Abstract
Kallmann syndrome is a neurological disorder characterized by various behavioral and neuroanatomical defects. The X-linked form of this disease is caused by mutations in the KAL-1 gene, which codes for a secreted molecule that is expressed in restricted regions of the brain. Its molecular mechanism of action has thus far remained largely elusive. We show here that expression of the Caenorhabditis elegans homolog of KAL-1 in selected sensory and interneuron classes causes a highly penetrant, dosage-dependent, and cell autonomous axon-branching phenotype. In a different cellular context, heterologous C. elegans kal-1 expression causes a highly penetrant axon-misrouting phenotype. The axon-branching and -misrouting activities require different domains of the KAL-1 protein. In a genetic modifier screen we isolated several loci that either suppress or enhance the kal-1-induced axonal defects, one of which codes for an enzyme that modifies specific residues in heparan sulfate proteoglycans, namely heparan-6O-sulfotransferase. We hypothesize that KAL-1 binds by means of a heparan sulfate proteoglycan to its cognate receptor or other extracellular cues to induce axonal branching and axon misrouting.
Kallmann syndrome is a genetic disease that was first recognized as such in 1944 by Franz Kallmann (1) and is defined as the association of hypogonadotropic hypogonadism and anosmia, i.e., the inability to smell. In addition, affected patients may display a variety of neurological abnormalities, including synkinesia (mirror movement) of the hands, as well as somatic defects such as unilateral renal agenesis (2, 3). Embryological studies suggested a common origin for the anosomic and hypogonadotropic phenotypes (4). During development, the olfactory receptor neurons born in the olfactory primordium project and extend their axons through the cribriform plate into the skull where they establish connections with secondary neurons in the olfactory bulb. Likewise, the neurons that later secrete gonadotropin-releasing hormone (GnRH) are born in the olfactory primordium and migrate along the olfactory tract to their final destination in the hypothalamus. Based on neuropathological studies on a human fetus affected by X chromosome-linked Kallman syndrome (5), it has been suggested that (i) the axons of the olfactory receptor neurons still project toward the forebrain; however, they fail to establish connections with their target cells), and (ii) GnRH-synthesizing neurons fail to reach the hypothalamus, thus disrupting the hypothalamic-pituitary hormonal axis and causing hypogonadotropic hypogonadism.
Genetically, Kallmann syndrome is heterogeneous with examples of autosomal recessive, autosomal dominant, as well as X-linked recessive modes of inheritance [see Online Mendelian Inheritance in Man (OMIM) at http://www.ncbi.nlm.nih.gov/omim/]. Although the molecular lesions responsible for the autosomal forms remain elusive, the gene for the X-linked form, named KAL-1, was identified more then 10 years ago (6, 7) and shown to code for a secreted protein with similarities to neural cell adhesion molecules. Expression of KAL-1 was observed in the target area of the axonal projections of the olfactory neurons in human fetal brain (8), and the protein was found to be associated with basement membranes and the interstitial matrix in neuronal tissues of the embryonic brain, e.g., in the olfactory bulb (9). A series of in vitro experiments revealed that recombinant KAL-1 can induce neurite outgrowth in cultured cerebellar mouse neurons but not in hippocampal or dorsal root ganglia neurons (10). The in vivo significance of these studies has remained unclear, however. The lack of a genetically tractable animal model to study KAL-1 prompted us to investigate the Caenorhabditis elegans ortholog of the human gene.
Materials and Methods
DNA Constructs and Transgenic Lines.
The kal-1∷gfp reporter gene construct contains a 5,270-bp fragment upstream of the ATG start codon of kal-1 that was amplified by PCR and subcloned into the gfp vector pPD95.75 (a gift from A. Fire, Carnegie Institute). C. elegans kal-1 and Drosophila kal-1 cDNAs were obtained from the respective expressed sequence tag (EST) sequencing consortia and completely sequenced (C. elegans EST: yk230c3; GenBank accession no. C40392; Drosophila EST: GH04611; GenBank accession no. AY060623). hst-6 cDNAs (GenBank accession no. AY081844) were isolated by PCR and 5′/3′ RACE from embryonic cDNA libraries. The mis/overexpression constructs, as well as construction of transgenic lines and chromosomal integration, are described in the supporting methods, which is published on the PNAS web site, www.pnas.org. Briefly, otIs35X, otIs76IV, and otIs77II express pttx-3∷kal-1; otIs78, otIs79, otIs80IV, and otIs81II express punc-119∷kal-1; otIs124 expresses pttx-3∷kal-1mWAP; and otIs83V expresses pgcy-8∷kal-1.
Scoring Neuroanatomy.
Transgenic gfp lines used to score neuroanatomy as well as detailed scoring criteria for axonal branches are described in the supporting methods.
Modifier Screen and Mapping of Modifier Mutants.
Modifier mutants were isolated from a clonal F1 screen of 1,310 haploid genomes by using ethyl methanesulfonate (EMS) as a mutagen (see supporting methods). ot17 was placed by three factor-mapping between lon-2 and unc-97 on LGX and further mapped between two newly identified polymorphic markers, otP3 and otP5. ot16 was placed between unc-79 and emb-5 on LGIII, whereas ot20 and ot21 were mapped close to unc-62 and to the right of stP3 on LGV, respectively.
Cell-Binding Assays.
