Summary
Protein turnover is a critical component of gene expression regulation and cellular homeostasis, yet methods for measuring turnover rates that are scalable and applicable to different models are still needed. We introduce an improved D2O (heavy water) labeling strategy to investigate the landscape of protein turnover in cell culture, with accurate calibration of per-residue deuterium incorporation in multiple cell types. Applying this method, we mapped the proteome-wide turnover landscape of pluripotent and differentiating human induced pluripotent stem cells (hiPSCs). Our analysis highlights the role of APC/C (anaphase-promoting complex/cyclosome) and SPOP (speckle-type POZ protein) degrons in the fast turnover of cell-cycle-related and DNA-binding hiPSC proteins. Upon pluripotency exit, many short-lived hiPSC proteins are depleted, while RNA-binding and -splicing proteins become hyperdynamic. The ability to identify fast-turnover proteins also facilitates secretome profiling, as exemplified in hiPSC-cardiomyocyte and primary human cardiac fibroblast analysis. This method is broadly applicable to protein turnover studies in primary, pluripotent, and transformed cells.
Keywords: protein turnover, turnover kinetics, heavy water, hiPSC, protein synthesis, degron, secretome, mass spectrometry
Graphical abstract

Highlights
-
•
D2O labeling measures protein turnover in primary, pluripotent, and transformed cells
-
•
D2O incorporates into multiple amino acids in vitro, including Ala, Glu, Asp, and Pro
-
•
Protein turnover analysis shows hiPSC differentiation alters fast-turnover proteins
-
•
We show application to secretome analysis in human cardiac myocytes and fibroblasts
Motivation
Dynamic stable isotope labeling by amino acids in cell culture coupled with mass spectrometry is commonly used to measure protein turnover in cell culture but requires altering culture medium composition and may not label some peptides. We describe a simple and convenient alternative for measuring protein turnover kinetics in cultured cells by adding low-volume D2O (heavy water) to standard tissue culture media. Addressing a critical gap, we determined the number of deuterium-accessible atoms on all 20 proteinogenic amino acids across multiple cell types. This allows accurate interpretation of D2O-labeled mass spectra to measure protein turnover kinetics and secretome flux on a proteome scale.
Alamillo et al. present a D2O labeling mass spectrometry method to measure protein turnover rates that is compatible with multiple cell cultures and medium formulations. The method reveals a parsimonious protein turnover landscape in human induced pluripotent stem cells and identifies hyperdynamic proteins that are unique to self-renewal states.
Introduction
Protein abundance in the cell is governed by a balance between synthesis and degradation. The half-life of a protein pool regulates its homeostasis and function, influencing diverse biological processes from cell cycle to gene regulation and stress response.1,2,3,4,5 Proliferating cells in culture can remove proteins through passive dilution to daughter cells, or active proteolysis.6,7 Proteins regulated by the latter undergo excess synthesis and degradation cycles beyond what is required for cell doubling, and are often targeted for degradation by E3 ubiquitin ligases via specific sequence motifs, which are known as degrons.8,9,10 Prior works have mapped hundreds of short-lived or rapidly degraded proteins in a few cell types to explore the roles of proteolysis in regulating protein abundance and disease pathogenesis.11,12 Nevertheless, the extent to which these fast-turnover proteins are dynamically synthesized and degraded in different cells and cell states remains unclear. However, active protein translation is known to be required to maintain open chromatin, a hallmark of pluripotent cells.13 The characterization of proteins that undergo rapid synthesis and degradation cycles in pluripotent cells remains an open goal that can shed light on intervention targets in cell fate and malignancy.
To measure protein turnover in cell culture, the common approaches are to monitor the depletion of existing proteins following cycloheximide inhibition of new protein synthesis11,12 or to use dynamic stable isotope labeling by amino acids in cell culture (SILAC) to label essential amino acids with multiple heavy stable isotope atom centers (e.g., 13C615N2-lysine).14,15,16,17 Despite their utility, these techniques have limitations that warrant the continued development of complementary methods. Cycloheximide can lead to unwanted effects on cellular physiology, including inhibition of protein degradation.18,19 Dynamic SILAC is constrained by the cost of synthetically labeled amino acids, the inability to label peptide sequences lacking targeted residues, and the need to modify culture media to use dialyzed serum and depleted basal media.
D2O (heavy water) provides a universal isotope label for multiple biomolecules. Upon D2O labeling, deuterium rapidly incorporates into stable C–H bonds in non-essential amino acids during their biosynthesis and metabolism.20,21,22 Labeled amino acids are subsequently incorporated into nascent protein chains to enable turnover quantification by mass spectrometry (MS). D2O labeling has been successfully applied to trace protein synthesis in rodents,20,21,22,23,24,25 non-human primates,20 and humans22,26,27,28; yet, its adoption to cell culture is underrealized. A key variable in analyzing D2O-labeled data to extract kinetics information is the number of deuterium-accessible sites in a peptide, which is established in mammals but remains unclear in cultured cells. Although D2O in culture media will equilibrate with hydrogen atoms in cellular amino acids,29,30 the extent to which it does so in each amino acid is unknown, given that many amino acids are already present in high concentration in culture media.30,31,32 As such, few studies have applied D2O to measure individual protein turnover in cell cultures. Where such studies existed, they were largely confined to measuring the total alanine rather than individual protein turnover rates29,33,34 or were focused on selected peptides of interest rather than proteome-scale investigations.23,31,35
Here, we describe an updated workflow using D2O labeling to measure proteome-wide individual protein turnover rates in cell culture. To enable interpretation of labeled MS data, we determined the extent of deuterium enrichment in all 20 proteinogenic amino acids following D2O enrichment by using machine learning strategies and experimental calibration with cells of known labeling proportions. The established per-residue enrichment values show clear differences from those used in animal studies or commonly assumed in earlier cell culture experiments and enable precise turnover calculations for virtually any protein sequence. We demonstrate the versatility and robustness of deuterium labeling in two separate applications—first, to map the protein turnover landscape of human induced pluripotent stem cells (hiPSCs) under maintained pluripotency and directed differentiation, and second, to characterize the secretome of human cardiac cells under baseline or stressed conditions. The results establish D2O labeling as a flexible and accessible method to assess protein degradation and homeostasis in multiple models.
