Abstract
Human CD34 (hCD34)-positive cells are used currently as a source for hematopoietic transplantation in humans. However, in steady-state murine hematopoiesis, hematopoietic stem cells (HSCs) with long-term reconstitution activity are found almost exclusively in the murine CD34 (mCD34)-negative to low fraction. To evaluate the possible differences in hCD34 and mCD34 gene expression in hematopoiesis, we made transgenic mouse strains with human genomic P1 artificial chromosome clones spanning the entire hCD34 genomic locus. In all transgenic mouse strains, a vast majority of phenotypic and functional HSC populations including mCD34−/lo express the hCD34 transgene. These data strongly support the notion that hCD34+ human bone marrow cells contain long-term HSCs that can maintain hematopoiesis throughout life.
Hematopoietic stem cells (HSCs) give rise to all blood cells. CD34 is a sialomucin-like adhesion molecule that is expressed on a few percent of primitive bone marrow cells. In human bone marrow, virtually all colony-forming unit activity resides in the population expressing human CD34 (hCD34). In primates, CD34+ but not CD34− cells repopulate lethally irradiated baboons (1). Retrovirally labeled CD34+ cells are able to reconstitute multilineage hematopoiesis after transplantation in patients with myeloma or breast cancer (2). It has been reported also that hCD34+Thy-1+ cells are highly enriched for human HSCs and possess multilineage reconstitution activity in SCID-hu (severe combined immunodeficient-human) mice (3). Therefore, hCD34+ cells as well as hCD34+Thy-1+ cells are used currently to reconstitute hematopoiesis in patients receiving myeloablative conditioning in autologous and allogeneic transplantation settings (4–6).
In murine hematopoiesis, HSCs are found almost exclusively in the Thy1.1loLineage(Lin)−/loSca-1+ population (7). Within the Thy1.1loLin−/loSca-1+ cells, HSC activity is enriched in a population expressing receptors for the steel factor c-Kit (8, 9). Osawa et al. (10) reported that within the Lin−/loSca-1+c-Kit+ population, mouse long-term HSCs are highly enriched in the murine CD34 (mCD34)−/lo fraction. The fact that mouse long-term HSCs in steady-state bone marrow do not express significant levels of mCD34 raised an important question of whether hCD34+ cells contain self-renewing long-term human HSCs. Furthermore, recent studies show that Thy1.1loLin−/loSca-1+ HSC populations and Lin−mCD34+ hematopoietic cells in murine bone marrow might contain cells capable of differentiation into hepatocytes (11) and other organs (12). Thus, it is important to evaluate whether the expression of hCD34 mimics that of mCD34 in the hematopoietic system.
These data led us to evaluate differences in the regulation of the hCD34 and mCD34 genes. Normal transcriptional regulation of human genes has been reproduced in transgenic mice that carry large artificial chromosomes obtained from yeast, bacteria, or P1 clones (13, 14). Because combinatorial action of multiple, proximal and long-range, cis elements is necessary for proper regulation of hCD34 expression (15), we have established several mouse lines harboring human genomic P1 artificial chromosome (PAC) clones containing the entire hCD34 gene including all exons, introns, and more than 18 kb of flanking 5′ and 3′ genomic sequences. In the present experiments, we performed detailed phenotypic and functional analyses of long-term HSCs in the hCD34 transgenic mice and found that the hCD34 transgene is expressed in long-term mCD34−/lo HSCs. These data support the clinical use of hCD34+ cells as a source for hematopoietic transplantation in humans.
Methods
Mice.
hCD34 transgenic mice were made by transducing hCD34 PAC clones. PAC88L2, PAC7H11, and PAC54A19 were isolated from a PAC human genomic DNA library with an hCD34 probe as described (15). To remove vector sequences, PAC88L2 was digested with SfiI, and PAC7H11 and PAC54A19 were digested with MluI. To obtain a construct containing 48 and 31 kb of 5′-hCD34 sequence, PAC88L2 was digested with AscI plus PvuI and MluI plus PvuI, respectively. Digested DNA was separated with field inversion gel electrophoresis (FIGE, Bio-Rad) for 17 h. After cutting out gels containing genomic DNAs, gels were digested with β-agarase (New England Biolabs). DNAs were purified and then suspended in 1 mM Tris⋅HCl (pH 7.6)/0.1 mM EDTA. DNAs were injected into fertilized oocytes of C57B6 or FVB/N mice and implanted into uteri of pseudopregnant C57B6 or FVB/N mice according to standard procedures. All experiments were performed by using procedures described in the protocol approved by our Institutional Animal Care and Use Committee.
Flow Cytometric Analysis and Cell Sorting.