Assays were done as described previously (see supporting methods for details).
Results and Discussion
The Kallmann Syndrome Gene kal-1 in Invertebrates.
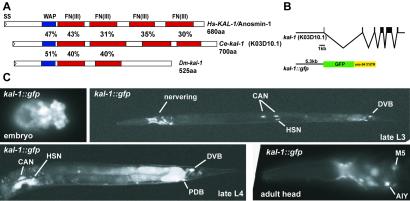
The completed genome sequences of C. elegans (11) and Drosophila melanogaster (12) allowed us to identify single orthologs of the human Kallmann syndrome gene in both organisms. By sequencing a full-length EST clone we found that the C. elegans gene consists of six exons and codes for a putative secreted protein of 700 amino acids that shares 36% overall similarity with its human ortholog (Fig. 1A). Like its human counterpart, it contains a WAP-type protease inhibitor domain followed by four consecutive FnIII motifs. No other homologs with a similar domain composition exist in C. elegans. Likewise, we sequenced a putative full-length EST clone representative of the only predicted KAL-1-like gene in Drosophila and found it to encode a protein that is smaller than its worm and vertebrate homologs, lacking the last two FnIII motifs (Fig. 1A). From here on, “kal-1 ” refers to the C. elegans ortholog of the human gene and “KAL-1” refers to the C. elegans protein, if not explicitly noted otherwise.
Figure 1.
The Kallmann syndrome gene kal-1 in C. elegans. (A) Schematic domain structure of the Kallmann syndrome proteins in humans (Hs), worms (Ce), and flies (Dm). Pairwise similarities of domains between proteins were calculated by aligning the domains as predicted by SMART. WAP, whey acidic protein domain; Fn(III), fibronectin III motif; SS, signal sequence. (B) Schematic drawing of the gene structure of C. elegans kal-1 (located on cosmid K03D10) and the transcriptional reporter fusion with green fluorescent protein (gfp) and the unc-54 3′ untranslated region. (C) Expression pattern of the kal-1∷gfp reporter construct in transgenic animals (otIs33IV).
A kal-1∷gfp Reporter Is Expressed in Neuronal and Nonneuronal Tissues in C. elegans.
To characterize potential sites of expression of the C. elegans kal-1 gene, we constructed a transcriptional reporter that fuses the gene encoding green fluorescent protein (gfp) to 5.3 kb of potential 5′ upstream regulatory sequence of kal-1 (Fig. 1B). Expression of the reporter is first detected in embryos at around the 50-cell stage in 2–3 cells (not shown) and widens during embryonic development (Fig. 1C). At the comma stage, expression is seen in a set of cells whose position is consistent with neuroblasts in the tail as well as in the head where they form a ring-like structure (Fig. 1C). In larval stages and throughout adult stages, expression is largely restricted to a set of neurons in several head ganglia (AIY, AIZ, RID, M5, ASI, and subsets of labial sensory neurons), motor neurons in the ventral nerve cord, neurons in the midbody region (HSN, CAN, and PVM) and in the tail ganglia (DVB, DVC, and PDB) (Fig. 1C). In the AIY interneurons, kal-1 is a downstream target of the ttx-3 transcription factor (13). Consistent nonneuronal expression could be observed in the excretory cell and uterine cells (Fig. 1C). Rat polyclonal antisera directed against portions of the worm KAL-1 protein were found to recognize KAL-1 in transgenic animals that overexpress KAL-1 in specific cells (Fig. 2A and Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org), but thus far failed to reveal endogenous KAL-1 protein, possibly because of limited levels of expression of the endogenous KAL-1 protein. In KAL-1-overexpressing animals, the KAL-1 protein remains associated with the surface of the overexpressing cell (see Fig. 6A) that is consistent with the reported cell surface-binding activity of vertebrate KAL-1 protein (9, 14) and consistent with the cell autonomous activity of the KAL-1 protein that we describe below.
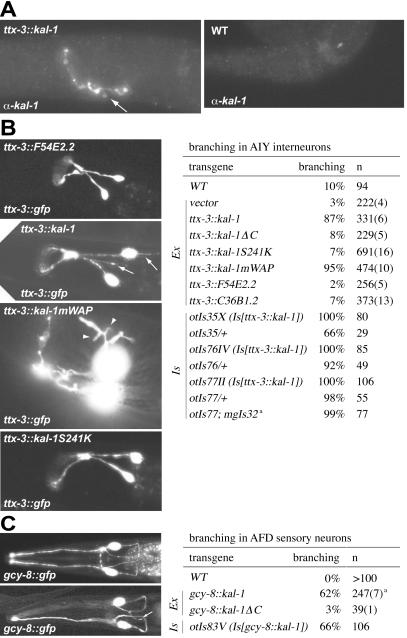
Figure 2.
