Abstract
A robust forward genetic model for Apicomplexa could greatly enhance functional analysis of genes in these important protozoan pathogens. We have developed and successfully tested a genetic complementation strategy based on genomic insertion in Toxoplasma gondii. Adapting recombination cloning to genomic DNA, we show that complementing sequences can be shuttled between parasite genome and bacterial plasmid, providing an efficient tool for the recovery and functional assessment of candidate genes. We show complementation, gene cloning, and biological verification with a mutant parasite lacking hypoxanthine-xanthine-guanine phosphoribosyltransferase and a T. gondii cDNA library. We also explored the utility of this approach to clone genes based on function from other apicomplexan parasites using Toxoplasma as a surrogate. A heterologous library containing Cryptosporidium parvum genomic DNA was generated, and we identified a C. parvum gene coding for inosine 5-monophosphate-dehydrogenase (IMPDH). Interestingly, phylogenetic analysis demonstrates a clear eubacterial origin of this gene and strongly suggests its lateral transfer from ɛ-proteobacteria. The prokaryotic origin of this enzyme might make it a promising target for therapeutics directed against Cryptosporidium.
Keywords: purine salvage‖lateral gene transfer‖Cryptosporidium parvum‖ Toxoplasma gondii
The phylum Apicomplexa includes a large number of obligate intracellular parasites, among which are the human pathogens Plasmodium (malaria), Toxoplasma (AIDS-related encephalitis), Cryptosporidium, and Cyclospora (severe enteritis) as well as many parasites of substantial veterinary importance (Eimeria, Theileria, Sarcocystis, and Babesia). Toxoplasma gondii has emerged as a versatile genetic model organism to study the basic biology of this group of parasites (1). T. gondii is safe and easy to culture in vitro, and excellent animal models are available; it is also amendable to molecular genetic experimentation. The high transfection efficiencies, which can be achieved in this organism, have spurred the development of a powerful set of reverse genetic tools (2–4).
Apicomplexa, as with most protists, diverged relatively early in the eukaryotic lineage and have many biological features not shared by major eukaryotic model systems (e.g., intracellular parasitism or the possession of secondary plastids). Several large scale sequencing efforts have increased the number of known apicomplexan genes drastically (5). Assigning biological functions to many of these genes, however, remains a major challenge. A forward genetic approach could bridge this gap and permit functional analysis of genes unique to Apicomplexa. The parasites maintain a haploid genome over most of their life cycle, which should greatly facilitate the generation of loss of function mutants. Indeed chemical as well as insertional mutagenesis has been applied successfully in several screens targeting nonessential genes involved in nucleotide biosynthesis (6–11). Parasites harboring conditional temperature-sensitive mutations are also obtained readily by chemical mutagenesis and are an attractive tool to study essential parasite pathways (refs. 12 and 13; M. J. Gubbels and B.S., unpublished data); however, a robust tool to identify the genes affected in these mutants is lacking. An episomal library derived from parasite genomic DNA has been developed for complementation cloning (14), but the high level of random integration of DNA into the Toxoplasma genome complicates recovery of episomes. Here we describe the development of a strategy based on high-frequency insertion of a library instead of episomal maintenance. We show that this approach enables the complementation of a T. gondii mutant using libraries from both T. gondii itself and from another apicomplexan, Cryptosporidium parvum.
Materials and Methods
Host Cells and Parasites.
RH strain T. gondii tachyzoites and transgenic lines derived from this strain were maintained by serial passage in primary human foreskin fibroblast cultures (3). C. parvum oocysts (Iowa isolate, originally obtained from C. Sterling, University of Arizona, Tucson, AZ) were obtained and processed as described (15).
Construction of Plasmids.
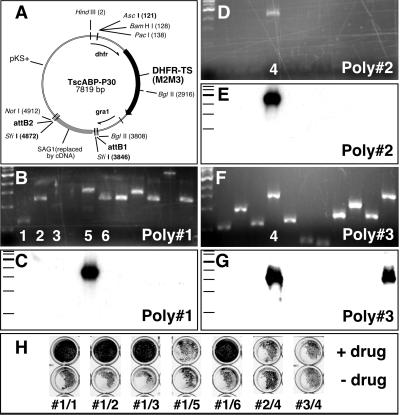
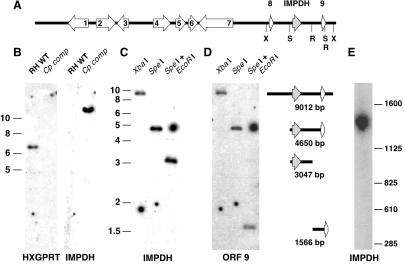
To test insert rescue, a cassette carrying a 2-kb minigene version of the T. gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene (HXGPRT; ref. 10) was amplified with or without flanking attachment B sites (attBs; ref. 16) and cloned into pDHFR*-TSc3ABP (4, 11). To build a plasmid vector for library construction, the coding sequence of the major surface protein P30/SAG1 was amplified by PCR using primers containing attBs and asymmetric SfiI sites as well as BglII and AvrII sites to permit cloning into plasmid pgraPCNA-YFP replacing the PCNA1 coding sequence (17). The P30-YFP expression cassette together with the upstream gra1 promoter region was excised from the resulting plasmid and introduced into the high-frequency insertion vector pDHFR*-TSc3ABP (4) downstream of the dihydrofolate reductase-thymidylate synthase (DHFR-TS) gene. Yellow fluorescent protein (YFP) was removed from this plasmid by restriction and religation, and the resulting vector TscABP-P30 (Fig. 2A) was used to construct the T. gondii cDNA library. A second library vector (TscABP-RV) was constructed by replacing the P30 sequence with a linker carrying an EcoRV site, facilitating the cloning of blunt-end genomic fragments. The T. gondii destination vector pdhfrDEST was obtained by introducing a cassette carrying the ccB as well as the CAT gene [amplified from plasmid pDONOR201 (Invitrogen) flanked by attRs] into plasmid pCAT-GFP (18), replacing CAT-GFP.
