Abstract
YgbQ is a cell division protein in Escherichia coli and Vibrio cholerae. In E. coli the ygbQ gene was discovered as a result of a computer search of the E. coli genome designed to find potential interacting partners for cell division protein FtsL. In V. cholerae, ygbQ was identified as an essential gene by using a transposon that fuses genes to an arabinose promoter. The role of YgbQ in cell division is supported by the following. Cells depleted of YgbQ in both organisms form long filaments, but DNA segregation is not affected. YgbQ localizes to the constriction site in wild-type E. coli cells. Localization of E. coli YgbQ to the constriction site depends on cell division proteins FtsQ and FtsL but not FtsW and FtsI, placing YgbQ in the sequential dependency order of proteins localizing to the division site. Localization of green fluorescent protein-FtsL also depends on YgbQ, indicating that FtsL and YgbQ colocalize to the division site in E. coli. Our results show colocalization of proteins to the bacterial midcell in E. coli and raise the possibility that these proteins interact in a coiled-coil structure.
Cell division in bacteria takes place at the midcell and occurs after the DNA has been duplicated and segregated into two daughter nucleoids. In Escherichia coli, this process requires a set of at least nine proteins that localize to the constriction site or septum. These proteins coordinate invagination of the cell membrane, inward growth of the peptidoglycan layer, and, finally, separation of daughter cells. The nine proteins, FtsZ, FtsA, ZipA, FtsK, FtsQ, FtsL, FtsW, FtsI, and FtsN, have been identified largely through genetic approaches. Because no systematic genetic approach for identifying such proteins has been performed or devised, there could be a number of thus-far-undiscovered proteins that play a role in cell division. Information is available regarding function for only a few of these proteins (reviewed in refs. 1 and 2).
Studies from a number of laboratories have led to a sequential dependency model for the assembly of this group of proteins at the cell septum (3). FtsZ arrives first at the cell septum, providing a scaffold for the recruitment of subsequent proteins. FtsA and ZipA localize to the septum independently of each other, but each depends on the presence of FtsZ for localization. The remaining proteins assemble at midcell in a strictly sequential dependency order as follows: FtsK–FtsQ–FtsL–FtsW–FtsI–FtsN. The mechanisms for this order of recruitment are not understood.
The work reported here arose from projects with different goals in the Beckwith and Mekalanos laboratories. The Beckwith laboratory has studied cell division in E. coli by focusing attention on three proteins, FtsQ, FtsL, and FtsI. These proteins have similar membrane topologies, i.e., a short amino-terminal domain facing the cytoplasm, a single transmembrane segment, and a carboxyl-terminal domain located in the periplasm. FtsL has a leucine zipper-like motif in its periplasmic domain, indicating that it may form coiled-coil homo- or heterooligomers. It forms homodimers in vitro but likely only weakly dimerizes in vivo (ref. 4 and N.B. and J.B., unpublished results). We suspected that there might be a partner protein that forms heterodimers or higher order heterooligomers with FtsL. A computer search of the E. coli genome for proteins with structural features similar to those of FtsL yielded several candidates, of which the YgbQ protein was most promising.
At the same time, the Mekalanos laboratory was working on an unrelated project to identify essential genes of the bacterium Vibrio cholerae. These studies led to the identification of a gene, the absence of whose product led to filamentation of the bacterium. A chance discussion in the hallway led to the discovery that we were studying homologous ORFs in the two organisms. Our combined studies, reported here, strongly indicate that the product of the ygbQ gene in E. coli and its homologue, VC0527, in V. cholerae are required for the cell division process. In addition, the results are consistent with the suggestion that FtsL and YgbQ interact and colocalize to the septum.
Material and Methods
Bacterial Strains, Plasmids, and Media.
Bacterial strains and plasmids are described in Table 1. Growth is in rich medium, NZY (E. coli) and LB (V. cholerae; ref. 5). When needed, ampicillin 200 μg/ml (plasmid) or 25 μg/ml (chromosome), chloramphenicol 30 μg/ml, kanamycin 40 μg/ml, spectinomycin 100 μg/ml, and streptomycin 500 μg/ml was added. Medium was supplemented with 0.1% (V. cholerae) or 0.2% l-arabinose (E. coli) and 0.2% d-glucose to induce or repress expression of genes controlled by the PBAD promoter, respectively. Standard techniques, PCR, electroporation, transformation, and P1 transduction, were used for cloning and analysis of DNA (6, 7). The enzymes for manipulating DNA were from New England Biolabs.
Table 1.
