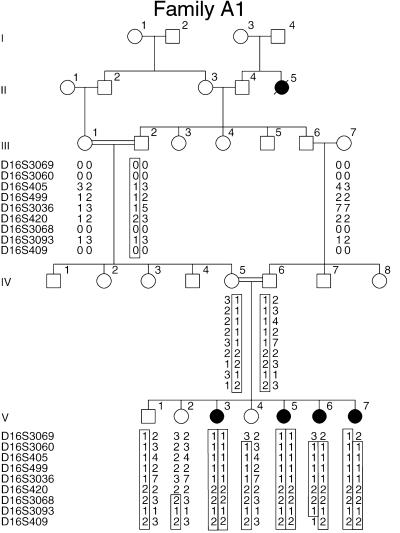
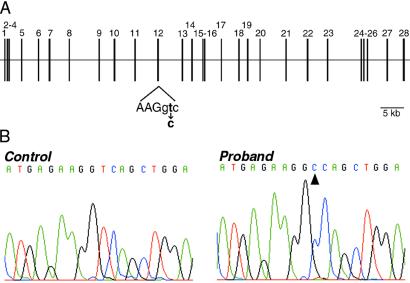
As the mouse gene encoding otoancorin (Otoa) is expressed only in the inner ear, it was considered to be an attractive candidate gene for isolated deafness. The corresponding human gene, OTOA, maps between markers D16S3046 and D16S412 on chromosome 16p12.2. Although no deafness locus had yet been reported in this region, we searched in our collection of 200 large affected families whether the involved deafness locus mapped to this region. Interestingly, in one consanguineous Palestinian family (family A1), the moderate to severe prelingual sensorineural recessive deafness was shown to segregate with a locus (hereafter termed DFNB22) on chromosome 16p13.1-q11.2. Family A1 supports a significant multipoint logarithm of odds (LOD) score (with a maximum of 3.56) for markers included in the region flanked by D16S3069 and D16S409 (Fig. 6). The DFNB22 interval contains OTOA (see Fig. 9, which is published as supporting information on the PNAS web site). We therefore established the exon/intron structure of the gene by comparing the mouse cDNA with BAC CIT987SK-A-270G1 sequences and by using a gene-prediction program (genscan). OTOA consists of 28 exons spanning approximately 82 kb (Fig. 7A). We designed primers to amplify and sequence the 28 exons and their flanking splice sites (see Table 1, which is published as supporting information on the PNAS web site). We identified a T to C transition at the exon 12/intron 12 junction in the homozygous state in one affected child of the family A1 (Fig. 7B). This mutation (IVS12+2T>C), which generates a recognition site for Cac81, affects the invariant T of the donor splice site GT dinucleotide (26). The IVS12+2T>C mutation was found to cosegregate with the hearing impairment in family A1, and was not detected in a total of 417 Jordanian, Lebanese, Palestinian, and Jewish (mainly from Sephardi and Yemenite ethnic groups) individuals with normal hearing. This mutation is expected to lead to aberrant splicing, such as exon 12 skipping (resulting in an in-frame deletion of 72 amino acids) or the use of a cryptic site. Although it could not be formally excluded that the mutation found in the single-family A1 may be nonpathogenic (27), the nature of the mutation found, a consensus splice site mutation, and the tissue specificity of the transcript strongly indicate OTOA is the causative gene for DFNB22. Mutations in consensus splice sites are one of the most common disease-causing mutations (28). Mutations in genes encoding two of the inner ear-restricted proteins, α-tectorin and otogelin, have already been shown to cause deafness (7, 29–31). Further evidence supports the rare occurrence of this form of deafness. None of 150 probands from multiplex families with nonsyndromic recessive deafness (mainly from Caucasian and Chinese origin) screened for mutations in the 28 coding exons of OTOA using SSCP, display mutation. Furthermore, no other families with deafness linked to the DFNB22 locus were found in additional collections of 150 large deaf families (from Israel and Spain) explored.
Figure 6.