Figure 2.
Genetic complementation of a T. gondii mutant. A library was constructed by using plasmid TscABP-P30 cloning cDNAs under control of the parasite promoter gra1 (A). RHΔHXGPRT tachyzoites were transfected with this library and selected under mycophenolic acid. Genomic DNA was isolated from emerging parasites without further cloning and subjected to BP-recombination cloning. Three independent experiments were analyzed by colony PCR (B, D, and F). DNA was hybridized with an HXGPRT-specific probe identifying HXGPRT-containing cDNAs in every population (C, E, and G). All HXGPRT-containing and four additional cDNAs were shuttled by a second recombination (LR, see Fig. 1A) into a T. gondii expression plasmid (pdhfrDEST) and used to retransfect RHΔHXGPRT parasites. All transformants completely lysed the host cell monolayer after 5 days in the absence of drug (H Lower; dark staining indicates protection of monolayer and parasite inhibition, and light staining indicates host cell lysis caused by parasite growth). Under mycophenolic acid selection only plasmids carrying HXGPRT cDNAs permitted parasite growth (H Upper).
Construction of Libraries.
To construct a T. gondii cDNA library poly(A)+ RNA was purified from RH strain tachyzoites (19) and converted to cDNA by using a directional strategy (20). The first strand product was converted to double-stranded cDNA in a three-cycle (72°C, 10 min; 95°C, 20 sec; three cycles of 95°C, 5 sec; 68°C, 8 min) primer extension by using the oligo(dT) and switch primers. The cDNA fragments were digested with SfiI, sized, and ligated into plasmid pTscABP-P30, replacing P30. The library was transformed into DH10B cells and plated (≈10,000 colonies per 150-mm plate). The library contained ≈0.5 million recombinants in a vector background of <1%, and the average insert size exceeded 1 kb. Genomic DNA was isolated from C. parvum oocysts, sheared into fragments by using a nebulizer, and prepared for cloning as described (15). Purified fragments (800–1,500 bp) were cloned into the EcoRV site of vector TscABP-RV. The C. parvum complementation library contained >1 million recombinants in a 10% vector background with an average insert size of 1.2 kb.
Parasite Transfection and Selection.
Parasite transformation was performed as described (3). For transformations using libraries, plasmids were linearized with AscI, and 50 μg of sterilized DNA as well as 50 units of NotI (21) were added before the electroporation of parasites with a pulse of 1,500 V, 25 Ω, and 25 μF by using a BTX ECM 630 electroporator (Gentronix, San Diego). Transfected parasites were cultured in medium supplemented with 50 μg/ml mycophenolic acid and 50 μg/ml xanthine to select for HXGPRT complementation (10).
Analysis of Complemented Parasites and Insert Rescue.
To rescue insertions, 0.5 μg of parasite genomic DNA was incubated with 75 ng of plasmid pDONOR201 and 2 μl of BP clonase (Invitrogen; see Fig. 1) for 2–12 h. Recombined plasmids were electroporated into DH10B cells, and transformed bacteria were selected on kanamycin. LR reactions (see Fig. 1) were performed by using 150 ng of pdhfrDEST (linearized with EcoRI; ref. 16). Probes for Southern blot analysis were labeled by random priming using a 600-bp SalI/EcoRI fragment of the T. gondii HXGPRT gene (not present in RHΔHXGPRT; ref. 10), a 1,400-bp SfiI/NotI fragment from pExpCpIMPDH, and linearized plasmid pKS+. The ORF 9/Cp expressed sequence tag 288 probe was a 631-bp fragment amplified by PCR from C. parvum genomic DNA using the primers 5′-ATGTTTAATTATTTGAAGTTGTTTTCGT-3′ and 5′-CACAATATTTGCAATGTTGTCTTTTTCC-3′.