Results
D2O incorporates deuterium into multiple non-essential amino acids in cell culture
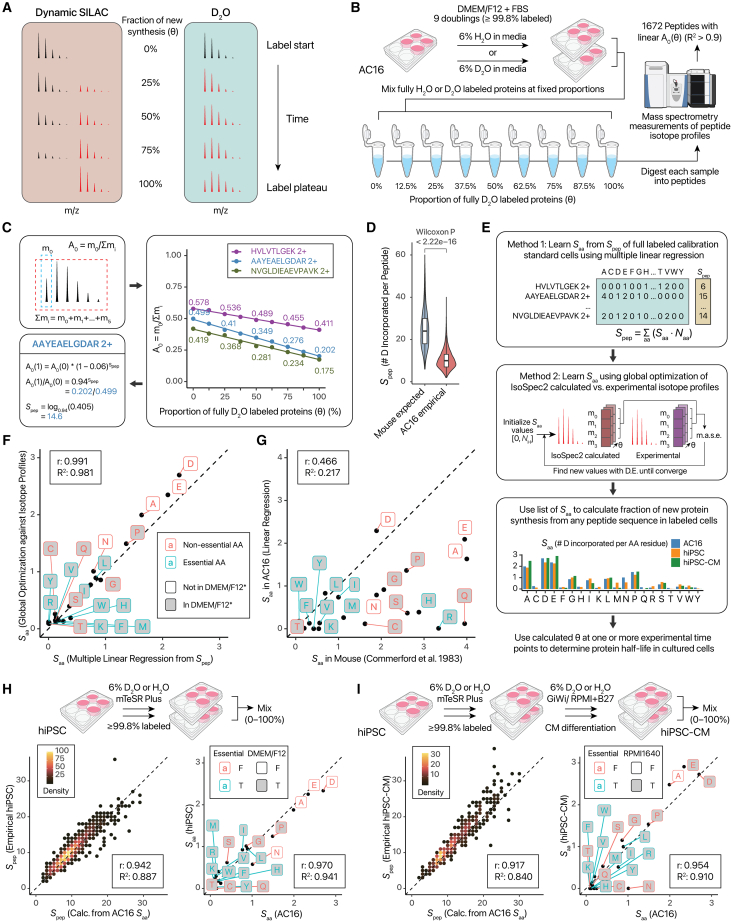
D2O labeling leads to a gradual shift in the isotope profiles of peptides, which is a function of the excess deuterium enrichment (p), the number of deuterium-accessible labeling sites in a peptide (Spep), and the fraction of newly synthesized peptide (θ) in the sample (Figure 1A). Finding the parameter of interest (θ) needed to calculate protein half-life therefore requires knowing the Spep of each peptide. In animal experiments, the conventional method calculates Spep of a sequence from the sum of per-residue labeling sites, commonly those measured from mouse tritium labeling.36 To acquire these values in cell culture, we generated a complete labeling calibration standard by expanding human AC16 cells in DMEM/F12-fetal bovine serum (FBS) media diluted with 6% D2O for 9 doublings37 (Figure 1B). This guarantees ≥99.8% complete labeling from cell division even in the absence of additional protein degradation (i.e., all proteins are fully labeled with 6% deuterium). This fully labeled pool is then mixed with unlabeled cell lysate at fixed proportions to establish the ground-truth fractional synthesis in each sample (Figure 1B). From the fully labeled samples, per-peptide labeling sites can be calculated by considering the proportion of peptide molecules that contain no label, even without knowing the individual amino acid labeling sites (Figure 1C). As expected, D2O incorporates deuterium into peptide sequences in cell culture, leading to a drop of the ratio of the monoisotopic peak over the entire isotope envelope (A0). However, considerably fewer atoms are incorporated than in animals (i.e., less A0 decreases from 0% to 100% labeling), with a median 9.7 sites per peptide vs. 24.6 when estimated using mouse in vivo labeling sites (Figure 1D). Training a multiple linear regression model, we learned the per-residue deuterium-accessible sites from the total labeling sites of each peptide and their amino acid compositions (Figure 1E; Table S1). In parallel, we corroborate the labeling sites with a complementary machine learning strategy that does not rely on the fitted A0 information by performing nonlinear global optimization to minimize the differences between the empirical isotope profiles and the predicted profiles calculated from a fine structure calculator38 (Figure 1E; STAR Methods). The two strategies returned near-identical values (r: 0.991; R2: 0.981), indicating that we are able to acquire internally consistent information on the extent of deuterium labeling in these cells (Figure 1F). The results show that Asp, Ala, Pro, Glu, and other amino acids are amenable to D2O labeling but often at a lower extent than in vivo; for example, both models predict ∼2 exchangeable hydrogens in Ala vs. 4 in animals (Table S1). Contrary to animals, Gln, Arg, Ser, and His are largely inaccessible to labeling in cell culture (Figure 1G).
Figure 1.
D2O labeling incorporates deuterium into proteins in cell culture with residue specificity
(A) Mass shifts of peptide isotope envelopes following SILAC and D2O labeling.
(B) Calibration standard cells are cultured in media with 6% v/v H2O or D2O for 9 doublings to yield unlabeled and fully labeled proteins, which are then mixed in fixed proportion and analyzed by liquid chromatography-tandem MS.
(C) Calculation of peptide labeling sites (Spep) from standards. Right: line plots of three peptides measured in our experiments. The initial value of A(0) is a function of peptide mass/length, whereas the decrease depends on numbers of D-accessible labeling sites.
(D) Violin/boxplot of peptide labeling sites calculated from the m0/mi ratio of the labeling standard samples, highlighting differences (Wilcoxon p < 2.2e–16) to animal labeling sites.
(E) Prediction of per-residue labeling sites (Saa). Method 1: multiple linear regression to learn Saa from empirical Spep. Method 2: global optimization against calculated isotopomer profiles using differential evolution (D.E.) (see STAR Methods). Bar charts: number of label-accessible hydrogens per amino acid across three cell types.
(F) Strong correlation of Saa values (r: 0.991) predicted from methods 1 and 2. Dashed line: 1:1. ∗Label shading denotes whether amino acids are supplemented in DMEM/F12 at appreciable concentration (≥10 mg/L, except l-tryptophan any concentration). The F12 nutrient mix contains low levels (<10 mg/L) of Ala, Asp, Asn, and Glu that are not marked.
(G) Comparison of learned Saa in AC16 against animal labeling sites.36
(H) Labeling sites in hiPSC cultured in mTeSR Plus. Left: AC16-derived Saa values accurately predict measured Spep in hiPSC. Right: similar predicted Saa (r: 0.97) between two cell types. Label shading denotes whether amino acids are supplemented in DMEM/F12.
(I) As in (H), but for hiPSC-CM. Label shading denotes whether amino acids are supplemented in RPMI-1640 (≥10 mg/L except l-tryptophan).
We next extended the fully labeled calibration standards to two additional cell types, hiPSC and hiPSC-derived cardiomyocytes (hiPSC-CM). Both are widely employed as physiologically relevant cell models for drug discovery and disease mechanism studies. Notably, hiPSCs are commonly cultured in specialized media (e.g., mTeSR Plus) for which SILAC-compatible depleted basal media is not commercially available, thus necessitating custom media formulations and providing a compelling use case for D2O labeling as a flexible alternative. To generate the calibration cells, hiPSC were cultured to ≥10 doublings in mTeSR Plus. To procure fully labeled non-proliferative hiPSC-CM, hiPSCs were passaged in D2O prior to differentiation. The empirical peptide labeling sites in hiPSCs are similar to those predicted using the AC16-derived cell culture values (Figure 1H) (Pearson’s r: 0.942; R2: 0.887) and similarly for hiPSC-CM (Figure 1I) (r: 0.917; R2: 0.840). The trained linear regression models likewise predicted similar labeling sites (hiPSC r: 0.970; hiPSC-CM r: 0.954) (Figures 1H and 1I). Notably different is the labeling of Asn, which incorporates deuterium in AC16 under DMEM/F12 with FBS, but to a lesser degree in hiPSCs under mTeSR and absent in hiPSC-CMs under RPMI-1640 with B27, which contains supplemented Asn. Conversely, multiple non-essential amino acids remain effectively labeled by D2O in hiPSC-CMs, despite RPMI-1640 being a complete medium that contains Asp, Glu, and Pro, indicating the primary source of these amino acids for protein production likely lies in endogenous biosynthesis rather than external uptake. The predicted labeling sites show considerable consistency, while reflecting some differences in cell types or culture medium compositions.