Genetic linkage analysis of the DFNB22-affected consanguineous family A1. Segregation analysis with polymorphic microsatellite markers located on chromosome 16 limited the DFNB22 locus interval to between D16S3069 and D16S409. Individuals with prelingual deafness are indicated by filled symbols and unaffected individuals by open symbols. “0” indicates ambiguous positioning of the allele on the gel. The haplotype associated with the mutated DFNB22 allele is boxed.
Figure 7.
Detection of the IVS12+2T>C mutation in family A1. (A) Schematic representation of the exon/intron structure of OTOA. Coding exons are indicated by vertical lines. The IVS12+2T>C mutation identified in the affected DFNB22 family A1 is indicated below. Exonic sequence is represented in uppercase letters, intronic sequence by lowercase letters. (B) Sequence analysis of the coding strand at the exon 12/intron 12 junction in a normal control and an affected child. The T to C transition in the affected child is indicated (arrowhead).
Discussion
All of the noncollageneous glycoproteins of the acellular gels of the inner ear that have been described so far in mammals, namely α-tectorin, β-tectorin, and otogelin, are molecules unique to the inner ear. Here, with the identification of otoancorin, we provide the first report, to our knowledge, of a protein specifically located at the interface between the apical surface of the sensory epithelia and the overlying acellular gel, and also entirely specific for the inner ear.
Sequence data show that otoancorin is probably synthesized as a GPI-linked membrane-bound protein. Motifs characteristic of endoproteinase cleavage sites were not found in the sequence, suggesting otoancorin remains membrane-bound. Furthermore, although transmembrane and/or secreted isoforms of GPI-linked proteins are often expressed as products of the same gene by alternative splicing (32), we found no evidence so far for alternatively spliced Otoa transcripts. Finally, results of immunofluorescence and immunoelectron microscopy, especially in α-tectorin mutant mice, show that otoancorin is present on the apical plasma membrane of epithelial cells and remains attached to the cell surface in the absence of a TM. Like GPI-anchored proteins (32), otoancorin displays a selective enrichment in the apical domain of polarized epithelial cells. Altogether, the present results qualify otoancorin as a GPI-linked membrane-anchored protein.
No homology with known protein domains was found in otoancorin. In particular, no homology was found with the noncollageneous glycoproteins of the acellular gels of the inner ear, all of which have a modular structure (2, 3). Otoancorin shares weak sequence homology with MPF/mesothelin precursors, which are poorly conserved between mouse, rat, and human. However, otoancorin and MPF/mesothelin share global similarity in their predicted secondary structure, being composed mainly of α-helices and random coils [Network Protein Sequence @nalysis (NPS@) Web Server, http://npsa-pbil.ibcp.fr/]. Furthermore, the two proteins have conserved cysteine residues that are likely to be involved in higher-order protein structure via disulfide bond formation. The role proposed for mesothelin in cell to substrate adhesion (23) suggests that otoancorin may act as an adhesion molecule. The relatively high interspecies divergence of murine and human otoancorin (77.3% identity) indicates that this molecule has evolved rapidly and may reflect the fact that this inner ear protein interacts with only a limited number of other molecules.
Otoancorin is first detected in the cochlea at embryonic day 16.5, on the upper surface of the SL, and persists thereafter in this zone where the limbal region of the TM remains permanently attached to the SL. Its transient site of expression on the surface of the GER is also strictly correlated with the transient presence in this region of the homogenous stripe, a mesh-like layer of material under the TM proposed to mediate a contact between the TM and the underlying neuroepithelium during development (33, 34). These correlations suggest otoancorin may mediate TM attachment at two sites, one permanent and another that is transiently maintained during a short period of development. Otoancorin is also detected on the apical surface of epithelial cells in the vestibule in all regions that lie beneath the otoconial membranes and the cupulae, suggesting it also plays a role in mediating the attachment of these acellular gels. Otoancorin may directly interact with otogelin and/or the tectorins at these attachment sites, as these molecules are present in the regions of the acellular gels that come into immediate contact with the epithelial surface (G.R., unpublished data). However, it is noteworthy that the acellular gels are attached not only to nonsensory cells but also to the hair bundles of the sensory hair cells. Because otoancorin has not been detected on hair bundles, the attachment of the acellular gels to the sensory cells must be mediated by other molecules, possibly integrins (35). Thus emerges the idea that a dual cell–matrix adhesion system is involved in the attachment of the acellular gels to the apical surface of the sensory epithelia.