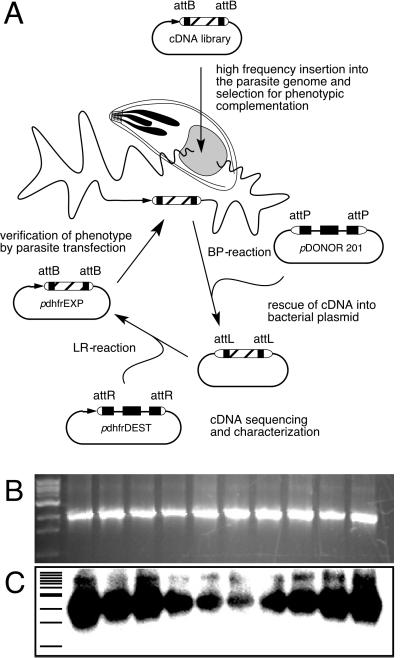
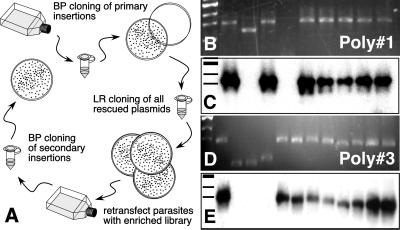
Figure 1.
Schematic diagram of the insert rescue strategy and the plasmid constructs involved. (A) Mutant parasites are transfected with a plasmid library carrying T. gondii cDNAs, which are depicted as hatched boxes flanked by λ phage atts. Parasites carrying complementing sequences are isolated by phenotypic selection, and inserted cDNAs are subsequently recovered into plasmid by BP-recombination cloning. To regenerate a parasite expression vector, rescued plasmids are recombined with a parasite-specific destination vector (pdhfrDEST) in an LR cloning reaction. The resulting plasmid is transfected into the original parasite mutant line to verify the complementation phenotype. (B) Parasites were transfected with a high-frequency insertion plasmid carrying a 2-kb HXGPRT minigene marker flanked by attBs. Genomic DNA was isolated from a clonal transgenic line and subjected to BP recombination. A uniform 2-kb insert was PCR-amplified from each bacterial colony. After transfer to nylon membrane this band hybridized strongly to a radioactive HXGPRT probe (C).
Phylogenetic Analysis.
Inosine 5-monophosphate dehydrogenase (IMPDH)-homologous sequences from eukaryotes, bacteria, and archaea were retrieved from GenBank as well as from unfinished microbial genome sequence databases from NCBI, The Institute for Genomics Research (Shewanella putrefaciens, Pseudomonas putida, Geobacter sulfurreducens, Desulfovibrio vulgaris, Staphylococcus aureus, Enterococcus faecalis, and Plasmodium yoelii; www.tigr.org and www.plasmoDB.org) (38) and from the Sanger Centre (Corneybacterium diptheriae and Bordetella pertussis; www.sanger.ac.uk). IMPDH amino acid sequences were aligned by using CLUSTALX 1.81 (22), and alignments were refined within MACCLADE 4.03 (23). The sequence alignments and species list are published as supporting information on the PNAS web site, www.pnas.org. Parsimony analysis was performed with PAUP* 4.0b8 (24), and distance analyses were done by using PHYLIP 3.6a2.1 (25) with the JTT substitution model (26) in PROTDIST. TREEPUZZLE 5.0 (27) was used to obtain maximum likelihood (ML) distances, using JTT and eight γ-distributed substitution rates and considering invariable sites; this ML distance matrix was used to construct a neighbor-joining tree with NEIGHBOR (PHYLIP 3.6a2.1). For the analyses shown in Fig 6B, TREEPUZZLE 5.0 was used for 10,000 quartet puzzling steps to obtain estimates of branch support, and MR BAYES 2.01 (28) was used for an ML analysis using the ML distance tree topology as the starting tree and the initial γ shape parameter α = 0.86. MR BAYES was run for 50,000 generations with four incrementally heated Markov chains and a sampling frequency of 50 generations. The JTT substitution model and nine rate categories (eight γ-distributed and one invariable) were used. Clade confidence values and a consensus ML tree were estimated after excluding the first 5,000 generations from the set of sampled trees with the best posterior probabilities.
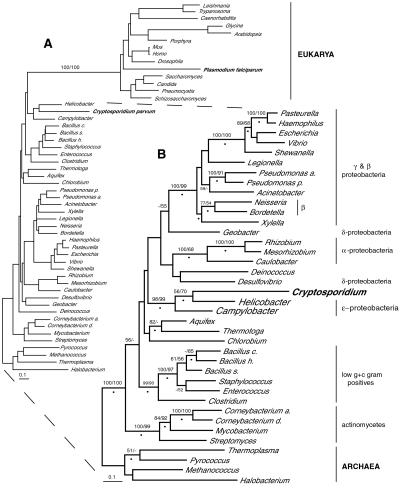
Figure 6.