Amino acid analysis corroborates the determined D2O labeling sites
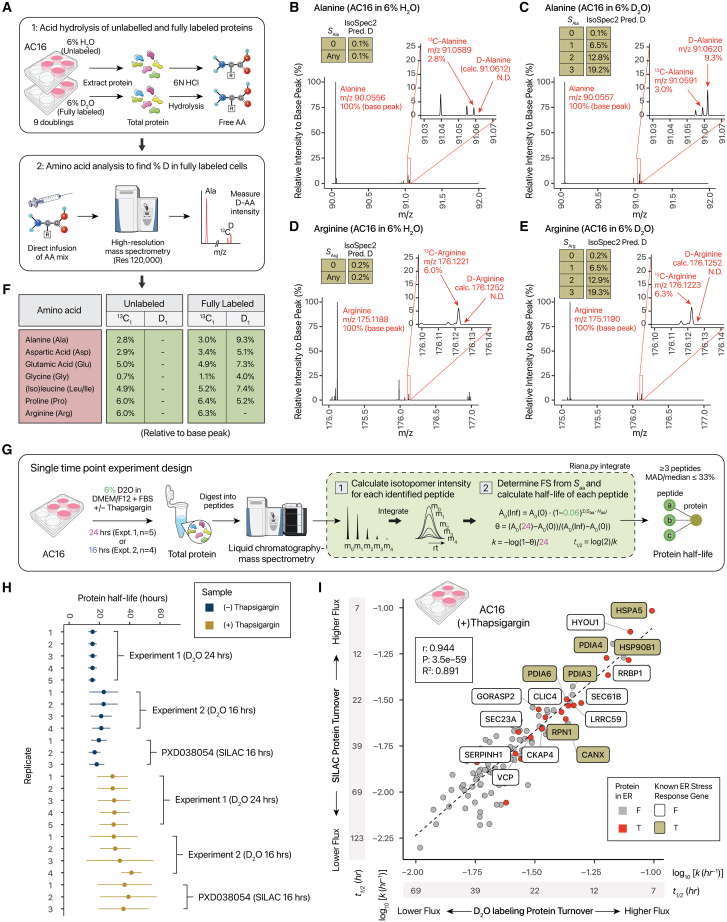
To experimentally verify the labeling sites, we performed acid hydrolysis to release free amino acids from proteins in unlabeled and fully labeled AC16 cells and then measured the deuterium-tagged amino acids using direct infusion high-resolution MS (Figure 2A). At 120,000 resolution, 13C- and D-containing amino acid peaks can be resolved by their mass differences from mass defects, allowing their relative intensities to the monoisotopic peaks to be compared with calculated isotope profiles under different D-accessible labeling site predictions. The relative intensity of the signals corresponding to D1-alanine (m/z 91.0621, +1 charge) is consistent with predictions in cell culture (1–2 labeling sites vs. 4 in adult mice in vivo) (Figures 2B and 2C). The measurements are not biased by transient time in the Orbitrap mass spectrometer, as similar values are obtained at different mass resolutions (Figure S1). Conversely, the absence of D1-arginine peaks (m/z 176.1252, +1 charge) is consistent with the predicted absence of deuterium-accessible arginine labeling sites (Figures 2D and 2E), whereas amino acid analysis also supports 1–2 labeling sites in proline and <1 site in glycine and leucine/isoleucine (Figure 2F; Table S2). We determined highly consistent labeling sites in three cell types (transformed AC16, pluripotent hiPSC, differentiated hiPSC-CM), supporting the use of D2O labeling for protein turnover studies in multiple cell types. The approach also provides a general strategy to find labeling sites in other cultures.
Figure 2.
Experimental validation of deuterium-incorporation sites and single-time-point kinetics analysis
(A) Experimental validation of predicted labeling sites using acid hydrolysis and high-resolution MS.
(B) Mass spectra from acid hydrolysis of unlabeled samples, showing no detectable d-alanine. N.D., not detected.
(C) As in (B), but for alanine in fully labeled samples, showing detectable d-Ala at m/z 91.0612 with 9.3% relative intensity to monoisotopic Ala (m/z 90.0550). Inset: IsoSpec2-predicted d-Ala relative intensity (0.1%, 6.5%, 12.8%, 19.2%) relative to the monoisotopic peak at SAla = 0–3, respectively.
(D) As in (B), but for Arg.
(E) As in (C), but for Arg, showing no detectable d-Arg (m/z 176.1252) in cell culture.
(F) Relative intensity of d-Ala, Asp, Glu, Gly, Leu/Ile, Pro, and Arg in unlabeled and fully labeled samples, using amino acid analysis with high-resolution MS. Asn and Gln are deamidated during acid hydrolysis39; hence, measured values are a mixture of Asp/Asn and Glu/Gln.
(G) Application of D2O labeling to proteome-wide turnover kinetics in normal and stressed AC16 cells.
(H) Point ranges of protein half-life measured by D2O and SILAC in AC16, showing comparable ranges between methods. D2O recapitulates a slowdown in cell proliferation and protein synthesis upon thapsigargin-induced ER stress. Error bars: SD.
(I) Scatterplot of D2O and SILAC measurements after 1 μM thapsigargin, revealing strong correlation (r: 0.94) and stress-response genes with high turnover flux upon ER stress.
D2O labeling enables single-point measurement of protein synthesis kinetics in cells
We next asked whether the labeling sites support interpretation of isotopomer envelopes to measure proteome-wide turnover rates. At baseline, AC16 cells proliferate quickly with a doubling time of ∼16–20 h, and protein isotope incorporation rates are expected to largely reflect proliferation rates under the particular culture conditions. Therefore, we labeled proliferating AC16 with 6% D2O for 24 h (n = 5) then quantified isotope incorporation from a single time point (Figure 2G). The quantified individual protein turnover rates returned an expected per-protein median half-life of 16.7–17.5 h (Figure 2H; Table S3), in agreement with the doubling time and with prior half-life measurements (∼16–20 h).40 We did not observe a change in proliferation rate upon diluting the media with low-dose (6%) D2O, consistent with prior reports of minimal effects on cell growth.31 In comparison, non-proliferative hiPSC-CM and primary human cardiac fibroblasts (hCFs) show considerably longer median half-lives, of 53.9 and 55.1 h, respectively (Figure S2; Table S3).
To evaluate measurement accuracy, we labeled AC16 under thapsigargin-induced endoplasmic reticulum (ER) stress and compared it to SILAC data. ER stress is known to halt cell proliferation, and accordingly we observed a nearly 2-fold reduction in protein half-life upon a single concomitant dose of thapsigargin with D2O, to 31.5–34.8 h. A second set of replicate experiments (n = 4), performed by a different experimentalist using a different AC16 stock labeled for 16 h, likewise recapitulates a nearly 2-fold decrease in turnover rates (Figure 2H). While overall protein turnover decreases, the span of measured turnover rates broadens as ER stress response pathways blunt protein synthesis except for some stress response proteins,40 allowing correlative comparison with SILAC measurements previously acquired at 16 h.39 The D2O labeling data show a robust correlation with dynamic SILAC (r: 0.94; p: 3.5e–59). In parallel, D2O labeling successfully captured the accelerated turnover of ER stress response proteins, including HSPA5/BiP, HYOU1, and HSP90B1 (Figure 2I; Table S3). Thus, we are able to measure proteome-wide turnover rates and interrogate cellular physiological responses using D2O labeling at a single time point.