Cell-specific expression of kal-1 causes axon branching. (A) Anti-KAL-1 antibody staining of transgenic animals that overexpress kal-1 in the AIY interneuron class (otIs35) and in wild-type N2 animals. Arrows point to axonal branches induced by KAL-1 protein (the unbranched nature of the AIY interneurons can be observed with gfp labeling; see B). See supporting information for more comments, micrographs, and technical notes on antibody staining. (B and C) Data were obtained from extrachromosomal (Ex) and integrated (Is) mis/overexpressing lines as indicated. Numbers in parentheses give the number of extrachromosomal lines. The number of defective animals of all extrachromosomal lines were added up to yield percentages; it is specifically noted if the spread of percentages of individual lines was significant. (B) Overexpression phenotype in AIY interneurons. Scoring criteria for the branching defects are detailed in the supporting methods. White arrows point to axonal branches and white arrowheads show arborized branches in lines overexpressing the kal-1mWAP construct. All experiments were performed in a mgIs18 (Is[ttx-3∷gfp]) gfp reporter background unless indicated otherwise. kal-1ΔC and kal-1S241K recapitulate mutations that were found in patients with Kallmann syndrome (18). kal-1mWAP contains two point mutations, C134S and C135S, that abolish two integral disulfide bonds in the WAP domain. Note that the branching effect is unrelated to the presence of gfp in the cell because KAL-1 antibody staining shows similar branches (A). a, results with a different Is[ttx-3∷gfp], termed mgIs32, are virtually identical. (C) Mis/overexpression phenotypes in AFD sensory neurons. A white arrow points to an axonal branch at the ventral portion of the main axon. All experiments were performed in an oyIs17 (Is[gcy-8∷gfp]) reporter background. a, results varied between 48% and 80% per line.
Forced Expression of KAL-1 Causes a Range of Specific Anatomical Defects.
We first attempted to examine the consequence of loss of kal-1 gene activity. The unavailability of kal-1 mutant animals prompted us to conduct RNA interference (RNAi) experiments (15). We observed no significant effect on overall appearance, morphology or neuroanatomy after injection of double-stranded RNA targeted against the kal-1 gene into wild-type animals (see supporting methods). This observation has to be taken with caution, however, because RNAi has been found to work less effectively in the nervous system (16). To functionally study kal-1 in C. elegans we thus used a mis/overexpression (“forced expression”) approach that was not only intended to observe the range of effects that kal-1 can exert but also to establish a gain-of-function (gf) phenotype that would allow us to conduct a genetic screen for interacting loci. To this end, we created transgenic lines that express the C. elegans kal-1 cDNA under the control of various promoters (including several different sensory neuron-, interneuron-, and motorneuron-specific promoters, a muscle-specific promoter, and a panneuronal promoter) and assessed the effects of such mis/overexpression with more than 10 gfp reporter strains that label the cells that heterologously express kal-1 and/or cells that are in proximity to the kal-1-expressing cells (see Table 2, which is published as supporting information on the PNAS web site). Below we describe the most penetrant effects observed after forced expression of kal-1.
Neuron-Specific Expression of kal-1 Causes Axonal-Branching Defects.
Expression of kal-1 in the AIY interneuron class under the control of an AIY-specific promoter fragment of the ttx-3 gene (17) led to a highly penetrant axon-branching defect (Fig. 2 A–C also see Fig. 4B). These branches emanate from variable positions along the main axon and are variable in length (see supporting methods). Like axon collaterals in vertebrate nervous systems, these branches are delayed in appearance relative to the outgrowth of the primary axon, because only 6% (n = 50) of kal-1-overexpressing animals show branches in late, pretzel-stage embryos (wild type = 0%, n = 48) whereas 100% (n = 80) of adult animals contain axonal branches (wild type = 10%, n = 94).
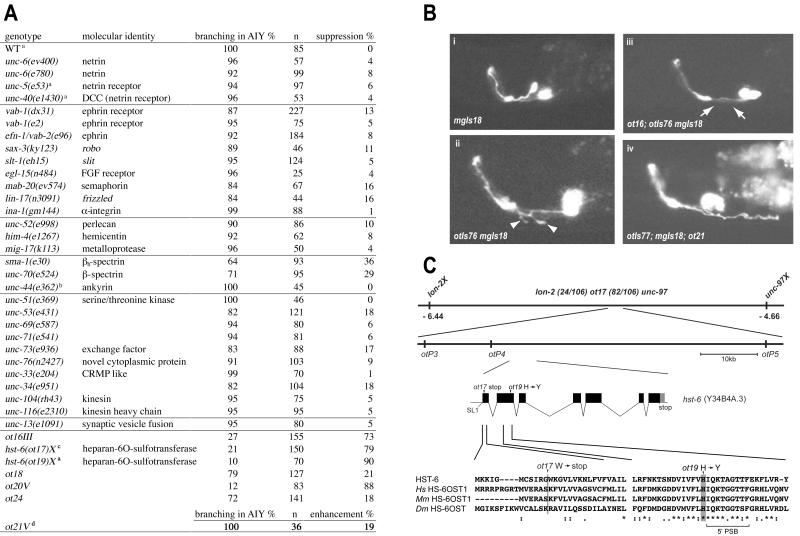
Figure 4.