Fetal liver and bone marrow cells of hCD34 C57B6 transgenic mice were stained with CD3 (KT31.1), CD4 (GK1.5), CD8 (53–6.7), B220 (6B2), Gr-1 (8C5), TER119, CD19 (1D3), IgM (R6–60.2), and IL-7Rα chain (A7R34, Bioscience, San Diego). Lin+ cells were removed partially with sheep anti-rat IgG-conjugated magnetic beads (Dynabeads M-450, Dynal, Oslo), and the remaining cells were stained with anti-Rat-Cy5-phycoerythrin (Tricolor, Caltag, Burlingame, CA). These lineage-depleted cells were stained with FITC-conjugated mCD34 (RAM34) (PharMingen), phycoerythrin-conjugated hCD34 (HPCA-2, Beckton Dickinson), biotinylated anti-Sca-1 (E13–161-7) and allophycocyanin (APC)-conjugated anti-c-Kit (2B8) monoclonal antibodies. Sca-1 was visualized by avidin-APC-Cy7 (Caltag). To analyze side population (SP) cells, cells were incubated with Hoechst 33342 (Sigma) before antibody stainings as reported (16). After staining, cells were resuspended with Hanks' balanced salt solution containing 10 mg/l of propidium iodide. Sorting methods for HSCs, common lymphoid progenitors (CLPs), common myeloid progenitors (CMPs), granulocyte/monocyte progenitors (GMPs), megakaryocyte/erythroid progenitors (MEPs), and other lymphoid progenitors have been described elsewhere (17, 18). CLP, proB, and preB cells were purified as IL-7Rα+Lin−Sca-1loc-Kit+, B220+ IgM-CD43+, and B220+IgM-CD43− bone marrow populations, respectively, and thymic precursor, proT, and preT cells were as CD4loCD8loCD25−c-Kit+, CD4loCD8loCD25+c-Kit+, and CD4loCD8loCD25+c-Kitlo thymocyte populations, respectively. All progenitor populations were sorted or analyzed by using a double laser (488 nm/350 nm Enterprise II + 647 nm Spectrum) high-speed cell sorter (Moflo-MLS, Cytomation, Fort Collins, CO).
Transplantation Assays.
HSC subpopulations purified from hCD34 transgenic mice (C57B6-Ly5.2) were injected into the retroorbital sinuses of lethally irradiated (920 rad delivered in a split dose) C57B6-Ly5.1 mice with 2 × 105 host-type bone marrow cells. Donor-derived cells were evaluated as Ly5.2+Ly5.1− cells after transplantation by using five-color flow cytometric analysis.
Genotyping of hCD34 Transgenic Mice.
Tail DNA was digested with EcoRI (New England Biolabs) and hybridized to a 1.5-kb SacI-digested fragment including hCD34 exon 1 and the 5′ end of intron 1. Several sets of PCR primers were used to confirm the transgene structures including one set at the 3′ end of the hCD34 coding sequence (5′-AGAAGAGATGAGGTGTGAGGAT-3′ and 5′-GGATCCACAAGAATGAGCATGTA-3′). To amplify a 500-bp genomic DNA fragment located at −55 kb, the primers used were 5′-ATCCCCTCTGCCTCTTTTTGGTG-3′ and 5′-AGCCGCTGCTACTGACTTATTGA-3′. To amplify a 500-bp genomic DNA fragment located at −38 kb, the primers used were 5′-ATGGGTGTAGGACCTGAAGTGGT-3′ and 5′-TGGATTGCATAGTTTTTGTTTCC-3′. To amplify a 500-bp genomic DNA fragment located at −10 kb, the primers used were 5′-GTGCTTTCATGGAGAGGGTTTTA-3′ and 5′-TAAGACCTCAAGGGGTTGGACTC-3′.
Results
The hCD34 Transgene Is Expressed in a Majority of Long-Term HSCs Phenotypically Defined by Different Criteria.
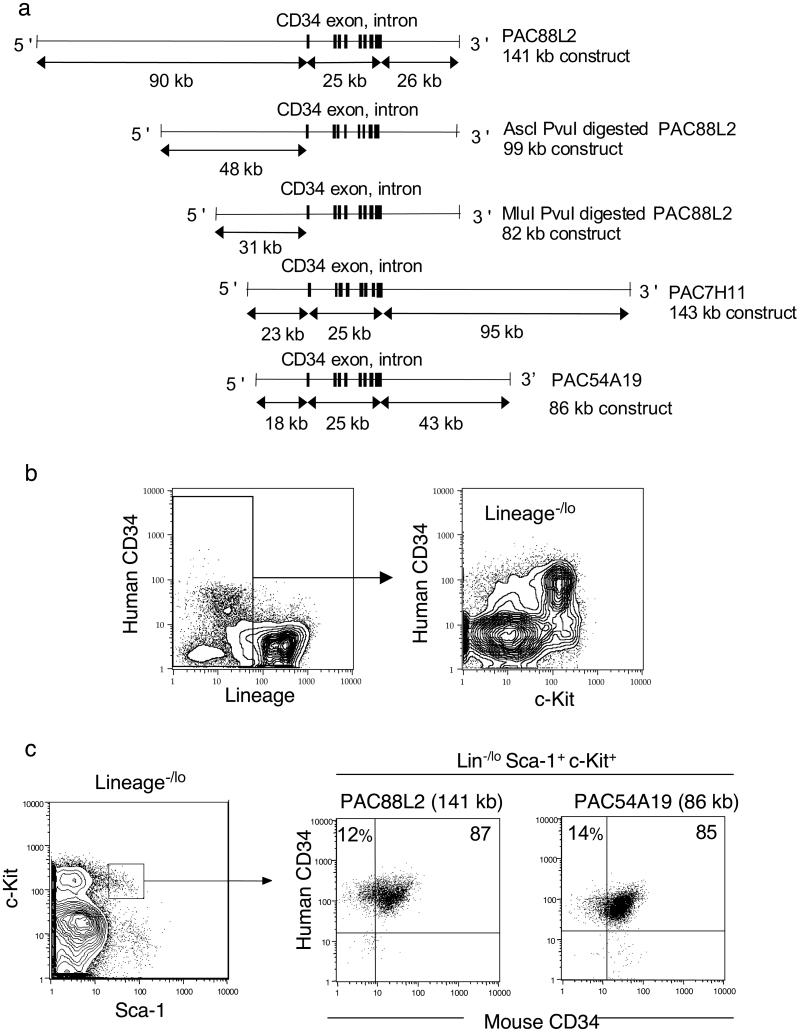