Strains and plasmids
| Strain or plasmid | Relevant genetic marker(s) or feature(s) | Source or reference |
|---|---|---|
| E. coli | ||
| JOE309 | MC4100 araD+ | 3 |
| NB974 | JOE309 Δ(λattL-lom)∷bla lacIqpBAD18-ygbQ | This study |
| NB946 | JOE309 ygbQ∷Kan Δ(λattL-lom)∷bla lacIqpBAD18-ygbQ | This study |
| NB805 | JOE309 Δ(λattL-lom)∷bla lacIq P207-gfp-ygbQec | This study |
| NB802 | JOE309 Δ(λattL-lom)∷bla lacIq P207-gfp-ygbQvc | This study |
| NB841 | JOE309 ftsL∷TnPhoA (Kanr) Δ(λattL-lom)∷bla lacIq P207-gfp-ygbQec/pLMG180 | This study, 5 |
| NB836 | JOE309 ftsQ∷TnPhoA50 (Kanr) Δ(λattL-lom)∷bla lacIq P207-gfp-ygbQec/pJC10 | This study, 26 |
| NB966 | JOE309 ftsI∷TnPhoAI137ΔIS50R (Kanr) Δ(λattL-lom)∷bla lacIq P207-gfp-ygbQec/pDSW262 | This study, 10 |
| NB991 | JOE309 ftsW∷Kan Δ(λattL-lom)∷bla lacIq P207-gfp-ygbQec/pDSW406 | This study, 27 |
| NB976 | JOE309 ygbQ∷Kan Δ(λattL-lom)∷bla lacIq pBAD18-ygbQ attφ80∷pJC115(P207-gfp-ftsI) | This study, 3 |
| NB977 | JOE309 ygbQ∷Kan Δ(λattL-lom)∷bla lacIqpBAD18-ygbQ attφ80∷pJC116(P207-gfp-ftsL) | This study, 3 |
| NB980 | JOE309 ygbQ∷Kan Δ(λattL-lom)∷bla lacIqpBAD18-ygbQ attφ80∷pJC118(P207-gfp-ftsQ) | This study, 3 |
| NB1000 | JOE309 ygbQ∷Kan Δ(λattL-lom)∷bla lacIqpBAD18-ygbQ P209-gfp-ftsW | This study |
| V. cholerae | ||
| N16961Sm | El Tor | 28 |
| NJ348 | N16961Sm with Tn10cm insertion pBSL181 | This study |
| NJ406 | YgbQ∷TnAraOut | This study |
| NJ600 | NJ406/pNJ25 | This study |
| Plasmids | ||
| pNB10 | pBAD18-ygbQ | This study, 29 |
| pNB11 | pBAD18-ygbQPBO | This study, 29 |
| pNB12 | pBAD33-ygbQPBO | This study, 29 |
| pNJ17 | TnAraOut delivery vehicle | 8 |
| pNJ25 | pDSW207-ygbQvc | This study, 10 |
| pBSL181 | Tn10cm | 9 |
Isolation of Arabinose-Dependent V. cholerae.
The introduction of TnAraOut into V. cholerae and the screen for insertion mutants that are arabinose-dependent for growth were done as described (8) with the following modification. NJ348 was isolated by mating Tn10cm (pBSL181; ref. 9) with N16961Sm to generate a chloramphenicol-resistant V. cholerae to reduce background during conjugation with pNJ17. NJ406 was isolated as a strain with an arabinose-dependent growth phenotype with a TnAraOut insertion 66 bp upstream of VC0527. This strain forms tiny colonies on LB plates and fails to grow in LB broth in the absence of arabinose.
Construction of ygbQ and the ygbQ-P-B-O Operon Under pBAD Promoter.
The ygbQ gene was amplified by PCR from chromosomal DNA of a wild-type E. coli strain using ygbQ5EcoRI (5′-CCGGAATTCTCGGATGCATGGGATGAT-3′) and ygbQ3XbaI (5′-CGCTCTAGATGTTAATTCCCGGGCTGA-3′) as primers. The PCR fragment was digested with EcoRI and XbaI and ligated into the same sites of pBAD18, and the resulting plasmid was named pNB10 (restriction sites are underlined in the primer sequences). The complete ygbQ-P-B-O operon was amplified by PCR from chromosomal DNA using ygbQ5EcoRI (5′-CCGGAATTCTCGGATGCATGGGATGAT-3′) and ygbO3XbaI (5′-CGCTCTAGACATTCACGCCAAGATAGACG-3′) as primers. The PCR fragment was digested with EcoRI and XbaI and ligated into the same sites of pBAD18, and the resulting plasmid was named pNB11. The ygbQ-P-B-O genes were recloned into a pBAD18 chloramphenicol vector to construct a ygbQ depletion strain (see below). pNB11 was digested with NheI and SalI. The 2.8-kb fragment containing the ygbQ-P-B and O genes was ligated into an NheI- and SalI-digested pBAD18 chloramphenicol vector, and the resulting plasmid was named pNB12.
