Abstract
Double-stranded (ds) RNA induces transcription of the 561 gene by activating IFN regulatory factor (IRF) transcription factors, whereas similar induction of the IFN-β gene is thought to require additional activation of NFκB and AP-1. In mutant P2.1 cells, dsRNA failed to activate NFκB, IRF-3, p38, or c-Jun N-terminal kinase, and transcription of neither 561 mRNA nor IFN-β mRNA was induced. The defect in the IRF-3 pathway was traced to a low cellular level of this protein because of its higher rate of degradation in P2.1 cells. As anticipated, in several clonal derivatives of P2.1 cells expressing different levels of transfected IRF-3, activation of IRF-3 and induction of 561 mRNA by dsRNA was restored fully, although the defects in other responses to dsRNA persisted. Surprisingly, IFN-β mRNA also was induced strongly in these cells in response to dsRNA, demonstrating that the activation of NFκB and AP-1 is not required. This conclusion was confirmed in wild-type cells overexpressing IRF-3 by blocking NFκB activation with the IκB superrepressor and AP-1 activation with a p38 inhibitor. Therefore, IRF-3 activation by dsRNA is sufficient to induce the transcription of genes with simple promoters such as 561 as well as complex promoters such as IFN-β.
Double-stranded (ds) RNA is a potent regulator of gene expression in mammalian cells (1). The addition of dsRNA to human cells in culture causes rapid induction of transcription of more than 100 genes, with a concomitant decrease in the mRNA levels of a different set of genes (2). The dsRNA-stimulated genes include those known to be induced by virus infection such as the genes encoding type I IFN as well as genes induced by cytokines such as IFN-α/β, IFN-γ, tumor necrosis factor, and IL-1. Thus, dsRNA, viruses, and different inflammatory cytokines induce the transcription of partially overlapping sets of cellular genes, the products of which mediate some of the common cellular responses to these agents.
The human IFN-β gene is one of the most well investigated dsRNA-inducible genes. Its promoter is complex, with several partially overlapping positive and negative regulatory elements (3, 4). Three families of transcription factors, all of which are activated by dsRNA, have been shown to participate in the induction process. Members of the IFN regulatory factor (IRF) family, most notably IRF-3 (5), bind to the cognate IFN response element site, NFκB binds to the κB site, and c-Jun/ATF-2 heterodimer binds to the AP-1 site (6–9). NFκB is activated by dsRNA, which causes degradation of IκB in response to its phosphorylation by the activated IκB kinase complex (10, 11). The dsRNA-dependent protein kinase (PKR) is required for activating this pathway, although its immediate substrate in this context remains unknown. Similarly, although IRF-3 is also activated by phosphorylation, the relevant protein kinase has not yet been identified (12, 13). dsRNA has been shown also to activate the stress-activated protein kinases p38 and c-Jun N-terminal kinase (JNK) (14, 15). Their actions lead to the activation of the transcription factors ATF-2 and c-Jun, respectively. Thus, treatment of cells with dsRNA causes the rapid activation of several cytoplasmic protein kinases, resulting in the activation and nuclear translocation of their target transcription factors. For activation of the complex IFN-β promoter, coordinate actions by several of these transcription factors are thought to be essential.
In contrast to the complex promoter of the IFN-β gene, the human 561 gene has a simple promoter with one perfect and another putative IFN-stimulated response element (ISRE) as the only recognizable cis elements (16). Recent microarray screens have identified 561 to be the human gene most strongly induced in response to type I IFN (17) or dsRNA (2). The encoded protein, p56, inhibits cellular protein synthesis by binding to the translation initiation factor, eukaryotic initiation factor-3 (18). Our previous studies have established that the induction of 561 mRNA by dsRNA is not mediated by IFN and does not require ongoing protein synthesis (16). The signaling pathways used by IFN-α/β and dsRNA are different, because dsRNA can induce the 561 gene in mutant cell lines that are defective in IFN-dependent signaling.
In the current study, we have taken advantage of the mutant cell line P2.1 to delineate further the dsRNA-mediated signaling pathways that lead to induced transcription of the 561 and IFN-β genes. Unlike the parental U4C cells, P2.1 cells are defective in the dsRNA response (19). Although PKR was activated by dsRNA in these cells, NFκB and IRF-1 were not. Here we report that p38, JNK, and IRF-3 also are not activated by dsRNA in these cells. Partial reconstitution was achieved by ectopic expression of IRF-3 in P2.1 cells; induction of the 561 and IFN-β genes were restored. However, dsRNA still failed to activate p38, JNK, and NFκB in the IRF-3-reconstituted cells, thus demonstrating the sufficiency of IRF-3 for driving transcription from both the simple 561 promoter and the complex IFN-β promoter.
Materials and Methods
Cell Culture and Generation of Cell Lines.
U4C, P2.1, and 2fTGH cells have been described previously (19, 20). U4C and P2.1 cells were cotransfected with the pCDNA3/hIRF-3 expression plasmid (7) and the pBABE/Puro selection plasmid. After selection in puromycin, individual clones were expanded and screened for IRF-3 expression by Western analysis.