The TM plays an important role in transmitting mechanical energy to the stereociliary bundles of the hair cells (7, 25), where sound is transduced into an electric signal. The identification of the OTOA mutation in family A1 strongly suggests that otoancorin plays an essential function in audition. The early developmental appearance of otoancorin is consistent with the prelingual onset of deafness resulting from a mutation in OTOA.
Supplementary Material
Acknowledgments
We thank the patients and their families for their participation. We thank I. Del Castillo for having kindly performed a screening of the DFNB22 locus in deafness families, M.-C. Simmler for critical reading of the manuscript, S. Chardenoux and S. Lainé for technical help, and P. Wincker and O. Jaillon for sequencing at the Genoscope. This work was supported by grants from the Fondation pour la Recherche Médicale (ARS 2000), the European Economic Community (QL G2-CT-1999–00988), the Wellcome Trust (057410/Z/99/Z), the National Institutes of Health (R01 DC05575, R03 DC04530, DC04293), and the French–Palestinian University Research Cooperation. I.Z. is supported by le Ministère de l'Education Nationale, de la Recherche et de la Technologie.
Abbreviations
- TM
tectorial membrane
- GPI
glycosylphosphatidylinositol
- GFP
green fluorescent protein
- MPF
megakaryocyte potentiating factor
- SL
spiral limbus
- GER
greater epithelial ridge
- IHC
inner hair cell
- RT
reverse transcription
- Pn
postnatal day n
Footnotes
References
- 1.Zheng J, Shen W, He D Z, Long K B, Madison L D, Dallos P. Nature (London) 2000;405:149–155. doi: 10.1038/35012009. [DOI] [PubMed] [Google Scholar]
- 2.Legan P K, Rau A, Keen J N, Richardson G P. J Biol Chem. 1997;272:8791–8801. doi: 10.1074/jbc.272.13.8791. [DOI] [PubMed] [Google Scholar]
- 3.Cohen-Salmon M, El-Amraoui A, Leibovici M, Petit C. Proc Natl Acad Sci USA. 1997;94:14450–14455. doi: 10.1073/pnas.94.26.14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verpy E, Leibovici M, Petit C. Proc Natl Acad Sci USA. 1999;96:529–534. doi: 10.1073/pnas.96.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Kowalski P E, Thalmann I, Ornitz D M, Mager D L, Thalmann R. Proc Natl Acad Sci USA. 1998;95:15345–15450. doi: 10.1073/pnas.95.26.15345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasko J A, Richardson G P. Hearing Res. 1988;35:21–38. doi: 10.1016/0378-5955(88)90037-8. [DOI] [PubMed] [Google Scholar]
- 7.Simmler M-C, Cohen-Salmon M, El-Amraoui A, Guillaud L, Benichou J-C, Petit C, Panthier J-J. Nat Genet. 2000;24:139–143. doi: 10.1038/72793. [DOI] [PubMed] [Google Scholar]
- 8.Richardson G P, Russell I J, Duance V C, Bailey A J. Hearing Res. 1987;25:45–60. doi: 10.1016/0378-5955(87)90078-5. [DOI] [PubMed] [Google Scholar]
- 9.Killick R, Richardson G P. Hearing Res. 1997;103:131–141. doi: 10.1016/s0378-5955(96)00174-8. [DOI] [PubMed] [Google Scholar]
- 10.Heller S, Sheane C A, Javed Z, Hudspeth A J. Proc Natl Acad Sci USA. 1998;95:11400–11405. doi: 10.1073/pnas.95.19.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson N G, Khetarpal U, Gutierrez-Espeleta G A, Bieber F R, Morton C C. Genomics. 1994;23:42–50. doi: 10.1006/geno.1994.1457. [DOI] [PubMed] [Google Scholar]