We reported that an hCD34 minigene including 4.5 kb of 5′+ and 3 kb of 3′-flanking sequence failed to express in murine multipotential lines or transgenic mice (15). In contrast, a 141-kb genomic fragment encompassing 90 kb of 5′- and 26 kb of 3′-flanking hCD34 sequences contained regulatory elements for CD34 mRNA expression in murine stable lines and transgenic mice. We found that regulatory elements located between −18 and −10 kb 5′ and/or between +17 and +26 kb 3′ downstream of hCD34 are required for transgenic expression (unpublished data). We have tested a number of PAC clones containing large genomic sequences containing hCD34 sequences in transgenic mouse lines to evaluate their potential for CD34 expression. All the mouse lines harboring the hCD34 transgenic constructs listed in Fig. 1a successfully expressed hCD34. In all transgenic mouse strains, hCD34+ cells were found in ≈3% of bone marrow cells and were enriched in the lineage antigen (Lin)−/lo fraction. Within the Lin−/lo cells, a majority of hCD34+ cells were found in the murine c-Kit+ fraction that contains ≈99% of total colony-forming unit activity in vitro (ref. 17; Fig. 1b). Therefore, the hCD34 transgene is expressed almost exclusively in immature hematopoietic cell populations in hCD34 transgenic mice.
Figure 1.
hCD34 expression in the HSC fraction in bone marrow cells of transgenic mice. (a) hCD34 genomic PAC constructs. These constructs were used to establish transgenic mouse lines. (b) Expression of hCD34 in mice harboring genomic hCD34 (PAC54A19). hCD34 was expressed almost exclusively in the Lin−/lo fraction of bone marrow cells. Within the Lin−/lo bone marrow cells, a majority of hCD34+ cells were found within the c-Kit+ fraction. (c) More than 98% of Lin−/loSca-1+c-Kit+ cells expressed hCD34 irrespective of mCD34 expression in both hCD34 (PAC88L2) and hCD34 (PAC54A19) transgenic mouse lines.
We tested the expression of the hCD34 transgene in HSC compartments. Fig. 1c shows the phenotype of HSC compartments by five-color fluorescence-activated cell sorter analysis in 10-week-old hCD34 (PAC54A19 and PAC88L2) transgenic mice. As reported previously (10), the Lin−/loSca-1+c-Kit+ HSC population was composed of mCD34+ and mCD34−/lo cells, and more than 95% of mCD34+ and mCD34−/lo HSCs express hCD34. Expression of hCD34 in the Lin−/loSca-1+c-Kit+ HSC population was observed in all transgenic mouse lines depicted in Fig. 1a (data not shown).
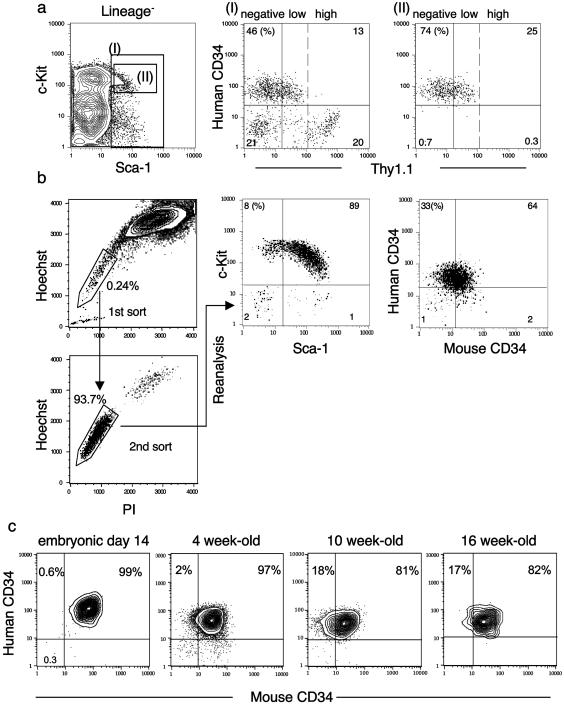
It has been reported that long-term HSCs express a low level of Thy1.1 (7, 19). We crossed hCD34 (PAC54A19) C57B6 (Thy-1.2) mice with C57B6-Thy1.1 mice to evaluate long-term Thy1.1loLin−Mac-1−Sca-1+ HSCs described by Morrison and Weissman (19). The Thy1.1loLin−(Mac-1−)Sca-1+c-Kit+ HSC population that contains long-term HSCs all were positive for hCD34 (Fig. 2a). We also purified cells that are capable of exporting the intracellular dye Hoechst 33342, called the SP, which reportedly contains the long-term HSC (ref. 16; Fig. 2b). The purified SP cells were Lin−Sca-1+c-Kit+ and contained both mCD34−/lo and mCD34+ cells. The mCD34−/lo fraction was enriched up to 33% in the SP fraction. hCD34 was expressed in >97% of the SP cells irrespective of mCD34 expression. These data suggest that the hCD34 transgene is expressed in long-term murine HSC populations defined by three independent groups (10, 16, 19).