Mutations that modify the KAL-1-dependent phenotype. (A) Genetic interaction of the kal-1(gf)-inducing axon-branching defect with mutants described as being involved in axonal patterning and other aspects of neuronal function (34, 35) and mutants retrieved from a modifier screen. Percent suppression does not refer to qualitative changes in branch appearance but penetrance of the defect. All experiments were done in a mgIs18 background to label AIY and, if not indicated otherwise, with the ttx-3∷kal-1 overexpressor otIs76IV. The moderately penetrant suppression by spectrins may be explained by their proposed role in localizing cell adhesion/signaling proteins (36). a, ttx-3∷kal-1 overexpressor otIs35X. b, ttx-3∷kal-1 overexpressor otIs77II. c, strain was marked with lon-2(e678). d, enhancement was arbitrarily defined as branches exceeding 20 μm (for quantification see Fig. 8, which is published as supporting information on the PNAS web site). (B) Representative example of the ttx-3∷gfp reporter control (mgIs18) (Upper Left), the ttx-3∷kal-1 overexpressor (otIs76 mgIs18) (Lower Left), the ot16 suppressor mutation in combination with the overexpressor (Upper Right), and the enhancer ot21 that shows considerably longer branches than the overexpressor alone (Lower Right). (C) hst-6 cDNA structure was experimentally determined by PCR and contained SL1 splice leader sequences. An alignment of two N-terminal portions of C. elegans HST-6 with its human (Hs), mouse (Mm), and fly (Dm) orthologs is shown. 5′PSB, binding site for the 3′-phosphoadenosine 5′-phosphosulfate cofactor (37).
To assess whether the phenotype was dosage-dependent we assayed the phenotype in animals that were heterozygous for each of three chromosomally integrated expression arrays. We found that in all cases, the lengths of the branches were shorter in the heterozygotes (data not shown), and at least in one heterozygous line (otIs35/+) the overall penetrance of the phenotype was significantly decreased whereas the converse was true for increased gene dosage (Fig. 2B and Fig. 7, which is published as supporting information on the PNAS web site).
Neither the expression vector alone nor expression of either a randomly chosen WAP domain-only protein present in the genome sequence (F54E2.2) or a randomly chosen FnIII-domain-only protein (C36B1.2) induced axon branching (Fig. 2B). Expression of ttx-3∷kal-1ΔC, a C-terminal deletion that deletes parts of the first FnIII motif and all consecutive FnIII motifs and which recapitulates truncations that have been identified in human Kallmann patients (18), also does not induce axonal branching. Similarly, introduction of a point mutation in the first FnIII domain of kal-1, S241K, which is at an equivalent position as the N267K missense mutation identified in a Kallmann patient (18), led to a complete loss of the ability to induce axonal branching (Fig. 2B). The loss of branching activity of the S241K mutant is unlikely the result of a loss of protein processing, stability, or secretion, because antibody staining shows similar expression levels of the S241K protein compared to overexpressed wild-type protein (see Fig. 6) and because it can be readily purified from the supernatant of transiently transfected cells (Fig. 5). Lastly, the disruption of two of the four disulfide bonds that define a WAP domain in the context of the full-length KAL-1 protein led to a qualitatively changed branching phenotype characterized by extensive arborization of the axonal branches (Fig. 2B). Taken together, these results establish that the axonal branching phenotype is a dosage-dependent KAL-1 protein- and domain-specific phenomenon.
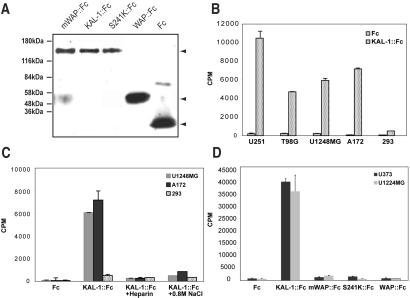
Figure 5.
Heparin-dependent binding of C. elegans KAL-1∷Fc protein fusions to cell lines. (A) Expression constructs. Supernatants of HEK-293T cells expressing the individual constructs were immunoblotted with anti-Fc antibodies. (B, C, and D) Binding KAL-1∷Fc to individual cell lines. U-25 and U-3731, human glioma; T986G, human glioblastoma; U-1248 MG, human glioblastoma; A172, human glioblastoma; 293, human kidney fibroblasts. (C) Binding of KAL-1∷Fc to cells can be inhibited by adding exogenous heparin or high salt. (D) Domain requirement for KAL-1 binding to cells. mWAP (C134S/C135S) and S241K are point mutations introduced into full-length KAL-1∷Fc (see text).
The protein- and domain-specificity of branch induction is a strong argument against the possibility that axon branching is merely a secondary reflection of an overall sickness of the neuron caused by aberrant kal-1 expression. Nevertheless, we tested the functionality of the AIY interneuron by subjecting ttx-3∷kal-1-expressing animals (otIs76) to two different behavioral assays that require AIY to function appropriately and found these animals to behave indistinguishably from wild type (E. Tsalik and O.H., data not shown). kal-1-induced axon branching thus seems to leave the overall functionality of the neuron intact.
kal-1 expression in the AFD sensory neuron class also caused a highly penetrant branching phenotype (Fig. 2C). Like in AIY, expression of KAL-1ΔC completely abolished the branch-inducing potential of KAL-1 protein. Intriguingly, the branching phenotypes in AIY interneurons and AFD sensory neurons were strictly cell-autonomous. In animals that express kal-1 under the control of the AIY-specific ttx-3 promoter, branching was observed in the AIY neurons, yet AFD, AWA, AWC, and ASER, the major presynaptic partners of AIY (19), remained completely unaffected. Conversely, expression of kal-1 in AFD neurons whose major synaptic output is to AIY caused branching in AFD, yet AIY interneurons as well as adjacent sensory neurons showed wild-type morphology (see Table 3, which is published as supporting information on the PNAS web site). The cell-autonomous role of the KAL-1 protein is consistent with its association with the cell surface of KAL-1-expressing cells (9) and our antibody staining (Fig. 6). The autocrine activity of KAL-1 also resembles the autocrine activity of several other extracellular axon outgrowth cues that act on the cells in which they are expressed (20–24).