Mapping the proteome-wide turnover kinetics in hiPSC with D2O labeling
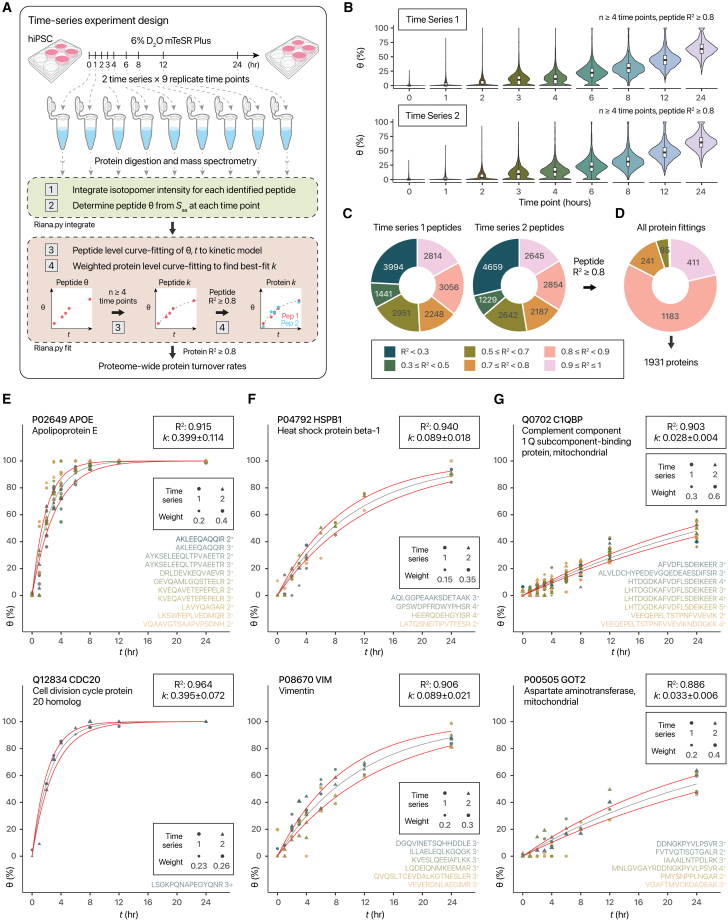
We next applied D2O labeling to a larger-scale time-course experiment, where hiPSCs are labeled in 6% D2O in mTeSR Plus at 9 biologically independent time points, each from 2 replicate time series (Figure 3A). Upon D2O labeling, hiPSC proteins show gradual increases in calculated fractional synthesis, as existing proteins are replaced by newly synthesized proteins over 24 h (Figure 3B). The samples were processed to quantify 42,865 distinct peptide sequences that are uniquely mappable to 6,905 proteins (median 3,599 proteins and 17,545 peptides per sample). We then performed curve fitting of all time points to a single exponential kinetic model in each peptide to find the best-fit isotope incorporation rate constant k of 18,392 peptides across 2,995 proteins. The majority of peptide time series showed high goodness-of-fit between data and kinetic model, with about half showing R2 ≥ 0.7 and over one-third with R2 ≥ 0.8 (Figures 3C and 3D). A second-pass protein-level fitting using peptides with peptide-level R2 ≥ 0.8 then yielded the high-confidence protein-level k of 1,931 proteins (Table S4). Nearly all proteins retained high R2 values, demonstrating that the labeling sites allow confident interpretation of spectra across many different peptides to derive fractional synthesis values (Figure 3E). Constituent peptides within the same protein give highly consistent k values (median intra-protein peptide geometric coefficient of variation [CV] 20.7%), comparable to precisions in SILAC or animal D2O experiments. Data quality is likewise reflected by good agreement in protein k when the two time series are fitted separately (r: 0.76); as well as a median dk/k (fitting error relative to k) of 24.9%. The data cover proteins with a wide range of turnover rates, from fast (k > 0.1 h−1; APOE and CDC20) (Figure 3E) to moderate (0.05 ≤ k < 0.1 h−1; HSPB1 and VIM) (Figure 3F) and slow (k < 0.05 h−1; C1QBP and GOT2) (Figure 3G).
Figure 3.
Application of D2O labeling to measuring proteome-wide turnover kinetics in hiPSCs
(A) Measurement of protein turnover in hiPSC-CM using D2O labeling in a multi-point time-course design.
(B) Fractional synthesis over the course of labeling in two replicate time series of 9 time points each.
(C) Proportion of quantified peptides that fit to a standard kinetics model at different R2 cutoffs.
(D) As in (C), but for a second-pass weighted protein-level fitting, after selecting peptides with R2 ≥ 0.8.
(E) Isotope incorporation kinetics of proteins selected to represent the dynamic range of turnover, highlighting two fast-turnover proteins APOE (top) and CDC20 (bottom). Black line: kinetic curve governed by best-fit k; red: standard error.
(F) Same as (E), but for moderate-turnover proteins HSPB1 and VIM.
(G) Same as (E), but for slow-turnover proteins C1QBP and GOT2.
Interestingly, the majority of hiPSC protein turnover rates occupy a relatively narrow band (Figure 4A). At the bottom 10th percentile, the measured protein turnover rate in hiPSC is 0.033 h−1 (Figure 4B), corresponding to a half-life of 21.2 h, which is in agreement with the typical hiPSC doubling time in the literature and our observations (24–36 h).41 The turnover rates have a heavily right-tailed distribution (Figure 4B) that deviates from the log-normal distribution typically observed in other proteomes in vivo42,43 (D’Agostino p: 3.7e–93; skewness 1.54). This distribution categorizes hiPSC proteins into three groups: (1) the majority of proteins in the bottom 70 percentiles showing little synthesis beyond that needed for cell doubling (half-life > 11.8 h); (2) proteins in the 10th–30th percentiles (half-life 6.6–11.8 h) with appreciable excess turnover cycles beyond cell doubling; and (3) proteins in the top 10 percentiles (half-life ≤ 6.6 h) with rapid synthesis-degradation kinetics. Fast-turnover proteins are significantly enriched in cell-cycle, proliferation signaling, and polyubiquitination processes (Fisher’s exact test q <0.01), whereas slow-turnover proteins are associated with translation and metabolism (Figure 4A; Table S5). These enrichments are corroborated by proteome-wide gene set enrichment analysis (GSEA), which shows that mitochondrial proteins tend to turn over slowly, whereas fast-turnover proteins are enriched in cell-cycle, extracellular space, and extracellular signaling terms (Figures S3A and S3B). Thus, the majority of hiPSC proteins are parsimoniously synthesized, even as proteins with different biological functions are synthesized and degraded to different extents.
Figure 4.
Features of hiPSC proteome-wide turnover dynamics
(A) Ranked protein turnover rates in hiPSCs. Red dashed lines show the turnover rates at the top 10th, 30th, 50th, and 90th percentiles. Top-10 percentiles are enriched in cell-cycle proteins. Colors: estimated absolute copy number per cell (see E).
(B) Histogram of turnover rates showing a right skewed distribution.
(C) Calculation of protein copy number and total energetic costs of protein turnover. [P], copy number; k, turnover rate constant; L, length.
(D) Correlation between protein copy number and turnover rates.
(E) Fast turnover is associated with high protein average energy cost per amino acid (ECPA).
(F) High-turnover proteins incur a disproportional energetics budget in ATP and amino acid biosynthetic cost. Proportional ATP usage and biosynthetic costs are calculated based on turnover beyond what is assumed to be minimally needed for cell doubling, at 90th percentile of all protein k.