Figure 2.
Expression of the hCD34 transgene in different defined HSC populations. (a) The Lin−/loSca-1+ fraction (I) in hCD34 (PAC54A19) transgenic mice contains both hCD34− and hCD34+ cells. hCD34+ cells are enriched in Lin−Sca-1+c-Kit+ fraction (II), which contains Thy1.1lo cells. (b) The Hoechst-exporting SP was purified, resorted, and subjected to five-color fluorescence-activated cell sorter analysis. A majority of SP cells were Lin−Sca-1+c-Kit+hCD34+. (c) Changes in expression of mCD34 and hCD34 in day 14 p.c. fetal liver and from bone marrow in 4-, 10-, and 16-week-old hCD34 transgenic mice. The percentage of mCD34− cells within the Lin−Sca-1+c-Kit+ fractions appeared to increase with age, comparing young 4-week to older 10- and 16-week-old mice. Nonetheless, more than 98% of Lin−Sca-1+c-Kit+ cells were positive for hCD34 irrespective of age.
The hCD34 Transgene Is Expressed in a Majority of Functional Long-Term HSCs.
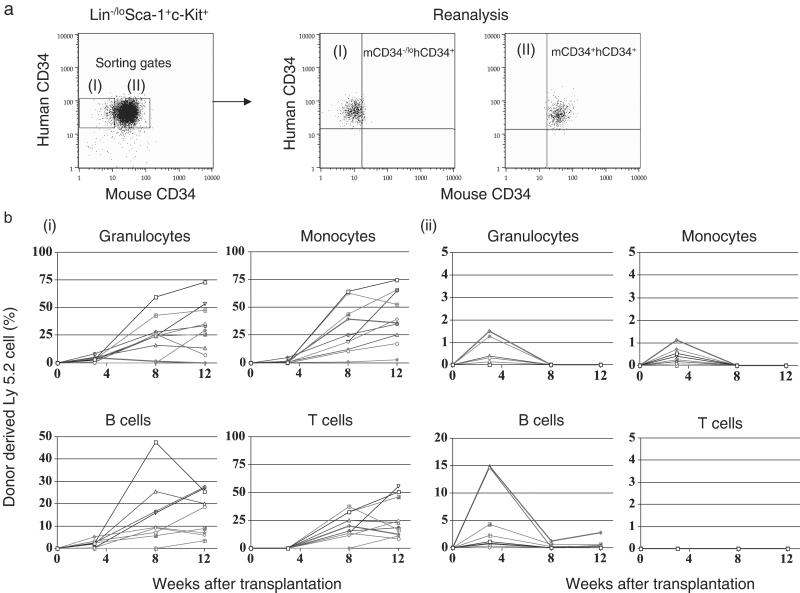
We then directly tested the reconstitution potential of purified mCD34−/lohCD34+ and mCD34+hCD34+ Lin−Sca-1+c-Kit+ HSCs from 4-, 10-, and 16-week-old C57B6 hCD34-Ly5.2 (PAC54A19) transgenic mice with a competitive reconstitution assay (7). In these mice, the frequencies of mCD34−/lo cells in the Lin−/loSca-1+c-Kit+ HSC population appears to increase with aging from the fetal liver stage to 16 weeks. However, hCD34 is expressed in a vast majority of Lin−/loSca-1+c-Kit+ cells irrespective of their ages (Fig. 2c). Fig. 3a shows sorting gates and results of reanalysis of purified mCD34−/lohCD34+ or mCD34+hCD34+ HSCs. In the reconstitution experiment, 35 cells from either mCD34−/lohCD34+ or mCD34+hCD34+ phenotypic HSCs of 4-week-old Ly5.2 mice and 2 × 105 C57B6-Ly5.1 unfractionated bone marrow cells were coinjected into lethally irradiated C57B6 Ly5.1 mice. In both groups, a majority of cases exhibited engraftment of Ly5.2+ donor-derived Mac-1+ granulocytes and monocytes and B220+ B cells 3 weeks posttransplant. The percentages of progeny from mCD34−/lohCD34+ HSCs continuously increased after transplantation and exhibited multilineage reconstitution including CD3+ T cells, B220+ B cells, Gr-1+Mac-1+ granulocytes, and Gr-1loMac-1+ monocytes. In contrast, progeny of mCD34+hCD34+ HSCs declined and disappeared by 12 weeks after transplantation in a majority of cases (Fig. 3). Table 1 summarizes the successful engraftment after competitive reconstitution. The long-term HSC potential was found almost exclusively in mCD34−/lohCD34+ HSCs purified from 10- and 16-week-old mice as well. These data indicate that long-term HSCs are highly enriched in the mCD34−/lohCD34+ fraction of the Lin−/loSca-1+c-Kit+ HSC population in mice ranging from 4 to 16 weeks in age. Thus, the expression of the hCD34 transgene has been initiated already at the stage of long-term HSCs.