Phylogenetic relationship of C. parvum IMPDH to ɛ-proteobacteria. Alignments of unambiguously aligned amino acid positions (441 sites in A and 452 in B) were analyzed by distance (Protdist + neighbor joining), maximum parsimony, and ML methods. The tree shown in A is the ML distance tree with α = 0.81 and proportion of invariable sites (iP) = 0. The tree shown in B is the ML consensus topology with mean lnL = −18,029 (−18,011.8 < lnL < −18,060.2), α =1.18 (0.89 < α < 1.49), and iP = 0.12 (0.08 < iP < 0.19). Numbers above the nodes represent the percentage of 1,000 bootstrap replicates of neighbor-joining and maximum parsimony (in that order) supporting the relationship. In A, only support for monophyly of the Eukarya excluding Cryptosporidium is shown, and in B, bootstrap support values >50% are shown, and nodes supported by at least 50% of 10,000 quartet puzzle steps are indicated by bullets (●).
Results and Discussion
Rescue of Inserted DNA from the Parasite.
Exceptionally high transformation frequencies have been reported in T. gondii using plasmids carrying the homologous DHFR-TS gene (29). This element inserts by nonhomologous recombination and has been used successfully to tag genes and promoters by saturation mutagenesis (4, 9–11). This high transformation frequency led us to explore the use of an insertional DHFR-TS based approach to achieve genetic complementation. Inserted sequences can be cloned by using marker rescues or PCR (4), but these methods lack the throughput needed for large-scale genetic screens. We tested whether λ phage atts and recombination cloning (16) could be used as a more efficient way to retrieve insertions. Plasmids were engineered carrying the HXGPRT marker (10) flanked by 25 bp of λ phage attBs in addition to the pyrimethamine-resistant DHFR-TS allele. The stable transfection of 1–5% obtained with this construct (data not shown) is consistent with the reported transfection efficiency of the parent DHFR-TS plasmid (4, 29). Stable drug-resistant parasites were the result of transgene integration rather than episomal maintenance as verified by Southern blot and lack of episome recovery by bacterial transformation (ref. 14; data not shown; detailed Southern analysis of other clones is shown in Fig. 3).
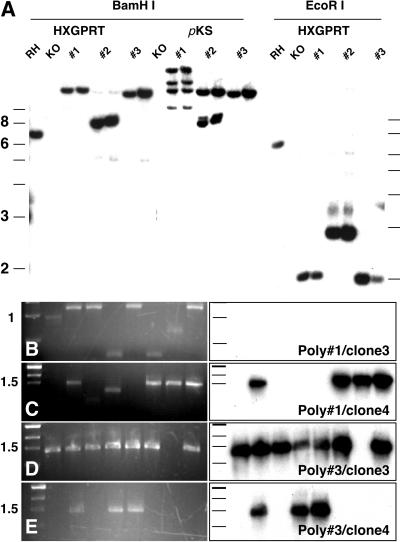
Figure 3.
Analysis of insertions from cloned complemented parasites. (A) Genomic DNA from two clones isolated from each of three primary populations were digested with BamHI or EcoRI before Southern blot analysis using labeled HXGPRT or pKS vector (after stripping) as probe. The RH strain shows the wild-type HXGPRT band, which is missing in RHΔHXGPRT (KO) and the various complements. In each complement clone, a single HXGPRT band is observed. Note that BamHI digestion results in a fragment bigger than the linearized vector, indicating that the transgene is inserted into the genome and not carried episomally. Reprobing with pKS reveals multiple insertions in #1 and #2. (B–E) Colony PCR and Southern blot (HXGPRT) of BP reactions using genomic DNA from cloned complemented parasites. Note the frequency of HXGPRT-containing inserts is lower in clones derived from population 1.
Genomic DNA isolated from these transgenic parasites was tested in BP cloning reactions by using commercially available enzymes (Gateway cloning, Invitrogen; ref. 16; see Fig. 1A for a schematic view of the cloning and recombination strategy). Several hundred bacterial colonies were recovered by using genomic DNA containing attBs. The background of colonies obtained by using DNA from parasites with no or an irrelevant insertion was ≈10%. Recovered inserts were analyzed by colony PCR using flanking att-specific primers. A 2-kb product was amplified uniformly. Restriction mapping and hybridization with an HXGPRT-specific probe showed that every colony contained the test gene (Fig. 1 B and C).
Complementation of a Parasite Mutant Using a T. gondii cDNA Library.
A cDNA library was constructed in the modified DHFR-TS high-frequency insertion vector TscABP-P30 by using T. gondii RH strain RNA (see Fig. 2 and Material and Methods for details on plasmid and library construction). We used the RHΔHXGPRT strain to explore the ability of this library to complement a lack of function mutant. This strain lacks HXGPRT activity because of a 1.5-kb deletion in the 3′ end of the corresponding gene introduced by gene targeting (10). These parasites grow at the wild-type rate under standard culture conditions but are highly sensitive to mycophenolic acid treatment, which inhibits IMPDH and thus prevents purine salvage. Expression of HXGPRT as a transgene in this background rescues purine salvage and restores parasite viability. RHΔHXGPRT tachyzoites were transfected in five independent electroporations. After 12 days of culture under mycophenolic acid, host cell monolayers from all five experiments had been lysed by parasite growth. Parasites from these populations retained drug resistance in subsequent passage into fresh monolayers, whereas drug resistance did not develop spontaneously in parasites transformed with a control plasmid lacking a functional HXGPRT gene. Genomic DNA was isolated from three primary drug-resistant parasite populations and subjected to BP-recombination cloning as described above. Colonies resulting from these reactions were analyzed by PCR (Fig. 2 B, D, and F), and the amplified fragments were blotted onto nylon membrane and hybridized with a 32P-labeled probe derived from the 3′ end of the T. gondii HXGPRT gene. Two fragments strongly hybridizing to the HXGPRT-specific probe were identified for population 3 and one each for populations 1 and 2 (Fig. 2 C, E, and G). The clones hybridizing to HXGPRT were sequenced and contained full-length HXGPRT cDNAs. The three cDNAs identified were independent from each other (starting at base pairs 1,126, 773, and 1,321 of the published genomic sequence GenBank accession no. U10247) and represented both previously described alternatively spliced forms (10). Sequencing also showed the presence of the 5′ template-switching primer introduced during cDNA synthesis, thus confirming that these fragments were derived from parasite RNA and not a laboratory contamination.