The dominant negative S32/36 IκBα super repressor (IκBαSR) was kindly provided by Inder Verma (21). The IκBαSR cDNA was cloned into the selectable retroviral vector pLXIN (CLONTECH). 2fTGH cells were transfected with the pLXIN/IκBαSR plasmid, and a pool of G418-resistant cells stably expressing the IκBαSR was obtained. These cells, termed 2f-SR, were subsequently transfected with the IRF-3 and puromycin plasmids, and IRF-3-expressing clones were isolated and screened for IRF-3 and IκBα expression by Western analysis.
dsRNA Treatment.
dsRNA stocks (1 mg/ml) were prepared by suspending poly(I)⋅poly(C) (Amersham Pharmacia) in PBS and shearing the RNA by passage through a 26-gauge needle. Working dsRNA stocks were stored at −20°C. To treat cells, dsRNA (100 μg/ml) was added directly to the medium for the indicated times.
Ribonuclease Protection Assay (RPA).
RNA was isolated with RNAzol B (Tel-Test, Friendswood, TX). RPAs were performed as described (22) or with the RPA III kit (Ambion, Austin, TX). The 561 probe corresponds to nucleotides 1,342–1,511 of the 561 message. The IFN-β and IRF-3 probes protect nucleotides 354–563 and 1,027–1,287, respectively. The actin RPA probe has been described previously by Enoch et al. (23). mRNA levels were measured with a Molecular Dynamics PhosphorImager.
Electrophoretic Mobility-Shift Assay (EMSA).
Whole-cell extracts were prepared as described by Leaman et al. (19). Lysates were incubated on ice for 15 min and clarified by centrifugation. EMSAs were performed as described by Sizemore et al. (24) with the NFκB site of the IP10 gene serving as the probe.
Western Analysis.
Whole-cell lysates were prepared as described for EMSAs (19). Westerns analyses were performed as described by Guo et al. (25) with antibodies against IκBα (Santa Cruz Biotechnology), actin (Sigma), and IRF-3 (a gift from Michael David, ref. 26). To determine the activation of p38, Western analyses were performed as described by Goh et al. (27). Protein expression levels were determined by analyzing the Western transfers with NIH IMAGE software.
Immunofluorescence.
Cells were plated on coverslips in 6-well dishes at least 16 h before treatment. After dsRNA treatment, cells were washed with PBS, fixed for 30 min with 4% paraformaldehyde, and permeabilized with 0.2% Triton X-100 for 15 min. The cells were blocked in PBS containing 0.02% Tween 20, 3% BSA, and 3% goat serum overnight at 4°C or for 2 h at room temperature. The cells were incubated with the primary IRF-3 and secondary Alexa 488 goat anti-rabbit (Molecular Probes) antibodies for 1 h at room temperature. Coverslips were mounted with antifade agent containing 4′,6-diamidino-2-phenylindole (Vector Labs) and examined.
Determination of IRF-3 Half-Life.
U4C.2 and P2.1.17 cells were washed and preincubated in methionine/cysteine-free DMEM for 1 h. Cells then were pulse-labeled with 200 μCi/ml (1 Ci = 37 GBq) 35S-labeled methionine/cysteine (Expre35S35S protein labeling mix, Perkin–Elmer) for 2 h. Cells were washed, and the label was chased with complete medium for the indicated times. After the chase, cells were washed with cold PBS and lysed in radioimmunoprecipitation assay lysis buffer with 25 mM NaF/0.4 mM PMSF/0.5 mM sodium orthovanadate/10 μg/ml pepstatin A/10 μg/ml aprotinin/10 μg/ml leupeptin. Lysates were incubated on ice for 15 min and clarified by centrifugation. Protein concentrations were determined, and 125 μg of protein extract was precleared with 100 μl Pansorbin (Calbiochem) for 1 h. The Pansorbin was removed by centrifugation, and the remaining lysate was incubated with the anti-IRF-3 monoclonal antibody SL-12 (28) and Protein G-Sepharose (Amersham Pharmacia) for 4 h. The immunoprecipitates were washed extensively with radioimmunoprecipitation assay buffer and separated by SDS/PAGE. The labeled proteins were visualized and quantified by using storage phosphor technology (Molecular Dynamics).
Results
The Spectra of dsRNA-Signaling Defects.
P2.1 cells are defective in dsRNA-dependent signaling (19). The nature of these defects was characterized further in the experiments shown in Fig. 1. We have shown previously that the activation of NFκB by dsRNA is defective in P2.1 cells. To understand the mechanism of this defect, we examined IκB degradation. There was a transient degradation of IκBα in U4C cells that peaked at 1 h (Fig. 1A) and corresponds to the kinetics of NFκB activation, as measured by its ability to bind to DNA (data not shown). Again, no such degradation of IκBα was observed in P2.1 cells at 1 h (Fig. 1B) or later time points (data not shown). In contrast, there was no change in the level of IκBβ in either cell line in response to dsRNA (data not shown).