Panneuronal kal-1 Expression Causes Axon-Misrouting Defects.
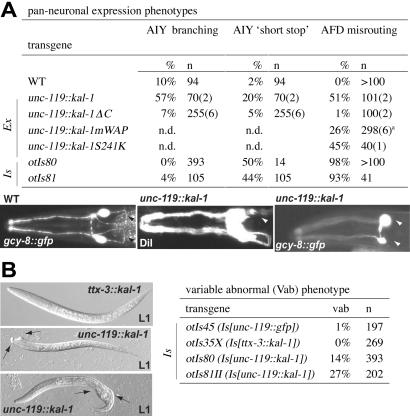
Expression of kal-1 under control of the panneuronal unc-119 promoter led to a broader set of phenotypes, yet the focus of kal-1 action was still very confined. The overall morphology of the nervous system, as assessed with a panneuronally expressed gfp reporter, did not reveal any obvious defects in unc-119∷kal-1 animals (data not shown). The effects of panneural kal-1 on AIY neuroanatomy only partly recapitulated the branching phenotypes observed in cell-specific kal-1 overexpression and, in addition, led to a partially penetrant axon outgrowth defect of AIY (“short stop”; Fig. 3A). Most conspicuously, however, a highly penetrant axon fascicle-misrouting defect was observed (Fig. 3A). Instead of entering the nerve ring through the amphid commissure, the axons of many sensory neurons entered the nerve ring laterally, thus bypassing the dorsoventral migration aspect of their normal trajectory. A similar amphid fascicle-misrouting phenotype is observed in unc-6/Netrin and vab-1/EphR mutants (25). By colabeling distinct subsets of amphid neurons with gfp and DiI, we found this to be an “all or nothing” effect, in which either all labeled sensory axons entered the nerve ring laterally or none of them did (data not shown), hinting that unc-119∷kal-1 may effect an as yet unknown nerve ring pioneer neuron. Consistent with this notion, axon misrouting is not induced by forced expression in subsets of sensory neurons (Table 2). All phenotypes were absent in lines that expressed the C-terminal KAL-1 deletion mutant (Fig. 3A).
Figure 3.
Panneuronal expression of kal-1 causes axon misrouting and hypodermal defects. (A) Panneuronal phenotypes in AIY interneurons and AFD sensory neurons. AIY “short stops” are defined as the failure of the two bilaterally symmetric axons to reach the dorsal midline. Shown Left are AFD wild-type morphology (black arrowheads point to the amphid commissure), and the amphid-misrouting phenotype exemplified by the absence of the amphid commissure [(Center) unilateral; (Right) bilateral absence, marked by white arrowheads)] as visualized with DiI staining (Center) or oyIs17, which exclusively labels AFD (Right). Animals were scored as defective if they displayed unilateral or bilateral defects, respectively. a, results varied between 4% and 44% per line. AIY morphology was examined in an mgIs18 background, and AFD morphology was examined in an oyIs17 background. Note that amphid commissure defects were 20% penetrant if otIs81 was scored in an N2 background with DiI filling (Table 1). (B) Variable abnormal (Vab) phenotypes as observed in lines overexpressing kal-1 panneuronally.
Experiments with the point mutants mWAP and S241K indicate that the branching activities and outgrowth activities are genetically separable. Although the mWAP mutation did not impair the overall branching propensity of kal-1 when expressed in AIY (it even enhanced it; Fig. 2B), it did impair the potential to misroute amphid axons when expressed panneuronally (Fig. 3A). The converse is true for the S241K mutation, which abrogates the branching activities while having no effect on the ability of kal-1 to misroute amphid sensory neurons. It is thus conceivable that the axon-branching and -misrouting activities are mediated through distinct receptor/binding proteins.
Panneuronal kal-1 Expression Causes Morphogenesis Defects.
In addition to the axon outgrowth phenotypes, we found that panneuronal kal-1 overexpressors display a moderately penetrant variable abnormal (Vab) phenotype, characterized by hypodermal morphological defects in the head and tail (Fig. 3B). Animals that overexpress kal-1 only in AIY do not show these defects, neither do animals that panneuronally express an unrelated protein. Similar, yet more severe, Vab phenotypes are observed in animals that are mutant for the ephrin receptor vab-1, which acts in neuroblasts to affect hypodermal cell morphogenesis (26). A kal-1 reporter is expressed in those neuroblasts at the time when vab-1 is required in these neuroblasts to ensure correct hypodermal cell morphogenesis. Taken together with the similar axon-misrouting phenotype of vab-1 mutants (25), these observations point to a potential intersection of kal-1 and ephrin-signaling systems, a notion that will require further genetic and biochemical tests.
Identification of Mutant Loci That Specifically Modify the KAL-1 Mis/Overexpression Phenotype.
To identify mutants that modify the described kal-1(gf) phenotypes, we took two parallel approaches. First, we tested various known mutations that were previously shown to play a role in axonal development and function for their effect on the kal-1(gf) phenotype and second, we conducted a screen to isolate new mutations in genes that are required for kal-1(gf) to exert its function. Such modifier mutations may define genes involved in processing, secretion, or localization of KAL-1, or genes coding for a receptor, coreceptor, otherwise interacting proteins, or signaling molecules that are genetically downstream of a KAL-1 receptor.