The ability to test potentially large numbers of putative complementations in a biological assay is essential to genetic analysis. To accomplish this goal we constructed a destination plasmid into which complementing sequences can be shuttled by a second recombination (Fig. 1A, LR reaction). Rescued plasmids were recombined with the pdhfr-DEST plasmid, and the resulting T. gondii expression vectors were used again to transfect RHΔHXGPRT parasites. Growth of transfected parasites in the presence and absence of mycophenolic acid was assayed by host cell monolayer disruption (3). Only parasites transfected with plasmids derived from the three HXGPRT-containing clones were able to lyse the host cell monolayer in the presence of drug (Fig. 2H). No signs of parasite development were observed in transformants that received cDNAs not hybridizing to HXGPRT; in contrast, all parasites grew equally well in the absence of drug.
We isolated clonal parasite lines from three drug-resistant populations obtained in the initial screen to characterize the library insertions in greater detail (two clones per population are shown in Fig. 3). Genomic DNA isolated from these clones, RH wild type and RHΔHXGPRT parasites, was analyzed by Southern blot using an HXGPRT-specific probe. The previously reported wild-type BamHI and EcoRI fragments of 7 and 6 kb, respectively (10) were detected in RH but were absent from the knockout as well as all complemented clones (Fig. 3A). A single new band of different size was found in each complemented clone. When restricted with BamHI (which cuts once in the vector) bands larger than linearized plasmid were observed, indicating that the vector indeed had inserted into the parasite genome. An EcoRI digest, which excises the HXGPRT cDNA and the adjacent promoter from the vector, shifts these bands to 2 and 2.9 kb, respectively, which is in agreement with the size of the cDNAs recovered by recombination cloning (Figs. 2 and 3 B–E). The blot was stripped and probed a second time with pKS vector sequence, and four insertions were detected for clones derived from population 1, two for population 2, and one for population 3. To assess the effect multiple insertions might have on the ability to recover a specific insertion by recombination, we performed BP reactions with genomic DNA from parasite clones of populations 1 and 3. For clones from population 3, which carry a single insertion, every productive colony PCR yielded the HXGPRT gene (Fig. 3 D and E). HXGPRT was not found in every recombination from clones from population 1, which likely is because of the multiple unrelated insertions in these clones (Fig. 3 B and C).
Multiple insertions are a commonly observed effect of restriction enzyme-mediated insertion enhancement in T. gondii (21). To be able to isolate complementing cDNA even in the presence of multiple insertions, we developed an enrichment protocol by bulk-recombination cloning and tested it by using primary polyclonal populations 1 and 3. Genomic DNA from each of these populations emerging from drug selection was used in a BP reaction, and all possible inserts were recovered en masse. The entire transformation then was plated, and inserts were shuttled by LR cloning into the pdhfrDEST vector, generating a library of recovered cDNAs in a T. gondii expression context (see Fig. 4A for a diagram of the strategy). We reasoned that using an enriched library we should be able to achieve stable transformation without restriction enzyme-mediated insertion enhancement, thus avoiding problems caused by multiple insertion. RHΔHXGPRT tachyzoites were transfected with 50 μg of enriched libraries without the addition of NotI. After 7 days the host monolayer was lysed by drug-resistant parasites, which were harvested for DNA isolation and recombination cloning. Regardless of the frequency of HXGPRT insertions in the primary drug-resistant population (Fig. 3), >75% of the bacterial colonies recovered from the secondary population now contained an HXGPRT cDNA (Fig. 4 B–E).
Figure 4.
Complementing cDNAs can be enriched by batch-recombination cloning. (A) Genomic DNA from primary polyclonal populations emerging from drug selection (see the Fig. 3 legend for further detail) was used in a BP-recombination reaction. The rescued inserts were transferred en masse into a parasite expression vector by LR recombination, amplified, and retransfected into RHΔHXGPRT. Recomplementing inserts from the secondary drug-resistant population were isolated by recombination cloning. The resulting bacterial colonies were analyzed by PCR (B and D) and hybridization with an HXGPRT-specific probe (C and E).
Heterologous Complementation with C. parvum DNA.