Figure 3.
Long-term reconstituting potential of mCD34−/lohCD34+ HSCs. (a) Gates for sorting and results of reanalysis of purified mCD34−/lohCD34+ and mCD34+hCD34+ cells within Lin−Sca-1+c-Kit+ HSCs. (b) Sequential changes in percentages of donor-derived blood cells after transplantation. Thirty-five mCD34−/lohCD34+ (I) or mCD34+hCD34+ (II) phenotypic HSCs purified from hCD34 (PAC54A19) C57B6-Ly5.2 transgenic mice were transplanted into lethally irradiated C56B6-Ly5.1 mice. Long-term reconstitution of all hematopoietic lineages was observed exclusively in mice transplanted with mCD34−/lohCD34+ HSCs. Summarized data are shown in Table 1.
Table 1.
In vivo reconstitution after mCD34−hCD34+ and mCD34+hCD34+ HSC transplantation
| Age, weeks | Injected cells | Cell no. | No. of mice | Number of mice with successful engraftment
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 Weeks posttransplant
|
8 Weeks posttransplant
|
12 Weeks posttransplant
|
|||||||||||||
| M | B | T | Total | M | B | T | Total | M | B | T | Total | ||||
| 4 | mCD34−hCD34+ | 35 | 10 | 1 (7) | 0 (2) | 0 (0) | 1 (7) | 8 (9) | 9 (9) | 9 (9) | 9 (9) | 10 (10) | 9 (10) | 10 (10) | 10 (10) |
| mCD34+hCD34+ | 35 | 10 | 0 (2) | 2 (5) | 0 (0) | 0 (6) | 0 (0) | 0 (1) | 0 (0) | 0 (1) | 0 (0) | 0 (1) | 0 (0) | 0 (1) | |
| 10 | mCD34−hCD34+ | 50 | 10 | 9 (10) | 1 (10) | 0 (0) | 9 (10) | 8 (9) | 9 (9) | 9 (9) | 9 (9) | 9 (9) | 9 (9) | 9 (9) | 9 (9) |
| mCD34+hCD34+ | 100 | 10 | 1 (10) | 4 (9) | 0 (0) | 4 (10) | 2 (2) | 1 (4) | 2 (3) | 4 (10) | 0 (0) | 1 (7) | 1 (3) | 1 (7) | |
| 16 | mCD34−hCD34+ | 50 | 10 | 8 (9) | 6 (8) | 0 (0) | 8 (9) | 8 (8) | 8 (8) | 8 (8) | 8 (8) | 8 (8) | 8 (8) | 8 (8) | 8 (8) |
| mCD34+hCD34+ | 50 | 10 | 0 (1) | 0 (6) | 0 (1) | 1 (6) | 1 (1) | 1 (2) | 0 (3) | 1 (3) | 1 (1) | 0 (3) | 0 (0) | 1 (3) | |
C57B6 (Ly5.1) mice were lethally irradiated and transplanted with mCD34+ and mCD34− HSCs purified from 4-, 10-, and 16-week-old hCD34 transgenic C57B6 (Ly5.2) mice. Percentages of donor-derived (Ly5.2+) cells within each lineage were evaluated 3, 8, and 12 weeks after transplantation. Numbers of mice that exhibited more than 5 and 1% (shown in parentheses) of donor-derived cells are shown. M, granulocytes plus monocytes; B, B cells; T, T cells.
Down-Regulation of hCD34 Transgene Expression at the Hematopoietic Progenitor Stage.