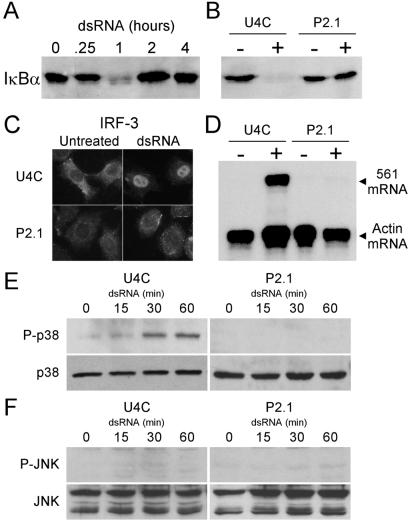
Figure 1.
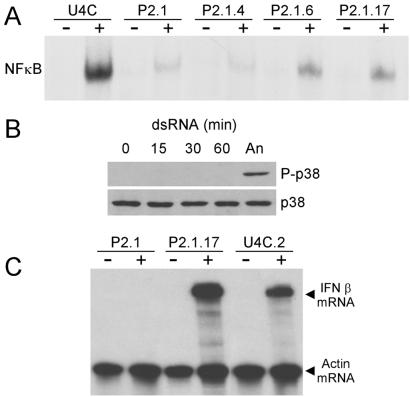
P2.1 cells display multiple dsRNA-mediated signaling defects. (A) Kinetics of degradation of IκBα in U4C cells. U4C cells were treated with dsRNA for the times indicated. Cell lysates were prepared, and Western analysis was performed with an antibody against IκBα. (B) Lack of IκBα degradation in P2.1 cells. U4C and P2.1 cells were left untreated (−) or treated with dsRNA for 1 h (+). Cell lysates were prepared, and Western analysis for IκBα was performed as described for A. (C) Failure of dsRNA-mediated IRF-3 translocation in P2.1 cells. U4C and P2.1 cells were left untreated or treated with dsRNA for 1 h. The cells were fixed, and immunofluorescence was performed to detect the nuclear translocation of IRF-3 in response to dsRNA. (D) Lack of induction of 561 mRNA in response to dsRNA in P2.1 cells. U4C and P2.1 cells were left untreated (−) or treated with dsRNA for 6 h (+). RNA was harvested, and an RPA was performed to detect the induction of 561 message. Actin mRNA served as an internal control. (E) Failure of p38 activation by dsRNA in P2.1 cells. U4C and P2.1 cells were treated with dsRNA for the times indicated. Cell lysates were prepared, and Western analyses were performed with antibodies against p38 or activated, phosphorylated p38 (P-p38). (F) Lack of JNK activation by dsRNA in U4C and P2.1 cells. U4C and P2.1 cells were treated with dsRNA for the times indicated. Cell lysates were prepared, and Western analyses were performed with antibodies against JNK or activated, phosphorylated JNK (P-JNK).
The IRF-3 transcription factor is also known to be activated by dsRNA (9, 29). As measured by its nuclear translocation, IRF-3 was activated after dsRNA treatment of U4C but not P2.1 cells (Fig. 1C). The lack of IRF-3 activation in P2.1 cells was reflected in their failure to induce 561 mRNA (Fig. 1D). Three protein kinases, PKR, p38, and JNK, are activated in dsRNA-treated cells (14, 15). We reported previously that PKR is activated in both U4C and P2.1 cells after dsRNA treatment (19). In contrast, p38 was phosphorylated only in U4C cells (Fig. 1E), thus revealing another difference in dsRNA-dependent signaling between the two cell lines. In P2.1 cells, the activation of p38 was not intrinsically defective, because it still could be activated by anisomycin (data not shown). Neither JNK-1 nor JNK-2 was activated by dsRNA treatment in either cell line (Fig. 1F), although both were activated by anisomycin (data not shown).
The above results showed that among the three kinases known to be activated by dsRNA, JNK is not activated in either line, p38 is activated in U4C but not P2.1 cells, and PKR is activated in both. At the level of transcription factor activation, IκB was not degraded in response to dsRNA treatment of P2.1 cells, and consequently NFκB was not released. Similarly, dsRNA failed to activate IRF-3 in P2.1 cells, and thus the transcription of IRF-driven genes such as 561 was not induced.
The Nature of the IRF-3 Defect.