The production of protein molecules accounts for a substantial part of the bioenergetic and biosynthetic budget of proliferating cells44,45 and is constrained by selective pressure.46 To explore the energy expenditure of protein synthesis in hiPSCs, we calculated the absolute copy numbers per cell of quantified proteins using the proteomic ruler method (Figure 4C; STAR Methods). The calculated absolute protein quantities are consistent across time points and correlate robustly with estimates in embryonic mouse fibroblasts14 (Pearson’s r: 0.76 [0.74–0.78]; p: 2.2e–313 in log-log scale) despite methodological differences (Figures S3C and S3D). As expected,47 protein abundance correlates negatively with k (Pearson’s r: −0.32; p: 2.2e–46) (Figure 4D), which may reflect a constraint on high-abundance protein synthesis due to energetic or ribosome capacity limits.45,48 Protein k is poorly explained by protein sequence (isoelectric points, hydropathy, molecular weights, lengths; Figures S4A–S4D) or structural features (disorder,49 helix and strand folds, turns; Figures S4F–S4I), consistent with prior observations.17 From the absolute protein abundance estimates, we calculated the peptide chain elongation ATP usage and amino acid biosynthetic cost50 of protein synthesis (Figures 4C and S4E). Surprisingly, fast-turnover proteins in hiPSC have a higher average amino acid biosynthetic cost than low-turnover proteins (Figure 4E). Although fast-turnover proteins represent a minor fraction (∼6%) of the total protein mass per hiPSC, they account for up to one-third of the energy budget needed for protein synthesis beyond the minimal levels required for cell proliferation (Figure 4F). Thus, this analysis reveals that hiPSC protein synthesis and usage are not constrained by the biosynthetic costs of amino acids, which deviates considerably from observations in aging animals, where proteins with metabolically expensive amino acids are synthesized parsimoniously.43
Hyperdynamic proteins in hiPSCs are associated with specific degron motifs
Proteins that are actively proteolyzed in proliferating cells have been referred to as rapidly degraded, short-lived, or unstable.11,12 Below, we use the terms fast-turnover or hyperdynamic proteins to highlight that they are not inherently unstable but synthesized and degraded quickly under presumed steady state. Despite previous studies aimed at mapping these proteins in multiple cell lines,11,12 we found a number of previously unreported fast-turnover proteins in hiPSCs, suggesting they are cell type specific (Figure 4C). Whereas some non-overlap exists because secreted and extracellular proteins were discarded in a prior study12 (as these proteins may be removed by media change rather than proteolysis; see below), we also identified >100 fast-turnover intracellular proteins that were not reported in prior studies, including KIF22, which functions in spindle formation in mitosis, BUB1B, which acts as a mitotic checkpoint protein, and PPP1CC, a nuclear phosphatase that participates in cell division, the nucleolar protein HEATR1, and splice factor SRSF5 (Table S4).
To investigate the proteolytic mechanisms that drive the synthesis-degradation cycles of these hyperdynamic proteins in hiPSCs, we performed a GSEA-style analysis to test whether particular sequence motifs targeted for protein degradation (degrons) confer faster turnover, which we call degron set enrichment analysis (DSEA; STAR Methods). Among 105 analyzed degron motifs that we retrieved from Degronopedia51 and that appear in ≥10 proteins with quantified turnover rates, 2 degrons showed a significant association (DSEA adjusted p, false discovery rate [FDR] < 5%) with higher protein turnover in hiPSCs, namely the anaphase-promoting complex/cyclosome (APC/C) KEN box (xKENx) and speckle-type POZ protein (SPOP) ([AVP]x[ST][ST][ST]) degrons, whereas a BAG6-type degron (LLLL) also shows nominal enrichment at FDR <10% (Figure 5A). The enriched degrons recognize partially overlapping sets of proteins, especially between APC/C and SPOP. Multiple cell-cycle proteins are present, enriched particularly in KEN box proteins, including CDC20, CENPF, CDCA2, and CDCA5 (Figure 5B), whereas multiple degron-containing hyperdynamic proteins also participate in DNA/RNA binding, DNA repair, and stress response (Figure 5B). Proteins that contain the KEN box or the SPOP degron showed significantly elevated turnover rates in hiPSCs than proteins that did not possess either, while we did not observe evidence of additive effects of the motifs (Figure 5C). Notably, the APC/C activator CDC20 itself possesses a KEN box sequence that is recognized by FZR1/Cdh1, as part of a feedback mechanism in the late cell cycle. This KEN box sequence moreover coincides with a predicted PEST motif (epestfind score: 8.85) at residues 97–109 (KENQPENSQTPTK) that is located within a predicted disordered region (MetaPredict52 scores >0.5). Thus, while no proteome-wide correlations were found between PEST motifs and turnover,17,53 our results here suggest they may contribute to degradation of specific hiPSC proteins. The enrichment of the APC/C KEN box contrasts with the lack of statistically significant enrichment of the other APC/C degrons D box (adjusted p: 0.25) or ABBA (adjusted p: 0.65) (Figure 5A); likewise, SCF motifs are not significantly enriched despite the central role of the SCF (Skp1-Cullin-F-box) complex in the cell cycle. Thus, cell-cycle-related ubiquitination processes are differentially active in hiPSCs.
Figure 5.
Fast-turnover proteins in hiPSCs possess degrons targeted by APC/C and SPOP
(A) Degron set enrichment analysis (DSEA) of proteins associated with various Degronopedia degrons (top 35 degrons shown), showing the significant enrichment of the APC/C (KEN) and SPOP degrons among fast-turnover proteins.
(B) Top 25 DSEA leading edges of significantly enriched degrons (APC/C KEN, SPOP, BAG6) and associated biological processes.
(C) Proteins containing KEN box or SPOP degron have significantly elevated turnover in hiPSCs.
(D) Kinetic curves of PRRC2B and PRRC2A in D2O labeled hiPSCs.
To further illustrate the role of degron motifs on turnover, we consider PRRC2B (proline-rich coiled-coil protein 2B), a poorly characterized RNA-binding protein with fast turnover in hiPSCs (Figure 5B). PRRC2B was not previously reported as a fast-turnover protein in four cell lines from two separate studies,11,12 but in our data, it is a top DSEA leading-edge protein of both the KEN box and SPOP degrons. As PRRC2B is not a known cell-cycle protein from annotations or large-scale discovery data,54 we focus on the potential effect of the SPOP degron. PRRC2B shares 35.7% sequence identity with its closest homolog PRRC2A (Figure S5A), with particular homology in the N-terminal disordered region (residues 1–269 in PRRC2B). Within the N-terminal region, we find a short linear motif55 recognized by SPOP only in PRRC2B (residues 96–100), not in PRRC2A (Figure S5B). This sequence also satisfies a more selective definition of the SPOP-binding consensus (SBC)56 motif in the literature (Φ-Π-S-S/T-S/T; Φ: non-polar; Π: polar) and is evolutionarily conserved (Figure S5C). Elsewhere in the sequence, PRRC2B features three additional SPOP motifs, two of which satisfy the SBC definition (residues 922–926; 996–1,000), vs. zero in PRRC2A. SPOP is a substrate-recognizing component of cullin-RING E3 ubiquitin ligases that localize to nuclear condensates.57 As predicted from the lack of the enriched degrons, PRRC2A is minimally turned over in hiPSCs (Figure 5D). SPOP has been characterized as both oncogenic or tumor suppressive in different contexts, including via the degradation of NANOG.58,59,60,61 While correlational in nature, our analysis prompts the hypothesis that SPOP functions to degrade multiple hiPSC proteins under baseline conditions, including PRRC2B but not its homolog PRRC2A, which can be readily tested via mutagenesis of degron sequences. More generally, we conclude that D2O labeling can be applied to a versatile human cell model to gain insights into the regulations of protein turnover.