Construction of Strains Containing Green Fluorescent Protein (GFP) Fusions.
ygbQ of E. coli was amplified by PCR using pNB10 as template and ygbQ5EcoRIgfp (5′-CGAGAATTCAACAACAACATGGGTAAACTAACGCTGCTGTTGC-3′) and ygbQ3HdIIIgfp (5′-TCGAAGCTTTTATCGATTGTTTTGCCCCGCAGAC-3′) as primers. The amplified product was digested with EcoRI and HindIII, ligated into the same sites of pDSW207 (10), and named pNB20. The GFP-YgbQ fusion protein has the linker sequence YKEFNNM, where Y and K are the last residues of GFP, and M is the first residue of YgbQ. ygbQ of V. cholerae was amplified by PCR with chromosomal DNA from N16961 using the primers ygbQF (5′-GCCGAATTCATGCGAGTATTTGCTTTGACC-3′) and ygbQR (5′-GGCTGCAGTTGGCGCGACTCCTCACCGAT-3′). The amplified product was digested with EcoRI and HindIII and ligated into the same sites of pDSW207 (10). The resulting plasmid was named pNJ25.
Stable introduction of the gfp-ygbQ fusions and gfp-ftsW into the E. coli chromosome at the λ attachment site were obtained with the InCh integration vector (11). The construction of gfp-ftsQ, gfp-ftsL, and gfp-ftsI fusion strains at the att site for the lambdoid phage φ80 are described by Chen and Beckwith (3).
Construction of a ygbQ Null Strain in E. coli.
To disrupt the chromosomal ygbQ gene we recombined a PCR product with chromosomal DNA (12). A DNA fragment was amplified by PCR using pKD4 (12) as template DNA and H1P1ygbQ (5′-GGATGTAGAGTCGTCTTCGGATGCATGGGATGATGATGCCGTTTTTCAGGTGTAGGCTGGAGCTGCTTC-3′) and P2H2Q (5′-GCGCAAACATCCAAATGAGTGGTTGCCATGTTAATTCCCGGGCTGATTTACATATGAATATCCTCCTTAG-3′) as primers. The resulting fragment was composed of a kanamycin-resistance gene and extensions that are homologous to the regions adjacent to each end of the ygbQ gene (underlined sequences). This PCR fragment was transformed into competent cells of strain NB885 carrying plasmids pKD46 (12) and pNB12. Cells were selected for growth on kanamycin in the presence of l-arabinose. Both the phage λ red recombinase gene on pKD46 and the ygbQ-P-B and O genes on pNB12 are under control of the pBAD promoter. The replacement of ygbQ by kanamycin was confirmed by PCR using kanamycin-, ygbE-, and ygbP-specific primers. Kanamycin-resistant colonies were tested for growth on ampicillin, to test for loss of pKD46, and both on l-arabinose and d-glucose to test for depletion of ygbQ. Strains with a disrupted ygbQ gene grew on d-glucose plates as small colonies, most likely because of basal expression from pNB12. The ygbQ∷Km was transduced into a strain that carried an extra chromosomal copy of only ygbQ under pBAD at the λatt site for complementation to avoid residual expression of ygbQ-P-B and O from pNB12 (NB946).
Growth Conditions for GFP-Fusion Protein Localization.
For localization of GFP fusions in wild-type background, cells were grown overnight at 30°C in NZY medium plus 25 μg/ml ampicillin, diluted 1:200 in fresh medium without ampicillin, and grown to an optical density at 600 nm (OD600) of 0.5. The cultures were diluted in medium containing 2.5 μM (gfp-ftsI), 5 μM (gfp-ftsQ), or 10 μM (gfp-ftsL and gfp-ygbQ) isopropyl β-D-thiogalactoside (IPTG). Cells were kept in logarithmic growth phase by serial dilutions in the same medium and grown for 4 h. Cells were fixed directly in growth medium and processed for microscopy as described below. For experiments involving depletion strains, the overnight culture was grown at 30°C in NZY medium containing chloramphenicol, kanamycin, and 0.2% l-arabinose. The culture was diluted 1:200 in the same medium and grown to an OD600 of 0.5. After washing the cells once in medium without sugar, the cells were resuspended in medium containing l-arabinose or d-glucose and IPTG. Cells were kept in logarithmic growth phase by serial dilutions in the same medium and grown for several hours. Cells were fixed when filaments of substantial length were observed. To study the localization of GFP-YgbQ in an FtsQ, FtsL, FtsW, or FtsI depletion strain, gfp-ygbQ was induced with 10 μM IPTG in all strains. Strains to study the localization of GFP-FtsQ, GFP-FtsL, GFP-FtsW, and GFP-FtsI in a ygbQ depletion strain were constructed by using gfp fusions integrated at the phage φ80 att site (3). The cultures were grown in the presence of 2.5 μM (gfp-ftsI), 5 μM (gfp-ftsQ), 10 μM (gfp-ftsL), or 1 mM (gfp-ftsW) IPTG.