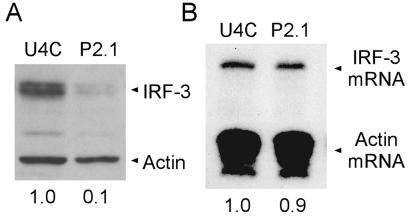
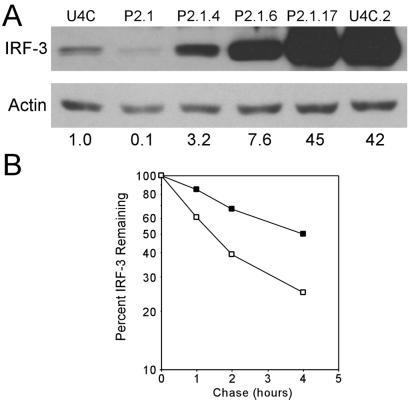
The lack of IRF-3 activation in P2.1 cells (Fig. 1C) probably was because of a 10-fold lower cellular level of IRF-3 in P2.1 cells as compared with U4C cells (Fig. 2A). However, this large difference was not reflected at the levels of IRF-3 mRNA, because both cell lines expressed similar mRNA levels (Fig. 2B). Thus, either IRF-3 synthesis or its turnover is defective in P2.1 cells. Pulse-labeling of newly synthesized proteins followed by immunoprecipitation of radiolabeled IRF-3 did not reveal any major difference in the rate of IRF-3 synthesis (data not shown). In contrast, measuring IRF-3 turnover by Western analysis of extracts of cells in which new protein synthesis had been blocked by cycloheximide treatment indicated enhanced degradation of IRF-3 in P2.1 cells (data not shown). However, it was difficult to compare accurately the turnover rates of IRF-3 in the two cell lines because of the low level of this protein in P2.1 cells (Fig. 2A). To circumvent this problem, cell lines expressing high levels of IRF-3 were established. P2.1.4 expressed about three times more IRF-3 than U4C cells, whereas P2.1.6 expressed about eight times more. In the P2.1.17 and U4C.2 lines, about 40 times more IRF-3 was expressed as compared with U4C cells (Fig. 3A).
Figure 2.
The level of IRF-3 protein but not mRNA is lower in mutant P2.1 cells. (A) The IRF-3 and actin protein levels in U4C and P2.1 cells were determined by Western analysis. The relative levels of IRF-3 in U4C and P2.1 cells, normalized against the actin levels, are shown beneath the figure. (B) IRF-3 and actin mRNA levels were determined by using RPA and quantified by using a Molecular Dynamics PhosphorImager. The relative normalized levels of IRF-3 mRNA are denoted beneath the figure.
Figure 3.
The degradation rate of IRF-3 is increased in P2.1 cells. (A) Establishment of U4C and P2.1 cell lines expressing IRF-3. P2.1 and U4C cells were transfected with an IRF-3 expression plasmid, and individual clones were isolated. Western analysis was performed to determine IRF-3 and actin expression levels. The normalized IRF-3 expression levels relative to that in U4C cells are given below the lanes. (B) Determination of IRF-3 half-life. U4C.2 (■) and P2.1.17 (□) cells were pulse-labeled with a mixture of 35S-labeled methionine and cysteine for 2 h. The label then was chased for the indicated times, and cell lysates were prepared. IRF-3 was immunoprecipitated from equal amounts of whole-cell extract and separated by SDS/PAGE. Radiolabeled IRF-3 was quantified with a Molecular Dynamics PhosphorImager.
Clones U4C.2 and P2.1.17, which express similar steady-state levels of IRF-3, were used to determine the turnover rates of the protein in the two lines. Cells were pulse-labeled with a mixture of [35S] methionine and cysteine, and the label was chased with excess unlabeled amino acids for various lengths of time. IRF-3 immunoprecipitated from cell extracts was analyzed by gel electrophoresis, and its radioactivity was determined. IRF-3 turned over much more rapidly in P2.1 cells (Fig. 3B). The half-life of the protein was ≈1.5 h in P2.1 cells and ≈4 h in U4C cells. These results suggest that in the parental P2.1 cells, IRF-3 turns over more rapidly than in U4C cells. This turnover was not caused by any change in the primary structure of IRF-3, because IRF-3 cDNA cloned from P2.1 cells by reverse transcription–PCR had the same sequence as the corresponding cDNA from U4C cells (data not shown).
Selective Restoration of dsRNA-Signaling Pathways.
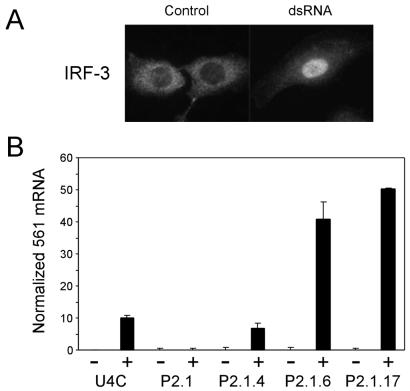
In the next series of experiments, we examined the status of dsRNA-dependent signaling in the IRF-3-expressing clonal derivates of P2.1 cells. In these clones, dsRNA can activate IRF-3, as monitored by its nuclear translocation (Fig. 4A). Consequently, the induction of 561 mRNA was restored as well (Fig. 4B). 561 mRNA was not detected in any clones before dsRNA treatment, although IRF-3 was highly overexpressed in some of them. However, dsRNA treatment led to 561 mRNA induction in all three clones tested. The levels of induction in P2.1.6 and P2.1.17 clones were similar, but the level was lower in P2.1.4 cells. These results demonstrate that the expression of exogenous IRF-3 restores IRF-3-dependent signaling in P2.1 cells.