The apicomplexan parasite C. parvum is an important human pathogen for which currently no treatment is available (30). Experimental progress with this organism has been limited by the lack of a genetic model to assess gene function. We explored the possibility of using the related parasite T. gondii as a genetically amendable surrogate. Because C. parvum has a small genome (107 bp) with very few introns, we constructed a library by using genomic DNA rather than cDNA. RHΔHXGPRT tachyzoites were transfected with this heterologous library and cultured in the presence of mycophenolic acid. In 1 of 5 transfections, drug-resistant parasites emerged, and a 1,387-bp insertion was rescued from these parasites by recombination cloning (GenBank accession no. AF426177). The putative complement and the T. gondii HXGPRT gene were used to probe Southern blots of the drug-resistant parasites as well as RHΔHXGPRT and RH wild-type strains. None of the complemented parasite samples hybridized with the T. gondii HXGPRT probe, indicating that drug resistance was not caused by a T. gondii contamination, and a single insertion of the recovered gene was observed (Fig. 5B). The complementing gene sequence conferred stable resistance to mycophenolic acid after retransfection of the knockout mutant albeit at lower efficiency than the T. gondii HXGPRT gene (data not shown).
Figure 5.
IMPDH is a genuine C. parvum gene. Drug-resistant T. gondii parasites emerging from complementation with a heterologous library carry a single insertion of C. parvum IMPDH (B). (A) IMPDH is located on a 40-kb genomic sequence contig carrying two previously identified C. parvum genes, protein disulfide isomerase (ORF2; ref. 43), and histone deacetylase (ORF3; ref. 33) as well as several previously unidentified ORFs. Flanking IMPDH are two genes sharing identical sequence with C. parvum expressed sequence tags 226 and 349 (ORF8) and 288, 420, and 654 (ORF9; ref. 33). This arrangement was verified by Southern blot using C. parvum genomic DNA and IMPDH (C) and ORF 9 (D) as probes. (E) IMPDH also hybridized to a 1.4-mb molecule (chromosome 6) after pulse field analysis of undigested C. parvum DNA. X, XbaI; S, SpeI; R, EcoRI; WT, wild type; Cp comp, complemented with C. parvum library.
The conceptual translation of the ORF contained in the recovered sequence was compared with GenBank, revealing clear similarity to inosine 5-monophosphate dehydrogenase. IMPDH is the target of mycophenolic acid and one of two possible genes that might be expected in this complementation screen. No HXGPRT sequence confirmed as genuine C. parvum gene was recovered in this screen. Surprisingly, the similarity of the isolated IMPDH was much greater to prokaryotic than to eukaryotic proteins, which prompted us to perform further experiments to ensure that the complementing sequence was indeed a genuine C. parvum gene and not a contaminant. Undigested C. parvum DNA was separated by pulse-field electrophoresis, blotted to membrane, and probed. The IMPDH sequence hybridized to a molecule of ≈1.4 mb, suggesting IMPDH to be present on chromosome 6 (1.44 mb; ref. 31; Fig. 5E). By searching the data from the ongoing C. parvum genome project we localized the isolated IMPDH gene to a 40,432-bp genomic contig (contig 1,083, http://www.cbc.umn.edu/ResearchProjects/AGAC/Cp/index.htm) that harbors the chromosome 6 marker HAPPY#43 (32). This contig also carries several previously identified C. parvum genes in addition to uncharacterized ORFs. In particular, the IMPDH gene is flanked by two ORFs, each with identical sequences to C. parvum expressed sequence tags (33). This genomic arrangement was verified further by Southern blot using a C. parvum genomic DNA sample obtained from a source independent of the DNA used for initial library construction (kindly provided by Dr. S. Upton, Kansas State University, Manhattan, KS). The observed fragments are in complete agreement with the contig-based predictions (Fig. 5 C and D).
Phylogenetic Analysis of C. parvum IMPDH.
All phylogenetic methods support the specific grouping of C. parvum IMPDH with the ɛ-proteobacterial clade of bacteria (Fig. 6). C. parvum IMPDH never grouped with other eukaryotic IMPDHs (e.g., Fig 6A) including the IMPDH from the apicomplexan Plasmodium falciparum in particular. The grouping of C. parvum IMPDH with ɛ-proteobacteria, specifically with Helicobacter pylori, was recovered in every phylogenetic analysis, and a careful series of such analyses, systematically removing problematic (e.g., highly divergent) sequences, clearly demonstrated the significance of this result. Our findings, which are based on rigorous phylogenetic analyses rather than pairwise comparisons such as BLAST scores, indicate that C. parvum IMPDH is not of eukaryotic nuclear descent or even mitochondrial origin (because it is not specifically related to the α-proteobacteria) but instead was obtained by lateral transfer from an ɛ-proteobacterium. This result is evidence of a lateral gene transfer (34–36) from a bacterium to an apicomplexan. Indeed, because C. parvum and members of ɛ-proteobacteria such as H. pylori share their habitat (the intestinal tract of mammals), this may have provided an opportunity for such a transfer.