Fluorescence Microscopy.
Cells were fixed in a mixture of paraformaldehyde and glutaraldehyde before microscopic analysis as described (13). After fixation, cells were stained with 0.2 μg/ml 4′,6-diamidino-2-phenylindole in PBS for 10 min at room temperature, washed twice in PBS, and immobilized on agarose slides as described (14). To examine cells we used an Orca 12-bit cooled charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan) mounted on an Axioskop 2 microscope (Zeiss). Images were taken by using Zeiss AXIOVISION software and analyzed with the public domain program OBJECTIMAGE 1.62p by Norbert Vischer (University of Amsterdam, Amsterdam, http://simon.bio.uva.nl/object-image.html). Images were processed in Adobe PHOTOSHOP for presentation.
Preparation of Membranes and Detection of FtsL by Western Blotting.
Membranes were prepared by sonicating cells in 100 mM Tris⋅HCl, pH 8.0, containing 1 mM PMSF. Whole cells were removed by centrifugation at 5,000 rpm for 5 min. Membranes were pelleted at 100,000 × g in a Beckman TL100 ultracentrifuge for 60 min and resuspended in 100 mM Tris⋅HCl, pH 8.0. A total of 10 μg of membranes was applied per lane on SDS/PAGE. Electrophoresis and immunoblotting were performed as described by Laemmli (15) and Towbin et al. (16), respectively. The membrane was probed with FtsL-specific polyclonal antibodies and subsequently with horseradish peroxidase-conjugated goat anti-rabbit antibodies and developed with chemiluminescence reagents (Amersham Pharmacia).
Results
Identification and Characterization of ygbQ in V. cholerae.
The mariner-based transposon, TnAraOut, causes genes to be transcriptionally fused to the arabinose-inducible promoter PBAD after transposition into a genome (8). Essential genes can be identified, because strains with TnAraOut insertions fused to essential genes depend on the presence of arabinose for growth. We found a TnAraOut inserted (strain NJ406) in V. cholerae 66 bp upstream of the predicted translational start of VC0527 that depended on arabinose for growth. The gene VC0527 is most similar to the E. coli gene ygbQ (and hereafter will be referred to as ygbQ).
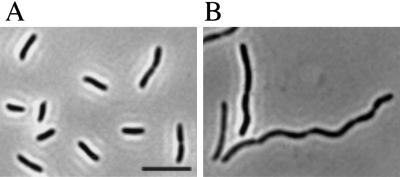
Growth of strain NJ406 in LB broth in the presence of arabinose showed a wild-type growth phenotype. With cells grown in the absence of arabinose, long filaments of V. cholerae formed (Fig. 1). The genomic organization of this region in V. cholerae and E. coli is similar. In E. coli it is known that the two genes downstream of ygbQ, ygbP, and ygbB are essential (17). To determine if the inability to grow in the absence of arabinose was caused by ygbQ or the downstream genes, a plasmid containing only ygbQ, pNJ25, was constructed. NJ406 could be complemented by IPTG-inducible expression of ygbQ from pNJ25, indicating that this gene alone could complement the filamentation phenotype. A discussion of cell division phenotypes between N.J. and J.B. led to the realization that we were studying the same gene.
Figure 1.
Phenotype of ygbQ depletion strain of V. cholerae. (A) Image of cells grown with arabinose to express ygbQ. (B) Image of cells grown with glucose to deplete YgbQ. (Scale bar, 10 μm.)
A Computer Search for Potential Interacting Partners of FtsL of E. coli.
The unique features of cell division protein FtsL and the fact that FtsL homodimers seem to be very weak or exist only temporarily in vivo led the Beckwith lab to look for potential partner proteins that interacted with FtsL. We sought a protein that would share many of the features of the structure and membrane topology of FtsL. A computer search was developed by using the entire E. coli chromosome to look for proteins that (i) had unknown function, (ii) contained a leucine zipper-like motif predicted to form a coiled-coil structure, and (iii) were predicted to be integral cytoplasmic membrane proteins. We found six such proteins including a putative tyrosine kinase, three proteins with considerably higher molecular weight than FtsL, and two small proteins.
The two small proteins are YqhH (SwissProt accession no. Q46860), an outer membrane lipoprotein precursor, and YgbQ (Q46894), a hypothetical protein conserved across bacterial species that shows some weak homology (20 of 59 residues are identical) with a protein of Bacillus halodurans. The B. halodurans protein itself is a homologue of the cell division initiator protein DivIC of Bacillus subtilis. (Homology searches did not show YgbQ to be a direct homolog of the B. subtilis DivIC.) In B. subtilis, FtsL and DivIC might function as a complex in cell division (18).