Figure 4.
Expression of IRF-3 in P2.1 cells restores IRF-3 activation and 561 induction. (A) Restoration of dsRNA-induced activation of IRF-3. P2.1.6 cells were treated with dsRNA and stained for IRF-3 immunofluorescence as described for Fig. 1C. (B) Restoration of 561 mRNA induction by dsRNA in IRF-3-expressing cells. Cells were left untreated (−) or treated with dsRNA for 6 h (+) and examined for the induction of 561 mRNA by RPA as described for Fig. 1D. The normalized 561 mRNA level in treated U4C cells was set to 10. The results of three separate experiments were averaged, and the graph represents the mean ± SD.
In contrast to IRF-3-mediated gene induction, the defects in other dsRNA-dependent signals persisted in P2.1 clones overexpressing IRF-3. NFκB activation by dsRNA, as measured by DNA-binding activity (Fig. 5A), was not restored to the level present in U4C cells, although there was a hint of activation in the P2.1.6 and P2.1.17 clones. However, examination of NFκB activation by nuclear translocation showed none in P2.1.6 and P2.1.17 cells (data not shown). Similarly, p38 was not activated by dsRNA in P2.1.17 cells (Fig. 5B). These results demonstrate that restoration of dsRNA-dependent signaling pathways was partial and selective in the P2.1-derived clones. Although IRF-3-mediated gene induction was restored, the other defects of the parental P2.1 cells persisted.
Figure 5.
Expression of IRF-3 does not restore other signaling defects of P2.1 cells but restores IFN-β mRNA induction. (A) dsRNA-induced activation of NFκB. U4C, P2.1, and the P2.1 clones expressing IRF-3 cells were left untreated (−) or treated with dsRNA for 60 min (+). An EMSA for NFκB was performed. (B) Lack of p38 activation. P2.1.17 cells were treated with dsRNA for the indicated times or anisomycin (An) for 15 min. Activation of p38 was determined by Western analysis as described for Fig. 1E. (C) Induction of IFN-β mRNA by dsRNA treatment. U4C, P2.1, and P2.1.17 cells were treated with dsRNA for 6 h (+) or left untreated (−). RNA was harvested, and an RPA was performed with probes for IFN-β and actin mRNAs.
IRF-3-Mediated Induction of IFN-β mRNA.
In contrast to the simple promoter of the 561 gene, containing only ISRE sites, the promoter of the human IFN-β gene is complex, and multiple transcription factors are needed for its activation (3, 8, 9, 15, 30, 31). Thus, it was surprising when we observed strong induction of IFN-β mRNA in response to dsRNA treatment of P2.1.17 cells (Fig. 5C). About half this level of induction was observed in P2.1.6 cells, which express a much lower level of IRF-3, whereas no induction was detectable in the P2.1.4 cells (data not shown). As expected, in the parental U4C.2 cells expressing a comparably high level of IRF-3, there was strong induction of IFN-β, and in neither cell line was IFN-β mRNA constitutively expressed. These results indicate that, similar to the induction of 561 mRNA, IRF-3 activation alone may drive the induction of IFN-β mRNA.
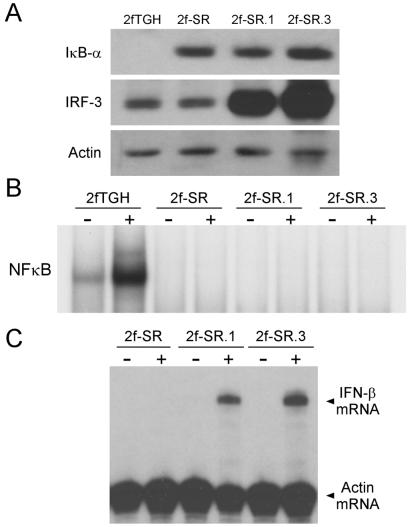
The conclusion that activation of a high level of IRF-3 by dsRNA may obviate the need for NFκB, p38, and JNK activation for IFN-β gene induction was confirmed in the experiment shown in Fig. 6. Because there was a slight activation of NFκB in the P2.1.17 cells (Fig. 5A), we established new cell lines in which NFκB activation was blocked completely. The parental 2fTGH cells, from which the U4C and P2.1 cells were derived, were used for this experiment. A new cell line, 2f-SR, expressing a high level of the IκBαSR, was established (Fig. 6A). Two other cell lines derived from the 2f-SR cells, 2f-SR.1 and 2f-SR.3, express different levels of transfected IRF-3 in addition to the IκBαSR (Fig. 6A). As expected, dsRNA activated NFκB in the parental 2fTGH cells but not in its super repressor-expressing derivatives (Fig. 6B). In both clones overexpressing IRF-3, dsRNA induced IFN-β mRNA efficiently in the complete absence of NFκB activation (Fig. 6C). Furthermore, pretreatment of the 2f-SR.3 cells with the p38 MAP kinase inhibitor (Calbiochem) did not block IFN-β induction by dsRNA (data not shown). Because JNK is not activated by dsRNA in these cells, we conclude that the activation of NFκB, p38, and JNK are dispensable for dsRNA-mediated induction of IFN-β mRNA in cells overexpressing IRF-3.