Interestingly, the IMPDH gene may have undergone multiple transfers between bacteria and protozoa. IMPDH genes from the parabasalid Tritrichomonas foetus and the spirochete Treponema denticola form a clade at the base of eukaryotes, to the exclusion of other spirochetes (which group within Bacteria as expected for a vertically transmitted gene; results not shown). Two putative IMPDH genes have emerged from the P. yoelii genome sequence (www.plasmoDB.org; ref. 37): one of them resembles α-proteobacterial IMPDH, and the other appears more eukaryotic (results not shown). This apparent variety in the phylogenetic origin of IMPDH genes (and possibly others in the purine salvage pathway) suggests that several evolutionary mechanisms including lateral gene transfer might be at work in shaping this essential pathway in parasitic protozoa.
Conclusion
An insertional complementation strategy was devised and successfully tested. We adapted recombination cloning to shuttle DNA fragments between the parasite genome and bacterial plasmid, greatly facilitating biological testing and enrichment of candidate sequences retrieved. This approach could be applied also to other insertion-based transfection systems including transposons or retroviruses. The possibility of complementation cloning opens the door to functional genetic analysis of parasite-specific pathways using conditional mutants and may provide a powerful tool for the investigation of the molecular basis of treatment failure and drug resistance. We also show that genetic complementation is feasible across species boundaries among Apicomplexa. Using this heterologous complementation, we have cloned a Cryptosporidium IMPDH gene involved in purine salvage; this pathway is essential to parasite survival and therefore is actively pursued for drug development (38–41). The finding that the parasite has obtained this gene by lateral gene transfer from a bacterial source might increase its potential as a target for drug development (41, 42).
Supplementary Material
Acknowledgments
We are grateful to Dr. Steve Upton (Kansas State University) for supplying C. parvum genomic DNA and the Sanger Centre and The Institute for Genomic Research for access to unfinished microbial genome data. Preliminary sequence and/or preliminary annotated sequence data from the P. yoelii genome was obtained from The Institute for Genomic Research web site (www.tigr.org). This sequencing program is carried on in collaboration with the Naval Medical Research Center and is supported by the U.S. Department of Defense. This work was funded in part by National Institutes of Health Grants AI 58475 (to B.S.), AI 48390 and 44600 (to M.W.W.), and AI 46397 (to M.S.A.) as well as additional support by Merck Research Laboratories and the University of Georgia Research Foundation (to B.S.). Partial support to J.M.L. came from a Georgia Tech/Emory seed grant.
Abbreviations
- HXGPRT
hypoxanthine-xanthine-guanine phosphoribosyltransferase
- att
attachment site
- DHFR-TS
dihydrofolate reductase-thymidylate synthase
- IMPDH
inosine 5-monophosphate dehydrogenase
- ML
maximum likelihood
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF426177).
References
- 1.Roos D S, Darling J A, Reynolds M G, Hager K M, Striepen B, Kissinger J C. In: Biology of Parasitism. Tschudi C, Pearce E, editors. Boston, MA: Kluwer; 1999. pp. 143–68. [Google Scholar]
- 2.Boothroyd J C, Kim K, Pfefferkorn E R, Sibley D L, Soldati D. Methods Mol Genet. 1994;6:1–29. [Google Scholar]
- 3.Roos D S, Donald R G, Morrissette N S, Moulton A L. Methods Cell Biol. 1994;45:27–63. doi: 10.1016/s0091-679x(08)61845-2. [DOI] [PubMed] [Google Scholar]
- 4.Roos D S, Sullivan W J, Striepen B, Bohne W, Donald R G. Methods. 1997;13:112–122. doi: 10.1006/meth.1997.0504. [DOI] [PubMed] [Google Scholar]
- 5.Tarleton R L, Kissinger J. Curr Opin Immunol. 2001;13:395–402. doi: 10.1016/s0952-7915(00)00233-8. [DOI] [PubMed] [Google Scholar]
- 6.Pfefferkorn E R, Pfefferkorn L C. J Parasitol. 1976;62:993–999. [PubMed] [Google Scholar]
- 7.Pfefferkorn E R, Pfefferkorn L C. Exp Parasitol. 1977;42:44–55. doi: 10.1016/0014-4894(77)90060-1. [DOI] [PubMed] [Google Scholar]
- 8.Pfefferkorn E R. In: The Biology of Parasitism. Englund P T, Sher A, editors. New York: Liss; 1988. pp. 479–502. [Google Scholar]