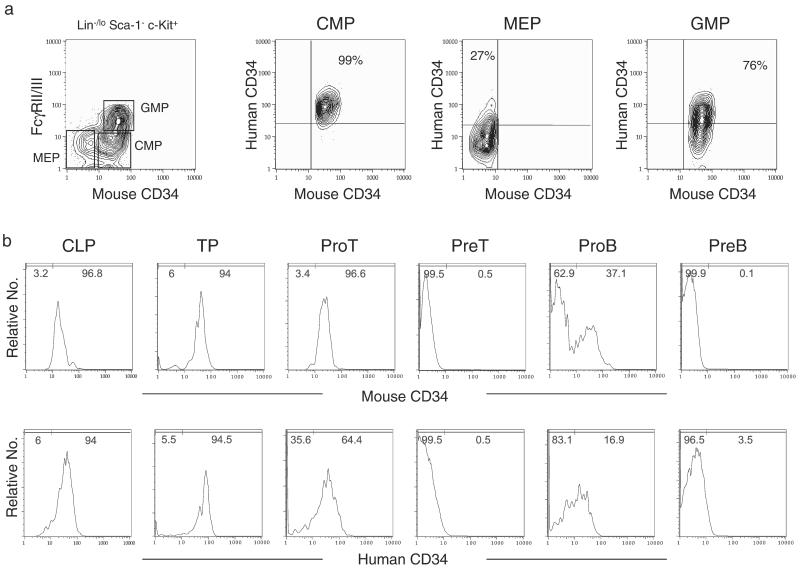
It is of importance to clarify the stages at which the hCD34 transgene is down-regulated in the mouse. We recently identified and purified Lin-restricted progenitors downstream of HSCs that represent critical hematopoietic branch points including CLPs (18) and CMPs (17), which are the earliest progenitors in the lymphoid and myeloid differentiation pathways, respectively. We also have identified MEPs and GMPs downstream of CMPs (17). We tested the expression of hCD34 and mCD34 in these populations as well as other T and B lymphoid-committed progenitors in hCD34 transgenic mice (Fig. 4). In myeloid differentiation pathways, all CMPs were mCD34+hCD34+. In contrast, MEPs contained ≈30% of mCD34−hCD34+ as well as mCD34−hCD34− cells, and GMPs contained ≈20% of mCD34+hCD34− as well as mCD34+hCD34+ cells. These data suggest that compared with mCD34, the down-regulation of hCD34 is delayed along the megakaryocyte/erythrocyte pathway, whereas it slightly preceded that of mCD34 along the granulocyte/macrophage differentiation pathway (Fig. 4a). In the lymphoid differentiation pathways, a majority of CLPs and the earliest thymic precursors expressed both hCD34 and mCD34. The expression of the hCD34 transgene in murine CLPs is reasonable, because we have found that the human bone marrow phenotypic counterpart of the murine CLP might be IL-7R+Lin−CD34+, and that this population is contained in the Lin−CD34+CD10+ common lymphoid progenitor fraction reported by Galy et al. (ref. 4; unpublished data). The down-regulation of hCD34 and mCD34 in lymphoid pathways was initiated at the proT and proB stages and completed at the preT and preB stages. In the proT and proB stages, percentages of hCD34+ cells were less than mCD34+ cells, suggesting that down-regulation of hCD34 in lymphoid lineages occur slightly earlier than that of mCD34 (Fig. 4b).
Figure 4.
Down-regulation of the hCD34 transgene in myeloid and lymphoid differentiation pathways. (a) Mouse myeloid progenitors including Lin−Sca-1−c-Kit+CD34+FcgRII/IIIlo CMPs, Lin−Sca-1−c-Kit+CD34+FcgRII/IIIhi GMPs, and Lin−Sca-1−c-Kit+CD34−FcgRII/IIIlo MEPs (17) purified from hCD34 (PAC88L2) C57B6 transgenic mice were reanalyzed to test expression of the hCD34 transgene. A vast majority of CMPs expressed hCD34. In contrast, ≈30% of mCD34− MEPs still maintain hCD34, whereas ≈20% of mCD34+ GMPs down-regulated hCD34. (b) Purified CLPs and the earliest thymic precursor (TP) were positive for both mCD34 and hCD34. In preT and preB stages, both mCD34 and hCD34 were down-regulated completely. The percentages of hCD34− cells were higher than those of mCD34− cells in both proT and proB stages, suggesting that the down-regulation of the hCD34 transgene occurs slightly earlier than that of mCD34.
Discussion
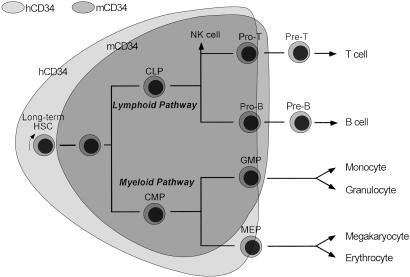
Our data demonstrate that the hCD34 transgene is expressed in phenotypically and functionally defined long-term murine HSCs. Furthermore, the down-regulation of hCD34 occurs slightly earlier than that in mCD34 along with granulocyte/macrophage and lymphoid differentiation pathways. These data strongly suggest that the regulation of CD34 expression is slightly but significantly different in human and murine hematopoiesis. The proposed differential regulation of hCD34 and mCD34 is shown in Fig. 5. In this model, a vast majority of HSCs as well as CMPs and CLPs express hCD34 in human hematopoiesis. The model is compatible with the use of hCD34-enriched cells in clinical applications requiring stem cells.