Figure 6.
NFκB activation is not required for the induction of IFN-β mRNA by dsRNA. (A) Western analysis of IκBαSR, IRF-3, and actin expression. 2fTGH cell lines expressing the IκBαSR and IRF-3 were established as described in Materials and Methods. Western analyses of the clones were performed with appropriate antibodies. (B) NFκB activation in response to dsRNA treatment. Cells were treated with dsRNA for 1 h (+) or left untreated (−). EMSA analysis of cell extracts was performed with the NFκB probe. (C) Induction of IFN-β mRNA after dsRNA treatment. Cells were treated with dsRNA for 6 h (+) or left untreated (−). RNA was harvested, and an RPA was performed with probes against IFN-β and actin mRNAs.
Discussion
In the current study, we have continued to investigate the nature of the defects of P2.1 cells in dsRNA-dependent signaling. As noted previously (19), transcription of neither the IFN-β gene nor the 561 gene was induced by dsRNA in these cells. The defects in gene induction were attributed to the lack of activation of the requisite transcription factors IRF-3 and NFκB in response to dsRNA treatment. IRF-3 activation in response to virus infection is mediated by extensive phosphorylation, which can be monitored because the phosphorylated form migrates more slowly during electrophoresis in a denaturing gel (8, 9, 32). In contrast, the activation of IRF-3 by dsRNA does not cause such a marked change in its electrophoretic mobility, probably because it is phosphorylated at fewer or different sites (K.L.P. and G.C.S., unpublished data). Nonetheless, IRF-3, activated by either dsRNA or virus infection, translocates from the cytoplasm to the nucleus (25, 33). Using nuclear translocation as an index of activation, we failed to detect activated IRF-3 in dsRNA-treated P2.1 cells. Further investigation revealed that this failure was caused by a low cellular level of IRF-3. Because restoration of IRF-3 to normal or higher levels by ectopic expression resulted in robust activation of IRF-3 by dsRNA, it seems that the dsRNA-elicited IRF-3 activation machinery is not defective in P2.1 cells. In contrast to the low level of IRF-3 protein, the level of IRF-3 mRNA was comparable in P2.1 and parental U4C cells. The mRNA also was translated equally well in the two cell lines, but the protein turned over more rapidly in P2.1 cells. To obtain quantitative values of the turnover rates of IRF-3 in the two lines, we had to use overexpressing lines to increase the quantities of radiolabeled protein. For this reason, the observed difference of a 2.7-fold shorter half-life in P2.1 cells (Fig. 3) could be an underestimate. At the normal level of IRF-3 synthesis in U4C and P2.1 cells, over 40-fold lower than those in P2.1.17 and U4C.2 cells (Fig. 3), the difference in the turnover rates could be higher, thus accounting for the 10-fold difference in the steady-state levels of the protein (Fig. 2). It seems that the enhanced degradation of IRF-3 in P2.1 cells is not caused by enhanced susceptibility to proteolysis because of any change in its primary structure.
Unlike IRF-3, NFκB and IκB were present at normal levels in P2.1 cells, and the pathway could be activated efficiently by tumor necrosis factor-α (19), indicating that the IκB kinase complex is functional in these cells. There was no nuclear translocation of NFκB and little DNA binding activity in extracts of dsRNA-treated P2.1 cells, suggesting that it had not been released from the IκB complex. The lack of NFκB release was caused by a lack of dsRNA-elicited degradation of IκBα. Thus, the defect in this pathway of signal transduction by dsRNA lies upstream of IκB degradation but downstream of PKR activation. A third transcriptional pathway activated by dsRNA is mediated by the transcription factor c- Jun/ATF-2 of the AP-1 family. Activation of this pathway in mouse embryo fibroblasts requires the protein kinase JNK2, which gets phosphorylated in dsRNA-treated cells in a PKR-independent manner (15). However, in the U4C and P2.1 cells used in our study, dsRNA did not activate either JNK1 or JNK 2, indicating that their activation could be cell type-dependent. dsRNA is known to activate another stress-activated protein kinase, p38 (14). Unlike JNK, p38 activation by dsRNA depends on PKR (27). We observed that PKR-mediated p38 activation by dsRNA was also defective in P2.1 cells, although other stresses such as inhibitors of protein synthesis could activate it. These observations suggest that the dsRNA-signaling defect in P2.1 cells lies upstream of p38 and IκB kinase activation, although PKR is activated normally. Activation of at least one other protein kinase that leads to dsRNA-mediated IRF-3 phosphorylation must be unaffected in these cells as well. With the available information, it is not apparent how mutation in a single gene in P2.1 cells could cause the observed multiple defects in dsRNA-dependent signaling. The lack of activation of IκB kinase and p38 may be connected at the level of an upstream kinase, the activation of which by a PKR-mediated pathway may be defective in P2.1 cells. Although how this defect leads to IRF-3 destabilization in P2.1 cells remains an open question, it is clear that excess IRF-3 supplied by ectopic expression can be activated efficiently by dsRNA. It is also evident that the low IRF-3 level is only a downstream defect resulting from the central lesion in P2.1 cells, because overexpression of IRF-3 restores only the IRF-3-signaling pathway without repairing other defects. Our study also formally established that activation of NFκB, p38, or JNK2 by dsRNA is not necessary for the activation of IRF-3. This conclusion is in tune with the results obtained by others (13, 34, 35) regarding the distinct properties of the as-yet-unidentified protein kinase responsible for activating IRF-3.