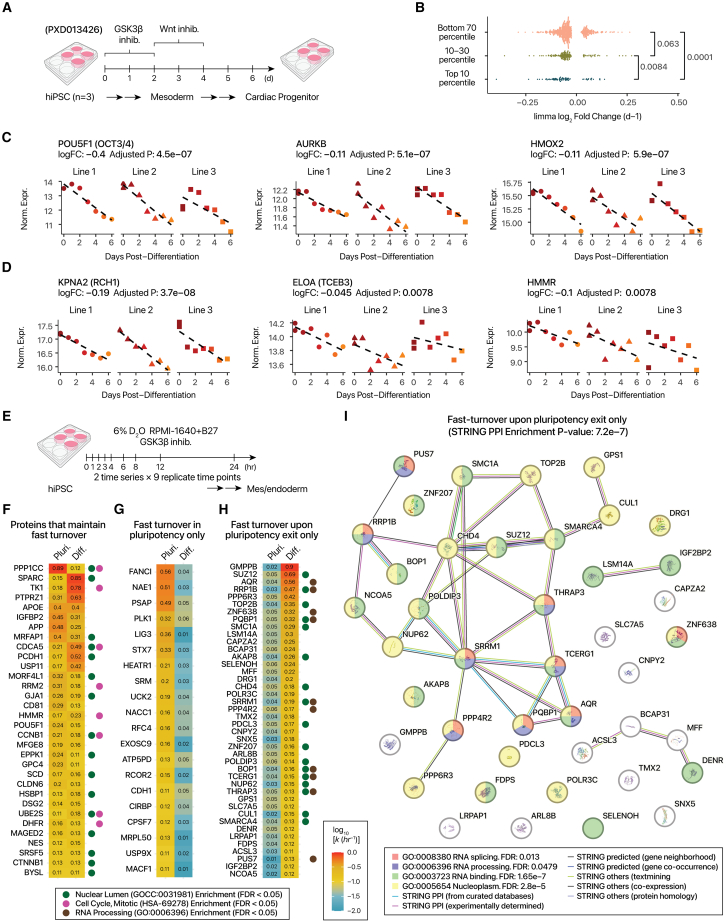
Hyperdynamic iPSC proteins are repressed upon exit of pluripotency
To further explore the roles of hyperdynamic proteins in pluripotency, we re-analyzed our previous data on hiPSC self-renewal exit and directed differentiation62 (Figure 6A). On a proteome level, fast-turnover hiPSC proteins have significantly more repressed expression during 6 days of directed differentiation to mesoderm and progenitor cells than slow-turnover proteins (Wilcoxon p: 5.6e–6), indicating that a number of fast-turnover proteins are preferentially depleted upon loss of pluripotency (Figure 6B). POU5F1 (OCT3/4) is a hyperdynamic protein and a master regulator of pluripotency, and its abundance decreased sharply upon differentiation (limma adjusted p < 0.01) (Figure 6C). Other hyperdynamic proteins, including aurora kinase B, which plays a role in chromosome segregation and chromatin remodeling, as well as heme oxygenase 2, likewise declined (Figure 6C). Moreover, several hyperdynamic proteins overlap with the list of genes required for permissive open chromatin in mouse embryonic stem cells reported by Bulut-Karslioglu et al.,13 including importin subunit alpha-1 (KPNA2), elongin-A, and hyaluronan mediated motility receptor. Thus, their excess turnover cycles may play a role in maintaining open chromatin in pluripotent cells, which become lost upon differentiation (Figure 6D).
Figure 6.
Regulation of fast-turnover hiPSC proteins upon differentiation and loss of pluripotency
(A) Comparison of fast-turnover hiPSC protein abundance upon loss of pluripotency. Data from three hiPSC lines (PXD013426) are analyzed.
(B) Decline in log2 abundance over days post-differentiation. Fast-turnover hiPSC proteins (top 10 percentile) are more likely to be depleted in mesoderm and progenitor stages. P: Wilcoxon rank-sum test.
(C) Example hyperdynamic proteins with decreased abundance upon hiPSC differentiation to mesoderm and progenitors (limma adjusted p < 0.05).
(D) Same as (C), but for hyperdynamic proteins known to maintain open chromatin in mouse pluripotent stem cells in Bulut-Karslioglu et al.13
(E) To examine protein turnover in early stages of pluripotency exit, hiPSC is labeled with 6% D2O concurrent with GSK3β inhibition in RPMI-1640 + B27 medium for mesendoderm induction. Samples are collected at 9 time points in 0–24 h post-labeling/differentiation.
(F) Heatmap of protein turnover rates among proteins that maintain fast turnover cycles (i.e., top 10 percentiles in pluripotent and differentiating cells). Colors denote log10 turnover rates in pluripotent or differentiating cells. Colored circles show proteins associated with significantly enriched terms (FDR < 0.05) in STRING.
(G) As in (E), but for proteins that become slow turnover upon differentiation (bottom 70 percentiles in differentiating cells).
(H) As in (F), but for proteins that become fast turnover upon differentiation.
(I) Gain of new fast-turnover proteins during early mesendoderm induction. STRING network showing a module of proteins that become hyperdynamic within hours of hiPSC-directed differentiation. Node colors: significantly enriched Reactome and Gene Ontology (GO) terms. Edge colors: STRING network relationships.
To interrogate how protein turnover changes during early pluripotency exit, we performed a new D2O labeling time course concurrent with directed mesendoderm differentiation over 24 h (Figure 6E). As the cells are in non-equilibrium, and some heterogeneity is expected in differentiating cell populations, we relaxed the peptide-level R2 cutoff to 0.7 and focused on 1,892 well-fitted proteins (Table S6). After classifying proteins as fast- (top 10 percentiles), moderate- (10th–30th percentiles), and slow-turnover (bottom 70 percentiles), we identified multiple distinct behaviors (Figures 6F–6H). Hyperdynamic proteins that retain fast turnover in differentiating hiPSCs include CDCA5 and CCNB1 (Figure 6F). These proteins form an interconnected STRING module significantly enriched in cell-cycle and nucleus proteins (Figure S6A). Other proteins are hyperdynamic in hiPSC but become slow turnover under differentiation, suggesting they are no longer synthesized and degraded in excess upon exit of pluripotency; while these proteins share no significantly enriched annotations (Figure S6B), we note a module of interconnected DNA repair and cell-cycle proteins FANCI, PLK1, RFC4, and LIG3 (Figures 6G and S8), indicating some cell-cycle regulators are no longer under active proteolytic regulation. A third category of proteins are minimally turned over (i.e., bottom 70 percentiles) in pluripotent hiPSCs but become hyperdynamic during differentiation to mesendoderm, which may be due to newly induced synthesis or rapid synthesis-degradation cycles (Figure 6H). Among these proteins are SRRM1, THRAP3, AQR, PQBP, and others enriched in RNA binding and splicing function (Figure 6I), suggesting protein regulation by active translation-degradation shifts rapidly from primarily regulating cell-cycle function toward post-transcriptional processes within hours of pluripotency exit. Thus, we identified cell-type and state-specific hyperdynamic proteins that may present prime targets for manipulating pluripotency and lineage determination.