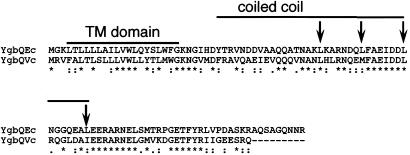
YgbQ is composed of 103 aa residues (11 kDa) and is predicted to be a bitopic membrane protein. A three-residue amino-terminal domain is predicted to face the cytoplasm, residues L4–G22 resembles a transmembrane segment, and the presumed periplasmic region, containing L46–L67, has a leucine zipper-like motif and is predicted to form a single coiled coil (Fig. 2). A hybrid protein in which alkaline phosphatase was fused to the carboxyl terminus of YgbQ resulted in dark blue colonies on 5-bromo-5-chloro-3-indolylphosphate (XP) indicator plates, suggesting that the carboxyl terminus of the protein is facing the periplasm (data not shown; ref. 19). This set of structural features resembles those of FtsL. The gene is in a gene cluster, including the downstream genes ygbP, ygbB, and ygbO, found in many Gram-negative and Gram-positive bacteria. Both ygbP and ygbB are essential genes involved in the pathway for isoprenoid biosynthesis in E. coli (17).
Figure 2.
CLUSTALW 1.81 alignment of YgbQ of E. coli (Ec) and V. cholerae (Vc). The leucine residues that form the leucine zipper-like motif in the periplasmic domain of YgbQ of E. coli are indicated with arrows. The overlined regions represent the putative transmembrane (TM) domain (residues 4–22) and the coiled-coil domain (residues 29–67) of YgbQ. Conserved amino acid residues are indicated by asterisks.
Because of the Bacillus information on the role of the homologous proteins and because YgbQ had a closely analogous domain structure to FtsL, we considered it to be an interesting candidate for an FtsL-interacting protein. We initiated studies to inactivate the ygbQ gene and characterize the phenotype of the null mutation. At this point we discovered that our two laboratories were working on homologous genes (50% identity) in E. coli and V. cholerae occupying the same chromosomal position in the two organisms. The V. cholerae protein, while not having a strict leucine-zipper motif, is predicted to form a coiled-coil structure by the COILS 2.2 program (20). Given that the inactivation of the V. cholerae gene led to a filamentous phenotype, we were encouraged further to pursue our study of the E. coli gene. While proceeding to construct a ygbQ null mutation, we decided also to determine immediately whether YgbQ localized to the cell septum.
E. coli YgbQ Localizes to the Division Site in a Wild-Type Background.
All known E. coli cell division proteins localize to the constriction site; unrelated membrane proteins that have been examined do not (see e.g. refs. 4, 10, and 13). Thus, localization of a protein to midcell provides strong suggestion that a protein is involved in cell division. We asked whether YgbQ localized to the cell septum. A gene fusion that expressed YgbQ with GFP at its amino terminus was constructed and inserted into the chromosome in single copy via λInCh (11). The strains are merodiploid for ygbQ, i.e., a wild-type copy of ygbQ is present at the normal chromosomal location, and a gfp-ygbQ copy is located at the λ attachment site expressed from the regulatable trc promoter. The strains were grown with different concentrations of IPTG in the media to induce different levels of expression of the GFP-YgbQ fusion protein. Using fluorescent microscopy, we looked for localization of GFP-YgbQ to the midcell with IPTG concentrations ranging between 10 and 50 μM. Most cells exhibited a bright band of fluorescence at their midpoints, indicating that GFP-YgbQ localizes to the division site (data not shown). GFP-FtsL localization to this region had been detected previously when 10 μM of inducer was used to induce a GFP-FtsL fusion protein.
We also looked for localization of the YgbQ of V. cholerae (YgbQvc) in E. coli by using a GFP-fusion protein analogous to the GFP-YgbQ of E. coli. We observed a fluorescent signal when expression of gfp-ygbQvc was induced with 50 μM IPTG compared with 10 μM for GFP-YgbQec; even with 50 μM IPTG, the fluorescent signal was less intense than with GFP-YgbQ of E. coli (data not shown). Perhaps the higher induction levels of GFP-YgbQvc are necessary to observe septal fluorescence because the nonnative protein competes less well with wild-type YgbQ of E. coli for localization to midcell.
Depletion of ygbQ in E. coli Is Correlated with Filamentation.
A YgbQ depletion strain of E. coli was constructed by chromosomal disruption of ygbQ (12). Because ygbQ is part of a cluster with essential genes involved in isoprenoid synthesis, disruption of ygbQ might affect not only ygbQ but also the expression of the downstream genes ygbP, ygbB, and ygbO. However, the system we used (12) is supposed to allow transcription of genes downstream of the disrupted gene.