Using 561 as a prototype IFN-stimulated gene, the transcription of which is stimulated also by dsRNA, we have shown previously that the ISRE is the responsive cis element in its promoter for both inducers (16). However, the signal transduction pathways are completely different, and the dsRNA-mediated induction does not require all the proteins needed for type I IFN-dependent signaling. The current study identifies IRF-3 as the crucial transcription factor for 561 gene induction by dsRNA; the restoration of IRF-3 restored 561 gene induction in P2.1-derived clones. Within a range, expression of a higher level of IRF-3 caused higher induction of 561 mRNA, indicating that the level of IRF-3 could be the limiting component in the signaling pathway. The above observation was true not only for the P2.1 cells but also for the U4C clones overexpressing IRF-3 (data not shown). The critical role of IRF-3 in ISRE-mediated signal transduction by dsRNA has been demonstrated by others, who have identified IRF-3 as a component of the corresponding ISRE-binding, dsRNA-activated, or virus-activated transacting factors (6, 8, 9, 29).
Restoration of the induction of IFN-β mRNA in P2.1.17 cells was unexpected. As the NFκB, the JNK, and the p38 pathways of dsRNA signaling remained defective in those cells (Fig. 5), this result indicated that these pathways are not essential for induction of the IFN-β gene if ample IRF-3 is available. This conclusion was confirmed in cells expressing the IκB super repressor. These observations compel us to modify the current model for human IFN-β gene induction, built on extensive studies by many laboratories (3, 31). Transcription of this gene requires the assembly of an enhanceosome on its promoter. The enhancer contains three partially overlapping positive regulatory elements to which NFκB, IRFs, and the c-Jun/ATF-2 transcription factors bind. Each of these factors is activated in cells in response to dsRNA or virus infection. In addition to the activated factors, the high mobility group protein HMG I(Y) binds to four sites within the IFN-β enhancer and unbends the DNA, thus facilitating enhanceosome formation (36). Previous studies have shown that inducers that can activate the individual transcription factors fail to induce transcription of the IFN-β gene, probably because of the need of the other factors for proper enhanceosome formation, although in transient transfection assays IFN-β-promoter driven reporters are induced partially (31).
Similarly, inducible expression of a constitutively active IRF-3 mutant did not cause IFN-β gene induction, indicating additional need for this process (37). However, the results presented here provide evidence suggesting that the assembly of the multicomponent enhanceosome may not be necessary for IFN-β gene transcription if one of the relevant transcription factors is available in excess. It was known that the individual cis elements, in the absence of the others, can drive dsRNA-induced transcription of transfected reporter genes, indicating that each element is capable of communicating with the basal transcription machinery and the coactivators (3). Our data show that the same is true for the complex promoter of the endogenous IFN-β gene in cells overexpressing IRF-3. It is conceivable that in cells containing limiting amounts of the transcription factors, none of the factors by themselves can bind stably to the cognate cis elements, but the coordinate assembly of the enhanceosome stabilizes binding of all factors to DNA and facilitates their interactions with the transcriptional machinery. In contrast, the same result could be achieved in IRF-3-overexpressing cells by promoting and stabilizing IRF-3/IFN-response element interaction, thus increasing the local concentrations of activated IRF-3. Therefore, the complex promoter of the IFN-β gene was functionally converted to a simple dsRNA-responsive ISRE-driven promoter similar to the ones present in IFN-stimulated genes such as 561.
Acknowledgments
We are grateful to Nywana Sizemore, Theresa Rowe, and David Wald for cell lines. We thank Peter Howley, Lucienne Ronco, Michael David, and Paula Pitha-Rowe for the gifts of reagents. This work was supported by National Institutes of Health Grants CA62220 and CA68782. K.L.P. is the recipient of a Cleveland Clinic Foundation Crile Fellowship.