Figure 5.
Proposed differential regulation of human and murine CD34 in steady-state hematopoiesis. In steady-state human bone marrow, we propose that a majority of long-term HSCs express hCD34. In contrast, in murine hematopoiesis, mCD34 is down-regulated in the long-term HSCs, although mCD34 may be up-regulated in some fraction of long-term HSCs because of a variety of activation signals. hCD34 expression is maintained at the CMP and CLP stages. The down-regulation of hCD34 occurs earlier as compared with mCD34 in the granulocyte/macrophage and lymphoid differentiation pathways but occurs later in the megakaryocyte/erythrocyte pathway.
It should be noted that the definitions of HSC phenotypes we used here may not cover all HSC populations in adult mouse bone marrow. It has been reported that some long-term HSCs may change their phenotypes according to alterations of the hematopoietic microenvironment, including hematopoietic stress, in both mice and humans. For example, the murine c-Kit− fraction in steady-state bone marrow reportedly contains some HSCs with long-term reconstitution activity at a very low frequency (20). It is possible that the HSC activity in the c-Kit− fraction may be caused by occasional down-regulation of c-Kit in a minor fraction of HSCs, because c-Kit down-regulation can occur in HSCs after 5-fluorouracil treatment (21, 22). Likewise, murine long-term HSCs may up-regulate mCD34 after transplantation (12) or after treatment with cytokines (23, 24) or 5-fluorouracil (25). It has been reported also that murine long-term HSCs are increasingly restricted to the mCD34− fraction with aging (26). Our data indicate that in the steady-state murine bone marrow, long-term HSCs reside almost exclusively within the mCD34− fraction irrespective of aging from 4 to 16 weeks and therefore suggest that hCD34+ human bone marrow cells should contain a majority of human counterparts for murine long-term HSCs in steady-state adult bone marrow.
Xenotransplantation assays have been developed to measure human HSC activity in vivo. These assays include reconstitution assays using immunodeficient nonobese diabetic/severe combined immunodeficient mice (27) and in utero transplantation assays in sheep (28). Although these systems are very useful for evaluating differentiation activity of human progenitor/stem cells, engraftment is considerably inefficient. The hCD34− fraction as well as the hCD34+ fraction reportedly contain cells of long-term differentiation potentials at a very low frequency evaluated by these systems (27, 28). The detailed phenotype of the hCD34− HSC population in humans is still not well described. Our data suggest that these hCD34− HSCs may not represent the human counterpart of mCD34−/lo HSCs, and that the HSC activity within the hCD34− population in humans may represent a minority of HSCs that are not included in the Lin−Sca-1+c-Kit+ HSC fraction in murine bone marrow.
These data suggest that hCD34 and mCD34 are regulated differently with respect to expression in long-term HSCs in steady-state bone marrow. Accordingly, it is reasonable to expect hCD34+ cells to possess the biological activities of murine HSCs defined as Thy-1.1loLin−/loSca-1+, Lin−Sca-1+c-Kit+, or SP cells that have been demonstrated in numerous previously published experiments. These activities might include the potential to differentiate into cells of other organs, because purified mouse Thy-1.1loLin−/loSca-1+, Lin−c-Kit+, or Lin−CD34+ populations contain cells that have been reported to differentiate into hepatocytes (11), myocytes (29), and multiple organs (12), respectively. Thus, CD34-enriched cells currently used for human hematopoietic transplantation should include a majority of long-term HSCs that can maintain hematolymphoid systems throughout life as well as a majority of myeloid and lymphoid progenitors and also may contain cells that have differentiation potential into cells of other organs.
Acknowledgments
We thank Joel Lawitts of the Beth Israel Deaconess Medical Center Transgenic Facility for production of mice harboring PAC clones, Maris Fenyus and Michele Buragas for assistance in mouse husbandry and genotyping, and Maris A. Handley for technical assistance in fluorescence-activated cell sorter operation. This work was supported by a U.S. Public Health Service grant (to D.G.T.) and grants from the Jose Carreras International Leukemia Foundation, Leukemia Research Foundation, and Claudia Adams Barr Program (to K.A.).