- 12.Verpy E, Leibovici M, Zwaenepoel I, Liu X-Z, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats Bronya J B, et al. Nat Genet. 2000;26:51–55. doi: 10.1038/79171. [DOI] [PubMed] [Google Scholar]
- 13.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 14.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Amraoui A, Sahly I, Picaud S, Sahel J, Abitbol M, Petit C. Hum Mol Genet. 1996;5:1171–1178. doi: 10.1093/hmg/5.8.1171. [DOI] [PubMed] [Google Scholar]
- 17.Goodyear R, Richardson G. J Neurosci. 1999;19:3761–3772. doi: 10.1523/JNEUROSCI.19-10-03761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, et al. Nature (London) 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 19.Gyapay G, Ginot F, Nguyen S, Vignal A, Weissenbach J. Methods. 1996;9:91–97. doi: 10.1006/meth.1996.0012. [DOI] [PubMed] [Google Scholar]
- 20.Kozak M. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 21.Englund P T. Annu Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- 22.Kojima T, Oh-eda M, Hattori K, Taniguchi Y, Tamura M, Ochi N, Yamaguchi N. J Biol Chem. 1995;270:21984–21990. doi: 10.1074/jbc.270.37.21984. [DOI] [PubMed] [Google Scholar]
- 23.Chang K, Pastan I. Proc Natl Acad Sci USA. 1996;93:136–140. doi: 10.1073/pnas.93.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boëda B, Weil D, Petit C. Hum Mol Genet. 2001;10:1581–1589. doi: 10.1093/hmg/10.15.1581. [DOI] [PubMed] [Google Scholar]
- 25.Legan P K, Lukashkina V A, Goodyear R J, Kossi M, Russell I J, Richardson G P. Neuron. 2000;28:273–285. doi: 10.1016/s0896-6273(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 26.Senapathy P, Shapiro M B, Harris N L. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 27.Bonné-Tamir B, Nystuen A, Seroussi E, Kalinsky H, Kwitek-Black A E, Korostishevsky M, Adato A, Sheffield V C. Am J Phys Anthropol. 1997;104:193–200. doi: 10.1002/(SICI)1096-8644(199710)104:2<193::AID-AJPA5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 28.Antonarakis S E, Krawczak M, Cooper D N. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. I. McGraw–Hill; 2001. pp. 343–377. [Google Scholar]
- 29.Verhoeven K, Van Laer L, Kirschhofer K, Legan P K, Hughes D C, Schatteman I, Verstreken M, Van Hauwe P, Coucke P, Chen A, et al. Nat Genet. 1998;19:60–62. doi: 10.1038/ng0598-60. [DOI] [PubMed] [Google Scholar]
- 30.Mustapha M, Weil D, Chardenoux S, Elias S, El-Zir E, Beckmann J S, Loiselet J, Petit C. Hum Mol Genet. 1999;8:409–412. doi: 10.1093/hmg/8.3.409. [DOI] [PubMed] [Google Scholar]
- 31.Simmler M-C, Zwaenepoel I, Verpy E, Guillaud L, Elbaz C, Petit C, Panthier J-J. Mamm Genome. 2000;11:961–966. doi: 10.1007/s003350010197. [DOI] [PubMed] [Google Scholar]
- 32.Lisanti M P, Rodriguez-Boulan E, Saltiel A R. J Membr Biol. 1990;117:1–10. doi: 10.1007/BF01871561. [DOI] [PubMed] [Google Scholar]
- 33.Lim D J. Hearing Res. 1987;28:9–21. doi: 10.1016/0378-5955(87)90149-3. [DOI] [PubMed] [Google Scholar]
- 34.Lim D J, Rueda J. In: Development of the Auditory and Vestibular Systems. Romand R, editor. Vol. 2. Amsterdam, The Netherlands: Elsevier; 1992. pp. 33–58. [Google Scholar]
- 35.Littlewood Evans A, Muller U. Nat Genet. 2000;24:424–428. doi: 10.1038/74286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.