The strain carrying the ygbQ deletion also carried an additional copy of ygbQ under PBAD control at the bacteriophage λatt site. This depletion strain is able to grow on medium supplemented with arabinose but not with glucose. The cells form extensive filaments when YgbQ is depleted (see Fig. 4). 4′,6-Diamidino-2-phenylindole staining indicated that the nucleoids in filaments are spaced regularly; thus, DNA segregation is not affected in a ygbQ null strain (data not shown). A plasmid expressing gfp-ygbQ complements the ygbQ deletion strain for growth. Depletion of ygbQ in a strain expressing ygbP, B, and O from a different promoter results in a filamentous phenotype, which indicates that in the ygbQ null, expression of the ygbP, B, and O genes was sufficient for growth (data not shown). Furthermore, the ygbP and ygbB depletion strains described by Campos et al. (17) have a depletion phenotype different from the ygbQ depletion strain; they stop growing immediately when the complementing pathway is inactivated, and cells do not filament. These results show that, as in V. cholerae, YgbQ in E. coli is essential, and its depletion results in inhibition of cell division.
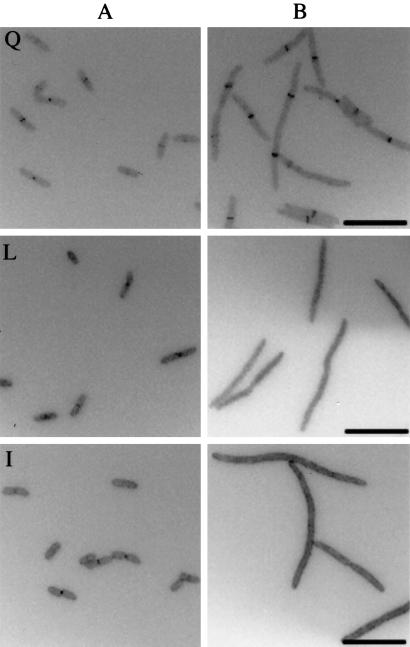
Figure 4.
Localization of GFP fusions to FtsQ (Q), FtsL (L), and FtsI (I) in a ygbQec depletion strain. (A) Localization of GFP-fusion proteins in cells grown with arabinose to express ygbQ. (B) Localization of GFP-fusion proteins in cells grown with glucose to deplete ygbQ. (Scale bars, 10 μm.)
Localization of YgbQ Depends on FtsQ and FtsL but Not on FtsW and FtsI.
The results from our studies on the V. cholerae and E. coli YgbQ homologues strongly suggested that YgbQ plays an important role in cell division. We wished to determine the position of YgbQ in the process of assembly of cell division proteins at midcell. To make this determination we studied the localization of GFP-YgbQ in mutants that could be depleted of cell division proteins that appeared late in the pathway, FtsQ, FtsL, FtsW, and FtsI, because the discovery of YgbQ in E. coli was based on a hunt for a partner of FtsL. Depletion strains for the cell division proteins each contained a null mutation for the protein to be depleted and a plasmid expressing the same cell division protein from a PBAD promoter. The cells grow normally in the presence of arabinose, but when shifted to glucose-containing medium, the cells eventually developed into long filaments. As cells are being depleted in glucose medium, we fix and prepare them while they still are exhibiting logarithmic growth.
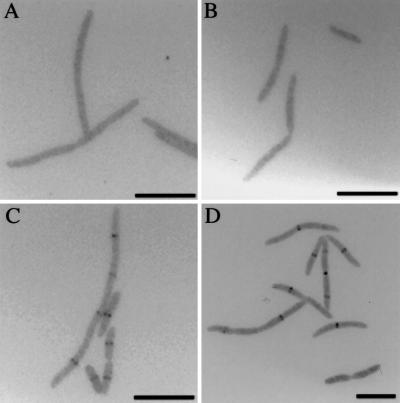
The cells of depletion strains for FtsQ, FtsL, FtsW, and FtsI grown in arabinose are of normal length, and GFP-YgbQ is found at the septum (data not shown). When these same cells are grown in glucose-containing medium, they all formed filaments. GFP-YgbQ localized to the septal region of filaments in the FtsW and FtsI depletion strains but not in the FtsQ and FtsL depletion strains (Fig. 3). As positive and negative controls, we showed that, as found previously (13, 21), GFP-FtsQ localizes to potential division sites in FtsL depletion filaments, and GFP-FtsI does not (data not shown). These results suggest that either (i) YgbQ localizes to the cell septum after FtsL and before FtsW or (ii) YgbQ colocalizes to the septum with FtsL.
Figure 3.
Localization of GFP fusions to YgbQ in FtsQ (A), FtsL (B), FtsW (C), and FtsI (D) depletion strains. (Scale bars, 10 μm.)
Localization of FtsQ, FtsL, FtsW, and FtsI in a YgbQ Depletion Strain.