Abbreviations
- ds
double-stranded
- IRF
IFN regulatory factor
- PKR
dsRNA-dependent protein kinase
- JNK
c-Jun N-terminal kinase
- EMSA
electrophoretic mobility-shift assay
- ISRE
IFN-stimulated response element
- IκBαSR
IκBα super repressor
- RPA
ribonuclease protection assay
References
- 1.Sen G C. Annu Rev Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 2.Geiss G, Jin G, Guo J, Bumgarner R, Katze M G, Sen G C. J Biol Chem. 2001;276:30178–30182. doi: 10.1074/jbc.c100137200. [DOI] [PubMed] [Google Scholar]
- 3.Maniatis T, Falvo J V, Kim T H, Kim T K, Lin C H, Parekh B S, Wathelet M G. Cold Spring Harbor Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi T. Annu Rev Immunol. 1988;6:439–464. doi: 10.1146/annurev.iy.06.040188.002255. [DOI] [PubMed] [Google Scholar]
- 5.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 6.Bovolenta C, Lou J, Kanno Y, Park B K, Thornton A M, Coligan J E, Schubert M, Ozato K. J Virol. 1995;69:4173–4181. doi: 10.1128/jvi.69.7.4173-4181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Au W C, Moore P A, Lowther W, Juang Y T, Pitha P M. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 9.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zamanian-Daryoush M, Mogensen T H, DiDonato J A, Williams B R. Mol Cell Biol. 2000;20:1278–1290. doi: 10.1128/mcb.20.4.1278-1290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin R, Genin P, Mamane Y, Hiscott J. Mol Cell Biol. 2000;20:6342–6353. doi: 10.1128/mcb.20.17.6342-6353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwamura T, Yoneyama M, Yamaguchi K, Suhara W, Mori W, Shiota K, Okabe Y, Namiki H, Fujita T. Genes Cells. 2001;6:375–388. doi: 10.1046/j.1365-2443.2001.00426.x. [DOI] [PubMed] [Google Scholar]
- 14.Iordanov M S, Paranjape J M, Zhou A, Wong J, Williams B R, Meurs E F, Silverman R H, Magun B E. Mol Cell Biol. 2000;20:617–627. doi: 10.1128/mcb.20.2.617-627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu W M, Ostertag D, Li Z W, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 16.Bandyopadhyay S K, Leonard G T, Jr, Bandyopadhyay T, Stark G R, Sen G C. J Biol Chem. 1995;270:19624–19629. doi: 10.1074/jbc.270.33.19624. [DOI] [PubMed] [Google Scholar]
- 17.Der S D, Zhou A, Williams B R, Silverman R H. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo J, Hui D J, Merrick W C, Sen G C. EMBO J. 2000;19:6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leaman D W, Salvekar A, Patel R, Sen G C, Stark G R. Proc Natl Acad Sci USA. 1998;95:9442–9447. doi: 10.1073/pnas.95.16.9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pellegrini S, John J, Shearer M, Kerr I M, Stark G R. Mol Cell Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 22.Kessler S P, Rowe T M, Blendy J A, Erickson R P, Sen G C. J Biol Chem. 1998;273:9971–9975. doi: 10.1074/jbc.273.16.9971. [DOI] [PubMed] [Google Scholar]
- 23.Enoch T, Zinn K, Maniatis T. Mol Cell Biol. 1986;6:801–810. doi: 10.1128/mcb.6.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sizemore N, Leung S, Stark G R. Mol Cell Biol. 1999;19:4798–4805. doi: 10.1128/mcb.19.7.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J, Peters K L, Sen G C. Virology. 2000;267:209–219. doi: 10.1006/viro.1999.0135. [DOI] [PubMed] [Google Scholar]
- 26.Navarro L, David M. J Biol Chem. 1999;274:35535–35538. doi: 10.1074/jbc.274.50.35535. [DOI] [PubMed] [Google Scholar]
- 27.Goh K C, deVeer M J, Williams B R. EMBO J. 2000;19:4292–4297. doi: 10.1093/emboj/19.16.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronco L V, Karpova A Y, Vidal M, Howley P M. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weaver B K, Kumar K P, Reich N C. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 31.Munshi N, Yie Y, Merika M, Senger K, Lomvardas S, Agalioti T, Thanos D. Cold Spring Harbor Symp Quant Biol. 1999;64:149–159. doi: 10.1101/sqb.1999.64.149. [DOI] [PubMed] [Google Scholar]
- 32.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar K P, McBride K M, Weaver B K, Dingwall C, Reich N C. Mol Cell Biol. 2000;20:4159–4168. doi: 10.1128/mcb.20.11.4159-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Servant M J, ten Oever B, LePage C, Conti L, Gessani S, Julkunen I, Lin R, Hiscott J. J Biol Chem. 2001;276:355–363. doi: 10.1074/jbc.M007790200. [DOI] [PubMed] [Google Scholar]
- 35.Smith E J, Marie I, Prakash A, Garcia-Sastre A, Levy D E. J Biol Chem. 2001;276:8951–8957. doi: 10.1074/jbc.M008717200. [DOI] [PubMed] [Google Scholar]
- 36.Falvo J V, Thanos D, Maniatis T. Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 37.Heylbroeck C, Balachandran S, Servant M J, DeLuca C, Barber G N, Lin R, Hiscott J. J Virol. 2000;74:3781–3792. doi: 10.1128/jvi.74.8.3781-3792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]