As a starting phenotype, we used the kal-1-induced branches in the AIY interneuron that represent the most penetrant kal-1(gf) phenotype. We found that mutations in various known extracellular axon-pathfinding cues (including Netrin, Slit, Ephrin, Semaphorin, and Integrin cues) had little to no effect on the expressivity of the kal-1-induced branches nor did mutations in many intracellular proteins required for axon outgrowth, synaptic vesicle transport, or synaptic transmission (Fig. 4A).
We next conducted a screen for ethyl methanesulfonate-induced mutations that would modify kal-1(gf)-induced branching as assessed by fluorescence microscopy of clonal populations of mutagenized animals. We identified six recessive mutations (ot16III, ot17X, ot19X, ot20V, ot21V, and ot24) and one semidominant mutation (ot18) that either enhance (ot21V) or suppress (all others) the kal-1-dependent axon-branching phenotype (Fig. 4 A and B and Materials and Methods).
We investigated the specificity of two suppressor loci (ot16III and ot17X) in more detail. Neither of these two loci suppressed axonal-branching defects observed in AIY after loss of the ttx-3 transcription factor (17) nor did they suppress ectopic axon outgrowth observed in AIY after loss of the sax-2 gene (27) (Table 1). In contrast, they suppressed the AFD-branching phenotype caused by expression of kal-1 in AFD and the amphid-misrouting phenotype caused by panneuronal expression of kal-1 (Table 1). ot17 does not suppress the axon-misrouting phenotype induced by unc-6, thus underscoring the kal-1 specificity of this modifier mutant (data not shown). The variable abnormal phenotype observed in animals that express kal-1 panneuronally is suppressed by ot16 but, interestingly, not by ot17 (Table 1). This observation suggests that the hypodermal and axonal defects caused by unc-119∷kal-1 are genetically separable. Taken together, the results show that the isolated modifier loci selectively interact with the kal-1-induced phenotypes.
Table 1.
Specificity of suppressor mutants ot16 and ot17
|
ttx-3(ks5)-induced branching
|
gcy8∷kal-1-‡ induced branching
|
sax-2(ot10)-induced ectopic neurites
|
unc-119∷kal-1-induced‡‡
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AIY†
|
AFD§
|
AIY†
|
Amphids¶
|
Misrouting amphids¶
|
Vab
|
|||||||
| % | n | % | n | % | n | % | n | % | n | % | n | |
| WT | 52% | 178 | 66% | 106 | 77% | 69 | 49% | 142 | 20%‖ | 146 | 27% | 202 |
| ot16 | 42% | 99 | 8% | 79 | ND** | ND** | 0%* | 141 | 11%* | 261 | ||
| ot17†† | 54% | 93 | 4% | 104 | 67% | 205 | 51% | 75 | 4% | 154 | 32% | 281 |
ND, not determined; Vab, variable abnormal; WT, wild type.
Strain is marked with unc-79(e1068).
Defects were visualized with mgIs18 (Is[ttx-3∷gfp]).
gcy-8∷kal-1 = otIs83V.
Defects were visualized with oyIs17 (Is[gcy8∷gfp]).
Defects were visualized with DiI filling.
The amphid commissure defect increases to 93% in an oyIs17 (Is[gcy-8∷gfp]) background (Fig. 4A).
Strain could not be built because of very close linkage.
unc-119∷kal-1 = otIs81II.
Strains are marked with lon-2(e678).
One of the Modifier Loci Encodes Heparan-6O-Sulfotransferase.
We positionally mapped one of the modifier loci, which is defined by two noncomplementing alleles, ot17 and ot19, to a 90-kb interval between the polymorphic markers otP3 and otP5 on the left arm of chromosome X. This region contains 14 predicted genes, one of which codes for the only worm ortholog of vertebrate heparan-6O-sulfotransferases, termed hst-6 by the worm nomenclature committee (“6” stands for the sugar ring position that is sulfated). Heparan-6O-sulfotransferases localize to the Golgi apparatus and catalyze the transfer of a sulfate moiety on position 6 of the glucosamine residue that is part of the characteristic disaccharide repeat unit of heparan sulfates, which are integral components of heparan sulfate proteoglycans (HSPGs) (28). Sequence analysis of the ot17 and ot19 alleles revealed point mutations in the coding region of hst-6, namely an early stop codon and a missense mutation in a highly conserved residue in the sulfate-binding pocket (Fig. 4C). Because of its molecular nature and the apparent absence of alternatively spliced forms, the ot17 allele is a likely molecular null. C. elegans HST-6 protein comprises 362 amino acids, shares between 49–54% overall sequence similarity with its three vertebrate orthologs (29), and is predicted to be a type II transmembrane protein.