- 9.Donald R G, Roos D S. Proc Natl Acad Sci USA. 1995;92:5749–5753. doi: 10.1073/pnas.92.12.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donald R G K, Carter D, Ullman B, Roos D S. J Biol Chem. 1996;271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan W J, Jr, Chiang C W, Wilson C M, Naguib F N, el Kouni M H, Donald R G, Roos D S. Mol Biochem Parasitol. 1999;103:1–14. doi: 10.1016/s0166-6851(99)00114-0. [DOI] [PubMed] [Google Scholar]
- 12.Pfefferkorn E R, Pfefferkorn L C. Exp Parasitol. 1976;39:365–376. doi: 10.1016/0014-4894(76)90040-0. [DOI] [PubMed] [Google Scholar]
- 13.Radke J R, Guerini M N, White M W. Exp Parasitol. 2000;96:168–177. doi: 10.1006/expr.2000.4568. [DOI] [PubMed] [Google Scholar]
- 14.Black M W, Boothroyd J C. J Biol Chem. 1998;273:3972–3979. doi: 10.1074/jbc.273.7.3972. [DOI] [PubMed] [Google Scholar]
- 15.Liu C, Vigdorovich V, Kapur V, Abrahamsen M S. Infect Immun. 1999;67:3960–3969. doi: 10.1128/iai.67.8.3960-3969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartley J L, Temple G F, Brasch M A. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radke J R, Striepen B, Guerini M, Jerome M J, Roos D S, White M W. Mol Biochem Parasitol. 2001;115:165–175. doi: 10.1016/s0166-6851(01)00284-5. [DOI] [PubMed] [Google Scholar]
- 18.Striepen B, He C Y, Matrajt M, Soldati D, Roos D S. Mol Biochem Parasitol. 1998;92:325–338. doi: 10.1016/s0166-6851(98)00011-5. [DOI] [PubMed] [Google Scholar]
- 19.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 20.Chenick A, Zhu Y Y, Diatchenko L, Li R, Hill J, Siebert P D. In: Gene Cloning and Analysis by RT-PCR. Siebert P, Larrick J, editors. Westborough, MA: BioTechniques; 1998. pp. 305–319. [Google Scholar]
- 21.Black M, Seeber F, Soldati D, Kim K, Boothroyd J C. Mol Biochem Parasitol. 1995;74:55–63. doi: 10.1016/0166-6851(95)02483-2. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddison W P, Maddison D R. macclade. Sunderland, MA: Sinauer; 2001. , Version 4.03. [Google Scholar]
- 24.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2001. , Version 4.0b8. [Google Scholar]
- 25.Felsenstein J. phylip (Phylogeny Interference Package), (Department of Genetics, Univeristy of Washington, Seattle), Version 3.6a2.1. 2001. [Google Scholar]
- 26.Jones D T, Taylor W R, Thornton J M. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 27.Strimmer K, von Haesler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 28.Huelsenbeck J P, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 29.Donald R G, Roos D S. Proc Natl Acad Sci USA. 1993;90:11703–11707. doi: 10.1073/pnas.90.24.11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fayer R. Cryptosporidium and Cryptosporidiosis. Boca Raton, FL: CRC; 1997. [Google Scholar]
- 31.Caccio S, Camilli R, La Rosa G, Pozio E. Gene. 1998;219:73–79. doi: 10.1016/s0378-1119(98)00376-x. [DOI] [PubMed] [Google Scholar]
- 32.Piper M B, Bankier A T, Dear P H. Genome Res. 1998;8:1299–1307. doi: 10.1101/gr.8.12.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strong W B, Nelson R G. Contrib Microbiol. 2000;6:92–115. doi: 10.1159/000060358. [DOI] [PubMed] [Google Scholar]
- 34.Andersson J O, Doolittle W F, Nesbo C L. Science. 2001;292:1848–1850. doi: 10.1126/science.1062241. [DOI] [PubMed] [Google Scholar]
- 35.de Koning A P, Brinkman F S, Jones S J, Keeling P J. Mol Biol Evol. 2000;17:1769–1773. doi: 10.1093/oxfordjournals.molbev.a026275. [DOI] [PubMed] [Google Scholar]
- 36.Doolittle W F. Trends Genet. 1998;14:307–311. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- 37.Bahl A, Brunk B, Coppel R L, Crabtree J, Diskin S J, Fraunholz M J, Grant G R, Gupta D, Huestis R L, Kissinger J L, et al. Nucleic Acids Res. 2002;30:87–90. doi: 10.1093/nar/30.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ullman B, Carter D. Int J Parasitol. 1997;27:203–213. doi: 10.1016/s0020-7519(96)00150-6. [DOI] [PubMed] [Google Scholar]
- 39.Whitby F G, Luecke H, Kuhn P, Somoza J R, Huete-Perez J A, Phillips J D, Hill C P, Fletterick R J, Wang C C. Biochemistry. 1997;36:10666–10674. doi: 10.1021/bi9708850. [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Hedstrom L. Biochemistry. 1998;37:11949–11952. doi: 10.1021/bi981132w. [DOI] [PubMed] [Google Scholar]
- 41.Zhang R, Evans G, Rotella F, Westbrook E, Huberman E, Joachimiak A, Collart F R. Curr Med Chem. 1999;6:537–543. [PubMed] [Google Scholar]
- 42.Zhang R, Evans G, Rotella F J, Westbrook E M, Beno D, Huberman E, Joachimiak A, Collart F R. Biochemistry. 1999;38:4691–4700. doi: 10.1021/bi982858v. [DOI] [PubMed] [Google Scholar]
- 43.Blunt D S, Montelone B A, Upton S J, Khramtsov N V. Gene. 1996;181:221–223. doi: 10.1016/s0378-1119(96)00460-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.