To clarify the position of YgbQ in the septal assembly sequence further, we studied localization of GFP-FtsL in a ygbQ null background by using GFP-FtsQ, GFP-FtsW, and GFP-FtsI as likely positive and negative controls. Because a complementing copy of pBAD-ygbQ was integrated at the λatt site in the YgbQ depletion strain, we integrated the GFP-fusion constructs into the chromosome at the att site for the lambdoid phage φ80 (3). Strains were grown in NZ medium supplemented with arabinose or glucose at 30°C, and expression of gfp-ftsQ, gfp-ftsL, gfp-ftsI, and gfp-ftsW was induced with 5 μM, 10 μM, 2.5 μM, and 1 mM IPTG, respectively. In cells grown in arabinose, we observed a fluorescent signal for GFP-FtsQ, GFP-FtsL, GFP-FtsW, and GFP-FtsI at the cell midpoint (Fig. 4). In filaments formed after depletion of YgbQ, we obtained results consistent with the order predicted above; GFP-FtsQ localizes to the septum, but GFP-FtsW and GFP-FtsI do not. However, we observed no localization of GFP-FtsL to midcell in YgbQ-depleted filaments (Fig. 4). Because YgbQ itself is not found at the septum in FtsL-depleted cells, our results suggest that FtsL and YgbQ localize to the division site in a codependent fashion.
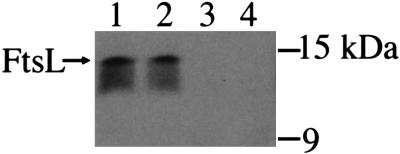
Results from B. subtilis suggest that its YgbQ analogue, DivIC, stabilizes FtsL. We asked whether depletion of YgbQ might affect FtsL stability in E. coli and assayed FtsL protein levels in a YgbQ depletion strain. We detected FtsL in a YgbQ depletion strain grown in arabinose but not in cells grown in glucose (Fig. 5). The likely instability of FtsL under these conditions suggests that YgbQ stabilizes FtsL, indicating an FtsL–YgbQ interaction.
Figure 5.
Detection of wild-type levels FtsL in membranes of a ygbQ null strain (NB946). Lanes 1 and 2, NB946 grown in arabinose to express ygbQ for 3 and 4 h, respectively; lanes 3 and 4, NB946 grown in glucose to deplete ygbQ for 3 and 4 h, respectively.
Discussion
In this article, we report the characterization of a gene, ygbQ, encoding a protein in the cell division pathway of E. coli and V. cholerae. In both organisms, depletion of YgbQ leads to a filamentous phenotype typical of null mutants in other cell division genes. Furthermore, YgbQ localizes to the midcell of E. coli, a feature known thus far to be specific only for those proteins directly involved in the cell division process. Finally, studies on the localization of YgbQ assign it a specific place in the septal assembly of those proteins required for cell division and are consistent with an interaction between it and FtsL.
We detected ygbQ of V. cholerae in a screen for essential genes. In E. coli, we focused on ygbQ after searching the genome for ORFs coding for proteins that might behave as partners of the cell division protein, FtsL. That we found this gene by searching for such a candidate and that we showed this gene to be involved in cell division in itself encouraged us to pursue its possible connection to FtsL. Furthermore, a pair of possibly related proteins in B. subtilis, FtsL and DivIC, seem to be interacting partners in the cell division process of that organism. YgbQ of E. coli shows only very weak identity with DivIC (14%), but it seems related through the homology of both to a protein in B. halodurans. The septal localization of DivIC in B. subtilis (22) depends on an FtsL homologue (23), and it is unstable in the absence of FtsL (23), suggesting that the two proteins interact (18). Localization of these Bacillus proteins has not been examined.
Our studies on the position of YgbQ in the protein assembly pathway at the E. coli septum are consistent also with an interaction of FtsL and YgbQ. YgbQ depends on FtsQ but not FtsW for its localization to the septum, as does FtsL. But neither of the two proteins, YgbQ and FtsL, are seen at the septum in the absence of the other. These results are consistent with a model in which FtsL and YgbQ colocalize to the septum. Such codependence also may be explained solely on the basis of YgbQ stabilization of FtsL. Our studies show a case of two proteins apparently codependent for their localization to the midcell of the bacteria.
FtsL and YgbQ of E. coli have a leucine zipper-like motif in their periplasmic domain. We previously found that the periplasmic domain of nearly all members of the FtsL family of proteins exhibited a high propensity to form coiled-coil structures in the predictions of the COILS 2.2 program. Although FtsL had the lowest coiled-coil propensity among its homologues according to the COILS 2.2 program, we pointed out why this sequence could still form such a structure (4). The periplasmic domain of YgbQ also is predicted to form a coiled-coil structure. Coiled coils are helical bundles of 2–5 α-helices that have a distinctive packing of amino acid side chains at the helix–helix interfaces and are found, for example, in transcription factors, skeletal and motor proteins, and membrane fusion proteins.