We further corroborated the heparan sulfate dependence of KAL-1 activity by establishing that C. elegans KAL-1 protein, purified from transfected mammalian cells, has the capability to bind to various cell lines in vitro and that this binding is competitively inhibited either by adding heparin or by high salt concentrations (Fig. 5). These observations are consistent with previous in vitro studies on human KAL-1 (10, 14). Extending these previous studies, we found that the heparin-dependent cell-binding activity is abrogated by both the mWAP mutation and the S241K mutation, corroborating our genetic finding that the WAP domain-dependent misrouting activity and the S241-dependent axon-branching activity of KAL-1 both require an HSPG accessory factor. Taken together, our in vivo and in vitro data on HSPG dependence of KAL-1 action conforms with the emerging theme, obtained from genetic analysis in Drosophila and in vitro binding assays, that specific receptor-ligand pairs require specific HSPG accessory factors for high-affinity interaction (30, 31).
Conclusions
We have shown here that heterologously expressed KAL-1 protein induces axon branching and axon misrouting in specific cellular contexts, either because of a hypermorphic, neomorphic, or antimorphic activity of the KAL-1 protein. Parallel in vitro and ex vivo studies revealed similar dual activities of recombinant human KAL-1 protein (32). The branching activity of KAL-1 is intriguing because only few extracellular proteins have been assigned such an activity, including for example the Slit2 protein (33). KAL-1 and Slit2 also share the intriguing feature of being able to induce branching as well as control axon navigation (33). KAL-1 may bind to a cognate receptor, which in turn activates cytoskeletal proteins to initiate an axon branch or to steer a growth cone. Alternatively, KAL-1 may not act through a specific receptor system but may localize a matrix-bound cue whose mislocalization or titration through KAL-1 mis/overexpression may indirectly cause axon branching. In either case, an HSPG accessory factor is likely to be required for KAL-1 to exert both effects on axogenesis.
Our finding of a locally restricted role of C. elegans kal-1 may lead to a reconsideration of the cellular defects in the olfactory system of patients with Kallmann syndrome. Perhaps the olfactory axon-targeting defect is not a direct consequence of the absence of a KAL-1 cue for attraction of olfactory receptor axons, but rather an indirect consequence of a cell-autonomous miswiring of target neurons in the olfactory bulb that normally express KAL-1. Such miswired target neurons may be incapable of attracting olfactory axons and/or be incapable of being recognized by olfactory axons to allow correct innervation. It should also be noted that human homologs of the hst-6 gene represent good candidates to be mutated in the non-X-linked forms of Kallmann syndrome.
Supplementary Material
Acknowledgments
We thank members of the worm community for providing gfp reporter constructs; the National Institutes of Health-funded Caenorhabitis Genetics Center for providing strains; Y. Kohara (National Institute of Genetics, Japan) for yk clones; E. Tsalik and T. Robakis (Columbia University) for providing thermotaxis data and for antibody staining, respectively; J. Zhu, L. Diamond, and Q. Chen for expert technical assistance; C. Petit for sharing data prior to publication; I. Greenwald, members of the Hobert lab, C. Petit, P. Sengupta, and an anonymous reviewer for suggestions on the manuscript; and members of the Hobert and Greenwald labs for discussion and support. This work was funded by a research grant from the Human Frontier Science Program Organization to the Hobert and Peles labs and by funds from the McKnight, Rita Allen, and Hirschl Foundations (O.H.). H.B. was partly funded by a scholarship from the Deutscher Akademischer Austauschdienst and K.B. was funded by a National Science Foundation predoctoral fellowship. E.P. is an Incumbent of the Madeleine Haas Russell Career Development Chair. O.H. is a Searle and Sloan Scholar.
Abbreviations
- EST
expressed sequence tag
- HSPG
heparan sulfate proteoglycan
Note Added in Proof.
After submission of this article, a kal-1(lf) allele was shown to display an ectopic axon-branching phenotype in neurons that have lost endogenous kal-1 function (38).
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY081844).
References
- 1.Kallmann F, Schoenfeld W A, Barrera S E. Am J Ment Defic. 1944;48:203–236. [Google Scholar]
- 2.Rugarli E I, Ballabio A. JAMA. 1993;270:2713–2716. doi: 10.1001/jama.270.22.2713. [DOI] [PubMed] [Google Scholar]
- 3.Hardelin J-P. Mol Cell Endocrinol. 2001;179:75–81. doi: 10.1016/s0303-7207(01)00462-2. [DOI] [PubMed] [Google Scholar]
- 4.Schwanzel-Fukuda M, Pfaff D W. Nature (London) 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- 5.Schwanzel-Fukuda M, Bick D, Pfaff D W. Brain Res Mol Brain Res. 1989;6:311–326. doi: 10.1016/0169-328x(89)90076-4. [DOI] [PubMed] [Google Scholar]