Abbreviations
- HSC
hematopoietic stem cell
- hCD34
human CD34
- mCD34
murine CD34
- Lin
lineage
- PAC
P1 artificial chromosome
- CLP
common lymphoid progenitor
- CMP
common myeloid progenitor
- GMP
granulocyte/monocyte progenitor
- MEP
megakaryocyte/erythroid progenitor
- SP
side population
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Berenson R J, Andrews R G, Bensinger W I, Kalamasz D, Knitter G, Buckner C D, Bernstein I D. J Clin Invest. 1988;81:951–955. doi: 10.1172/JCI113409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunbar C E, Cottler-Fox M, O'Shaughnessy J A, Doren S, Carter C, Berenson R, Brown S, Moen R C, Greenblatt J, Stewart F M, et al. Blood. 1995;85:3048–3057. [PubMed] [Google Scholar]
- 3.Baum C M, Weissman I L, Tsukamoto A S, Buckle A M, Peault B. Proc Natl Acad Sci USA. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galy A, Travis M, Cen D, Chen B. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- 5.Link H, Arseniev L, Bahre O, Kadar J G, Diedrich H, Poliwoda H. Blood. 1996;87:4903–4909. [PubMed] [Google Scholar]
- 6.Michallet M, Philip T, Philip I, Godinot H, Sebban C, Salles G, Thiebaut A, Biron P, Lopez F, Mazars P, et al. Exp Hematol (Charlottesville, Va) 2000;28:858–870. doi: 10.1016/s0301-472x(00)00169-7. [DOI] [PubMed] [Google Scholar]
- 7.Spangrude G J, Heimfeld S, Weissman I L. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 8.Ikuta K, Weissman I L. Proc Natl Acad Sci USA. 1992;89:1502–1506. doi: 10.1073/pnas.89.4.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogawa M, Matsuzaki Y, Nishikawa S, Hayashi S, Kunisada T, Sudo T, Kina T, Nakauchi H. J Exp Med. 1991;174:63–71. doi: 10.1084/jem.174.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osawa M, Hanada K, Hamada H, Nakauchi H. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 11.Lagasse E, Connors H, Al-Dhalimy M, Reitsma M, Dohse M, Osborne L, Wang X, Finegold M, Weissman I L, Grompe M. Nat Med. 2000;6:1229–1234. doi: 10.1038/81326. [DOI] [PubMed] [Google Scholar]
- 12.Krause D S, Theise N D, Collector M I, Henegariu O, Hwang S, Gardner R, Neutzel S, Sharkis S J. Cell. 2001;105:369–377. doi: 10.1016/s0092-8674(01)00328-2. [DOI] [PubMed] [Google Scholar]
- 13.Peterson K R, Clegg C H, Huxley C, Josephson B M, Haugen H S, Furukawa T, Stamatoyannopoulos G. Proc Natl Acad Sci USA. 1993;90:7593–7597. doi: 10.1073/pnas.90.16.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufman R M, Pham C T, Ley T J. Blood. 1999;94:3178–3184. [PubMed] [Google Scholar]
- 15.Radomska H S, Satterthwaite A B, Burn T C, Oliff I A, Huettner C S, Tenen D G. Gene. 1998;222:305–318. doi: 10.1016/s0378-1119(98)00491-0. [DOI] [PubMed] [Google Scholar]
- 16.Goodell M A, Rosenzweig M, Kim H, Marks D F, DeMaria M, Paradis G, Grupp S A, Sieff C A, Mulligan R C, Johnson R P. Nat Med. 1997;3:1337–1345. doi: 10.1038/nm1297-1337. [DOI] [PubMed] [Google Scholar]
- 17.Akashi K, Traver D, Miyamoto T, Weissman I L. Nature (London) 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 18.Kondo M, Weissman I L, Akashi K. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 19.Morrison S J, Weissman I L. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 20.Ortiz M, Wine J W, Lohrey N, Ruscetti F W, Spence S E, Keller J R. Immunity. 1999;10:173–182. doi: 10.1016/s1074-7613(00)80018-7. [DOI] [PubMed] [Google Scholar]
- 21.Doi H, Inaba M, Yamamoto Y, Taketani S, Mori S I, Sugihara A, Ogata H, Toki J, Hisha H, Inaba K, Sogo S, Adachi M, Matsuda T, Good R A, Ikehara S. Proc Natl Acad Sci USA. 1997;94:2513–2517. doi: 10.1073/pnas.94.6.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Randall T D, Weissman I L. Blood. 1997;89:3596–3606. [PubMed] [Google Scholar]
- 23.Tajima F, Sato T, Lever J H, Ogawa M. Blood. 2000;96:1989–1993. [PubMed] [Google Scholar]
- 24.Tajima F, Deguchi T, Laver J H, Zeng H, Ogawa M. Blood. 2001;97:2618–2624. doi: 10.1182/blood.v97.9.2618. [DOI] [PubMed] [Google Scholar]
- 25.Sato T, Laver J H, Ogawa M. Blood. 1999;94:2548–2554. [PubMed] [Google Scholar]
- 26.Ito T, Tajima F, Ogawa M. Exp Hematol (Charlottesville, Va) 2000;28:1269–1273. doi: 10.1016/s0301-472x(00)00535-x. [DOI] [PubMed] [Google Scholar]
- 27.Bhatia M, Bonnet D, Murdoch B, Gan O I, Dick J E. Nat Med. 1998;4:1038–1045. doi: 10.1038/2023. [DOI] [PubMed] [Google Scholar]
- 28.Zanjani E D, Almeida-Porada G, Livingston A G, Flake A W, Ogawa M. Exp Hematol (Charlottesville, Va) 1998;26:353–360. [PubMed] [Google Scholar]
- 29.Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson S M, Li B, Pickel J, McKay R, Nadal-Ginard B, Bodine D M, Leri A, Anversa P. Nature (London) 2001;410:701–705. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]