D2O labeling enables secretome flux measurement in hiPSC-CM
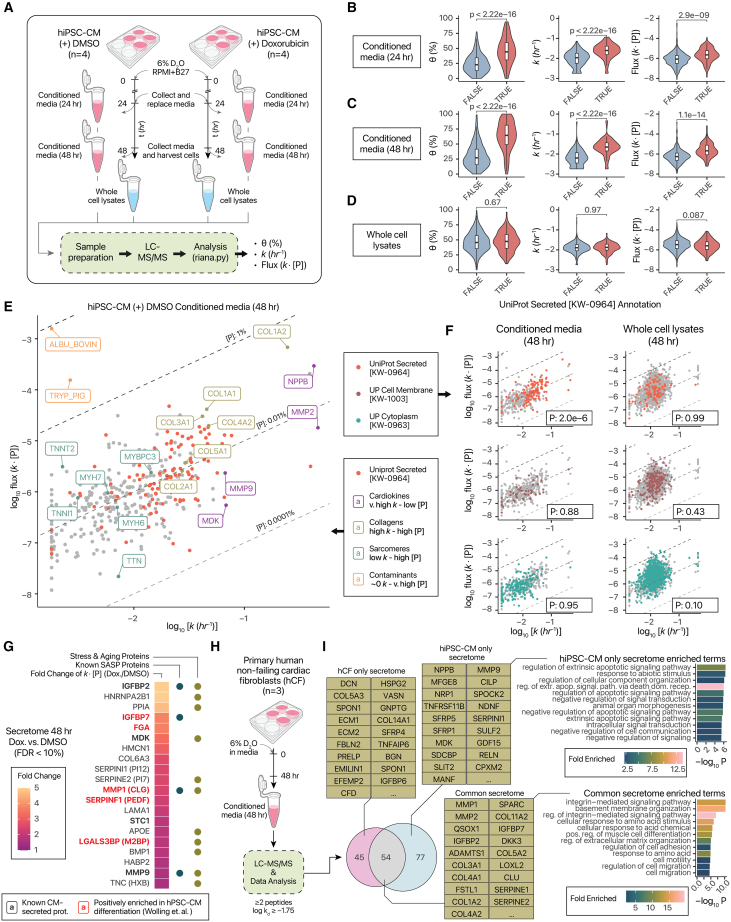
Lastly, we explored an application of D2O labeling to facilitate secreted protein analysis in cell culture. The secretome plays an important role in cellular communication and crosstalk, but existing MS methods are hindered by challenges of distinguishing true secreted proteins from background proteins such as those from culture media supplements, basement matrix, or passive leakage from cell lysis.63 Current strategies to circumvent these challenges may be incompletely bioorthogonal64 or require genetic manipulation65; thus, generalizable methods remain needed that can be applicable to different cell types and secretomes. Extracellular proteins often exhibit fast apparent k, not necessarily due to proteolytic degradation but because the existing protein pools are removed during media change, leading to immediate equilibration with background isotope enrichment. Fast apparent k should therefore distinguish secreted proteins from the bulk of high-abundance intracellular proteins that are not synthesized beyond cell-doubling needs.66,67,68 Hence, we applied D2O labeling to analyze cell culture secretomes, focusing on proteins secreted from terminally differentiated, non-dividing hiPSC-CMs at 24 and 48 h of labeling. The experimental and analysis workflow is then applied to calculate the fractional synthesis θ, turnover rate constant k of proteins, and flux (k × protein concentration [P]) (Figure 7A).
Figure 7.
Application of D2O labeling to secretome kinetics measurements
(A) Experimental plans to analyze the secretome of hiPSC-CM treated with doxorubicin or vehicle (DMSO) for 24–48 h.
(B) Violin/boxplots (interquartile range [IQR]/1.5-IQR) showing the fractional synthesis (left), k (center), and flux (right) of proteins in the conditioned media of hiPSC-CM after 24 h of labeling. Colors: UniProt Secreted (KW-0964) (red) vs. non-secreted proteins (blue).
(C) Same as (B), but for conditioned media collected at 48 h of labeling.
(D) Same as (B), but for whole-cell lysates at 48 h of labeling.
(E) Protein k vs. flux in the conditioned media of hiPSC-CM at 48 h. Red data points: UniProt Secreted (KW-0964). Label colors correspond to highlighted protein categories.
(F) Same as (E), but secreted (top), cell membrane (center), and cytoplasm (bottom) proteins are highlighted for conditioned media (left) and whole-cell lysates (right).
(G) Heatmap of proteins with higher flux following doxorubicin. Fill: fold-change (doxorubicin/DMSO). Bullets denote known CM-secreted, SASP, or stress proteins.
(H) Application to analyze the secretome of primary hCF.
(I) Overlap in secretomes of hCF and hiPSC-CM. Tables: example hiPSC-CM-only, hCF-only, and common secreted proteins. Bar charts: significantly enriched GO Biological Processes in hiPSC-CM-only vs. shared secretomes.
In conditioned media collected after 24 h, annotated secreted proteins (UniProt KW-0964) showed higher θ, k, and flux than non-secreted proteins (Figure 7B), a trend that continued in 48-h conditioned media (Figure 7C). Surprisingly, this difference is not apparent in whole-cell lysates, which may reflect cell-type or experimental design differences from hiPSCs (Figure 7D). Focusing on the 48-h conditioned media, we observed that turnover kinetics allowed ready distinction of extrinsic proteins bovine serum albumin (in the culture media) and porcine trypsin (used in protein digestion), both of which have very high [P] but ∼0 k, as no deuterium incorporation occurred (Figure 7E). Conversely, classically CM-secreted proteins, including NPPB, MDK, and MMP2, can be distinguished from intracellular sarcomeric proteins known to leak passively into the medium upon cell lysis, as the former have very high k and the latter high [P] but low k. Next, hiPSC-CM expresses multiple types of collagens,69 and we find that collagens have high k and [P] in the conditioned media (Figure 7E). The fast kinetics of secreted proteins is also reflected in the left-tailed distribution of annotated secreted proteins (bootstrap p: 2e–6), contrasted with annotated cell membrane and cytoplasmic proteins in conditioned media (Figure 7F). Thus, D2O labeling adds discriminant power to distinguish secreted proteins from intracellular or media contaminants, which can be exploited to identify high-flux secretome components. For instance, a heuristic cutoff of log10 k ≥ –1.75 covers ≥60% of UniProt secreted proteins found in the experiments while excluding sarcomeric proteins and non-cellular contaminants, leading to a 4.75-fold odds ratio in secreted vs. cytoplasm annotations. Using this cutoff, we compared the secretome flux of hiPSC-CM at baseline vs. treatment with doxorubicin (Figure S7), a chemotherapy agent that causes cardiac toxicity and senescence. We observe that doxorubicin treatment induced hiPSC-CM secretion of multiple proteins, including known CM secretomes,70 senescence-associated secretory phenotype (SASP) components IGFBP7 and MMP1, and stress-responsive proteins including MDK (FDR < 10%) (Figure 7G). Therefore, D2O labeling can facilitate the identification of perturbation-induced secretome flux in cell models.
Finally, we applied D2O labeling to primary hCFs, obtained from non-failing donor hearts not suitable for transplant, and labeled with D2O for 48 h (Figure 7H). In total, we identified 99 high-flux hCF-secreted proteins and 131 high-flux iPSC-CM proteins passing our criteria, 54 of which were common between two cell types (Table S7). Different classes of proteins are found between CM-secreted proteins and commonly secreted proteins. Proteins in cell death-regulating and signaling-related pathways are statistically overrepresented in hiPSC-CM-only secretomes, whereas common and hCF-only proteins are significantly enriched in extracellular matrix organization and integrin signaling-related proteins (Figure 7I). Thus, D2O labeling may also avail studies of cell-specific secretomes within an organ, so as to investigate cellular crosstalk and paracrine signaling.
Discussion
Recent-year methodological advances on analytical frameworks,37,71 alongside bioinformatics tools that enable automated data processing on a proteome scale,42,72,73,74 have enabled the use of D2O labeling for protein-wide turnover kinetics studies in animals53,75,76,77 that show comparable or higher performance than SILAC.73 In contrast, applications in cell culture have been less developed, with previous work focusing largely on the equilibration of D2O with alanine in cultured cells, in the assumption that labeling was limited to alanine and glycine and to an extent identical to that in adult animals (i.e., four deuterium atoms in alanine and two deuterium atoms in glycine).31 A commonly employed approach is to hydrolyze total proteins from cells or subcellular compartments, and then derivatize and measure the deuterium enrichment of released alanine using gas chromatography (GC)-MS.29,30,33,34 Although effective for total protein turnover, this method cannot account for individual protein-specific turnover rates. Alternatively, proteins of interest may be purified to measure their specific alanine incorporation,23 which reveals their individual turnover rates but does not scale easily to proteome-wide coverage. In landmark studies, Busch and colleagues demonstrated that peptide-specific turnover rates can be discerned in cell culture D2O labeling by using mass isotopomer distribution analysis to estimate per-peptide total labeling sites from their enrichment plateaus.31,35 Although these seminal works demonstrated the feasibility of analyzing individual peptide isotopomer data to derive protein-specific turnover rates, the analysis was limited to selected Ala/Gly-rich peptides within a handful of proteins of interest (isolated major histocompatibility complex class I/II proteins). Thus, proteome-wide investigations using D2O in cell culture remained to be established.