- 6.Legouis R, Hardelin J P, Levilliers J, Claverie J M, Compain S, Wunderle V, Millasseau P, Le Paslier D, Cohen D, Caterina D, et al. Cell. 1991;67:423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- 7.Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, Maestrini E, Pieretti M, Taillon-Miller P, et al. Nature (London) 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- 8.Lutz B, Kuratani S, Rugarli E I, Wawersik S, Wong C, Bieber F R, Ballabio A, Eichele G. Hum Mol Genet. 1994;3:1717–1723. doi: 10.1093/hmg/3.10.1717. [DOI] [PubMed] [Google Scholar]
- 9.Hardelin J P, Julliard A K, Moniot B, Soussi-Yanicostas N, Verney C, Schwanzel-Fukuda M, Ayer-Le Lievre C, Petit C. Dev Dyn. 1999;215:26–44. doi: 10.1002/(SICI)1097-0177(199905)215:1<26::AID-DVDY4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 10.Soussi-Yanicostas N, Faivre-Sarrailh C, Hardelin J P, Levilliers J, Rougon G, Petit C. J Cell Sci. 1998;111:2953–2965. doi: 10.1242/jcs.111.19.2953. [DOI] [PubMed] [Google Scholar]
- 11.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 12.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 13.Altun-Gultekin Z, Andachi Y, Tsalik E L, Pilgrim D, Kohara Y, Hobert O. Development (Cambridge, UK) 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- 14.Soussi-Yanicostas N, Hardelin J P, Arroyo-Jimenez M M, Ardouin O, Legouis R, Levilliers J, Traincard F, Betton J M, Cabanie L, Petit C. J Cell Sci. 1996;109:1749–1757. doi: 10.1242/jcs.109.7.1749. [DOI] [PubMed] [Google Scholar]
- 15.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 16.Timmons L, Court D L, Fire A. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 17.Hobert O, Mori I, Yamashita Y, Honda H, Ohshima Y, Liu Y, Ruvkun G. Neuron. 1997;19:345–357. doi: 10.1016/s0896-6273(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 18.Hardelin J P, Levilliers J, Blanchard S, Carel J C, Leutenegger M, Pinard-Bertelletto J P, Bouloux P, Petit C. Hum Mol Genet. 1993;2:373–377. doi: 10.1093/hmg/2.4.373. [DOI] [PubMed] [Google Scholar]
- 19.White J G, Southgate E, Thomson J N, Brenner S. Philos Trans R Soc London B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 20.Shepherd I, Luo Y, Raper J A, Chang S. Dev Biol. 1996;173:185–199. doi: 10.1006/dbio.1996.0016. [DOI] [PubMed] [Google Scholar]
- 21.Maina F, Hilton M C, Andres R, Wyatt S, Klein R, Davies A M. Neuron. 1998;20:835–846. doi: 10.1016/s0896-6273(00)80466-3. [DOI] [PubMed] [Google Scholar]
- 22.Yang X M, Toma J G, Bamji S X, Belliveau D J, Kohn J, Park M, Miller F D. J Neurosci. 1998;18:8369–8381. doi: 10.1523/JNEUROSCI.18-20-08369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sondell M, Sundler F, Kanje M. Eur J Neurosci. 2000;12:4243–4254. doi: 10.1046/j.0953-816x.2000.01326.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang K H, Brose K, Arnott D, Kidd T, Goodman C S, Henzel W, Tessier-Lavigne M. Cell. 1999;96:771–784. doi: 10.1016/s0092-8674(00)80588-7. [DOI] [PubMed] [Google Scholar]
- 25.Zallen J A, Kirch S A, Bargmann C I. Development (Cambridge, UK) 1999;126:3679–3692. doi: 10.1242/dev.126.16.3679. [DOI] [PubMed] [Google Scholar]
- 26.George S E, Simokat K, Hardin J, Chisholm A D. Cell. 1998;92:633–643. doi: 10.1016/s0092-8674(00)81131-9. [DOI] [PubMed] [Google Scholar]
- 27.Zallen J A, Peckol E L, Tobin D M, Bargmann C I. Mol Biol Cell. 2000;11:3177–3190. doi: 10.1091/mbc.11.9.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandtlow C E, Zimmermann D R. Physiol Rev. 2000;80:1267–1290. doi: 10.1152/physrev.2000.80.4.1267. [DOI] [PubMed] [Google Scholar]
- 29.Habuchi H, Tanaka M, Habuchi O, Yoshida K, Suzuki H, Ban K, Kimata K. J Biol Chem. 2000;275:2859–2868. doi: 10.1074/jbc.275.4.2859. [DOI] [PubMed] [Google Scholar]
- 30.Bernfield M, Gotte M, Park P W, Reizes O, Fitzgerald M L, Lincecum J, Zako M. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 31.Perrimon N, Bernfield M. Nature (London) 2000;404:725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 32. Soussi-Yanicostas, N., de Castro, F., Julliard, K., Perfettini, I., Chédotal, A. & Petit, C. (2002) Cell, in press. [DOI] [PubMed]
- 33.Brose K, Tessier-Lavigne M. Curr Opin Neurobiol. 2000;10:95–102. doi: 10.1016/s0959-4388(99)00066-5. [DOI] [PubMed] [Google Scholar]
- 34.Antebi A, Norris C, Hedgecock E M, Garriga G. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 583–610. [PubMed] [Google Scholar]
- 35.Rand J B, Nonet M L. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 611–644. [Google Scholar]
- 36.Hammarlund M, Davis W S, Jorgensen E M. J Cell Biol. 2000;149:931–942. doi: 10.1083/jcb.149.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kakuta Y, Pedersen L G, Pedersen L C, Negishi M. Trends Biochem Sci. 1998;23:129–130. doi: 10.1016/s0968-0004(98)01182-7. [DOI] [PubMed] [Google Scholar]
- 38.Rugarli E I, Di Schiavi E, Hilliard M A, Arbucci S, Ghezzi C, Facciolli A, Coppola G, Ballabio A, Bazzicalupo P. Development (Cambridge, UK) 2002;129:1283–1294. doi: 10.1242/dev.129.5.1283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.