FtsL and YgbQ of V. cholerae do not have a strict leucine zipper motif; however, they do exhibit the same predictions for coiled-coil properties as their E. coli homologues. Our current hypothesis is that FtsL and YgbQ form a coiled-coil heterodimeric or multimeric structure that plays a role in the cell division process.
What function might such a complex of FtsL and YgbQ perform? Because coiled-coil structures are formed between pairs of partner proteins that are involved in membrane fusion events in eukaryotic cells (24, 25), we raise the possibility that these two E. coli proteins might form a multimeric complex that promotes a membrane fusion event at the cell septum. For example, it would seem that the increasing constriction of the midcell eventually might require a system catalyzing the fusion of the cell membranes leading to separation into daughter cells, a step for which a protein complex promoting such fusion would be needed.
Acknowledgments
We are grateful to members of the Beckwith and Mekalanos laboratories for assistance and encouragement. We also thank Dr. Boronat for providing bacterial strains. This work was supported by National Institute of General Medical Sciences Grant GM-38922 (to N.B. and D.B.) and National Institute of Allergy and Infectious Diseases Grant AI-26289 (to J.J.M. and N.J.). J.B. was supported by an American Cancer Society Research Professorship.
Abbreviations
- GFP
green fluorescent protein
- IPTG
isopropyl β-D-thiogalactoside
References
- 1.Rothfield L, Justice S, Garcia-Lara J. Annu Rev Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- 2.Margolin W. FEMS Microbiol Rev. 2000;24:531–548. doi: 10.1111/j.1574-6976.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen J C, Beckwith J. Mol Microbiol. 2001;42:395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- 4.Ghigo J-M, Beckwith J. J Bacteriol. 2000;182:116–129. doi: 10.1128/jb.182.1.116-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guzman L-M, Barondess J, Beckwith J. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 6.Miller J H. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 7.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 8.Judson N, Mekalanos J J. Nat Biotechnol. 2000;18:740–745. doi: 10.1038/77305. [DOI] [PubMed] [Google Scholar]
- 9.Alexeyev M F, Shokolenko I N. Gene. 1995;160:59–62. doi: 10.1016/0378-1119(95)00141-r. [DOI] [PubMed] [Google Scholar]
- 10.Weiss D S, Chen J C, Ghigo J-M, Boyd D, Beckwith J. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd D, Weiss D S, Chen J C, Beckwith J. J Bacteriol. 2000;182:842–847. doi: 10.1128/jb.182.3.842-847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko K A, Wanner B L. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J C, Weiss D S, Ghigo J-M, Beckwith J. J Bacteriol. 1999;181:521–530. doi: 10.1128/jb.181.2.521-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Helvoort J M L M, Woldringh C L. Mol Microbiol. 1994;13:577–583. doi: 10.1111/j.1365-2958.1994.tb00452.x. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli E K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campos N, Rodríguez-Concepción M, Sauret-Güeto S, Gallego F, Lois L-M, Boronat A. Biochem J. 2001;353:59–67. [PMC free article] [PubMed] [Google Scholar]
- 18.Sievers J, Errington J. Mol Microbiol. 2000;36:846–855. doi: 10.1046/j.1365-2958.2000.01895.x. [DOI] [PubMed] [Google Scholar]
- 19.Manoil C, Beckwith J. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 20.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 21.Ghigo J M, Weiss D S, Chen J C, Yarrow J C, Beckwith J. Mol Microbiol. 1999;31:725–737. doi: 10.1046/j.1365-2958.1999.01213.x. [DOI] [PubMed] [Google Scholar]
- 22.Katis V L, Harry E J, Wake R G. Mol Microbiol. 1997;26:1047–1055. doi: 10.1046/j.1365-2958.1997.6422012.x. [DOI] [PubMed] [Google Scholar]
- 23.Daniel R A, Harry E J, Katis V L, Wake R G, Errington J. Mol Microbiol. 1998;29:593–604. doi: 10.1046/j.1365-2958.1998.00954.x. [DOI] [PubMed] [Google Scholar]
- 24.Jahn R, Südhof T C. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- 25.Eckert D M, Kim P S. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 26.Carson M J, Barondess J, Beckwith J. J Bacteriol. 1991;173:2187–2195. doi: 10.1128/jb.173.7.2187-2195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer K L N, Weiss D S. J Bacteriol. 2002;184:904–912. doi: 10.1128/jb.184.4.904-912.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fullner K J, Mekalanos J J. Infect Immun. 1999;67:1393–1404. doi: 10.1128/iai.67.3.1393-1404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzman L-M, Belin D, Carson M, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]