This study presents an updated method to perform proteome-wide turnover kinetics studies in cell culture using low-dose D2O medium dilution. Building on precedents of D2O labeling to study protein turnover, we address a critical gap to greatly expand its applicability in cell culture by determining the number of deuterium-accessible labeling sites across all proteinogenic amino acids in multiple cell lines. This yields a general strategy to acquire fractional protein synthesis values to calculate turnover rates from virtually any peptide in D2O-labeled proteomics data. Compared to dynamic SILAC, D2O offers the advantages of convenience and compatibility with standard cell culture conditions, eliminating the possible secondary effects of cycloheximide or the need for specialized SILAC medium formulations. As we show that D2O labels multiple non-essential amino acids, it can cover a wider selection of peptide sequences, especially where specific modification sites or alternative proteases are considered. The low cost of D2O also facilitates prolonged or large-scale labeling in certain experimental designs. For example, some primary cells have a long protein half-life approaching the protein lifetimes in animals,16 which require longer labeling to accumulate sufficient isotope signatures. Other studies, such as focusing on protein turnover in subcellular components,40 may require up to 108 cells per experiment that will likewise benefit from the scalability of D2O. Finally, the development and application of next-generation stem cell and organoid models rely heavily on commercial media and differentiation kits for batch consistency and reproducibility, which can be easily compatible with D2O labeling without formulation of multiple SILAC-compatible media.
Using this method, we mapped the proteome-wide turnover kinetics of pluripotent and differentiating hiPSCs. We identified a parsimonious protein synthesis landscape in hiPSCs, where the majority of hiPSC proteins are not synthesized beyond cell-doubling demand and are primarily removed via passive dilution to daughter cells. This contrasts with observations in animals, where median protein half-life (∼4 days in the mouse liver77) is far shorter than DNA half-life (>200 days), indicating that a typical protein pool is turned over many times in a cell’s lifetime. Mitochondrial proteins appear to have particularly minimal turnover in hiPSCs, consistent with hiPSCs’ having a high glycolytic rate while limiting the use of mitochondrial oxidative metabolism to maintain pluripotency.78 Conversely, our analysis also uncovers a number of fast-turnover proteins with half-lives as short as 1–3 h, indicating they are synthesized and degraded in excess beyond what is required by cell doubling. These hyperdynamic hiPSC proteins are enriched in cell-cycle regulation, DNA repair, and stress response. Many are not previously reported as short-lived and are quickly depleted upon the loss of hiPSC pluripotency. Our analysis further highlights that SPOP and APC/C degrons play a role in the synthesis-degradation cycles of fast-turnover hiPSC proteins that function in pluripotency and genome and proteome integrity. These observations echo prior work showing active translation and degradation of cell-cycle and chromatin regulators are required to maintain a permissive open chromatin, a hallmark of mouse PSCs.13 Fast-turnover hiPSC proteins may thus present prime targets for manipulating pluripotency-related processes via protein degradation.
In differentiating hiPSCs, we identified a shift in the hyperdynamic turnover of post-transcriptional regulators from primarily DNA-binding to RNA-binding proteins. Our observations are reminiscent of the prevalent translational/post-translational regulations of nuclear proteins previously described in differentiating mouse embryonic stem cells (i.e., their protein level changes were not well mirrored by RNA changes79), and suggests a mechanism of rapidly rewiring gene expression networks in differentiating cells that merits further exploration. Finally, we applied D2O labeling to analyze cellular secretomes in primary and differentiated cells. Fast label kinetics helps distinguish secretome candidates from contaminants and allows comparison of secretion flux across conditions. Such kinetic information may be useful for elucidating the biological contexts of protein secretion events (e.g., delayed kinetics may reflect stalling across secretory pathways).
Limitations of the study
The first limitation is that a low dose of D2O was used, which is within the range of previous works (4%–15%).30,31,32,34,77 While no effect on proliferation was observed, effects on cellular phenotypes need to be evaluated further, such as using gene expression profiling to ensure minimal perturbation.
Second, we assume rapid and constant equilibration rates of D2O with amino acids. This is established in animals, but in cell culture, it might be influenced by cell feeding, where media change can flood the cells with unlabeled amino acids.30 The influence of cell feeding time and schedule is not investigated. Minor differences in labeling sites exist across cell types that may be due to cellular biochemistry and media compositions. In slow-growing or non-immortalized cells, it may be infeasible to perform fully labeled calibration standards. Sites from other cells may be used that only approximate the true labeling extent. MS analysis in D2O labeling requires accurate measurement of the relative abundance of mass isotopomers within a peptide isotope envelope, which is sensitive to the spectral accuracy of instruments. More work is needed to examine accuracy and precision in different experimental setups.
Lastly, our calculation of protein k assumes constant protein abundance, where protein synthesis and degradation rates are equal. This assumption does not hold in the hiPSC differentiation or hiPSC-CM stressor experiments; hence, only the apparent kinetics of label incorporation is reported. Protein abundance may also drift during prolonged culturing, which was not investigated. The theoretical treatment and experimental measurements of non-equilibrium protein turnover is an emerging area.80,81 These limitations present promising areas of future development.
Resource availability
Lead contact
Requests for further information and resources should be directed to and will be fulfilled by the lead contact, Edward Lau (edward.lau@cuanschutz.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Raw MS data are available on ProteomeXchange under accession numbers ProteomeXchange: PXD048321, PXD060287, PXD060295.
-
•
Riana is open-source and available on GitHub at http://github.com/ed-lau/riana. A visualization web application for isotopomer envelopment is also available at http://heart.shinyapps.io/D2O_Isotope/. Archival DOIs for the versions used in this paper are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We thank Dr. R.J. Beynon at the University of Liverpool for helpful discussions and insights. We thank Drs. M.R. Bristow, J.A. Schwisow, and E. Jonas for assistance with procuring human heart samples and Drs. P. Buttrick, A. Ambardekar, and B. Kopecky and the University of Colorado’s Division of Cardiology for maintenance of the human cardiac tissue biobank. This work was supported in part by the National Heart, Lung, and Blood Institute (NHLBI) grant R01HL171711 and an American Heart Association Collaborative Sciences Award (24CSA1255857) to T.A.M.; NHLBI K99/R00HL166708 to J.G.T.; the National Institute of General Medical Sciences (NIGMS) R35GM146815 and NHLBI R00HL144829 to E.L.; and NHLBI R01HL141278, NIGMS R01GM144456, and the University of Colorado School of Medicine Advanced Proteomics Infrastructure (API) funds to M.P.Y.L. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Conceptualization, M.P.Y.L. and E.L.; methodology, M.P.Y.L. and E.L.; investigation, L.A., D.C.M.N., J.C., A.B., B.P., J.P., P.S., and J.G.T.; writing – original draft, M.P.Y.L. and E.L.; writing – review & editing, M.P.Y.L. and E.L.; funding acquisition, T.A.M., M.P.Y.L., and E.L.; resources, T.A.M., M.P.Y.L., and E.L.; supervision, M.P.Y.L. and E.L.
Declaration of interests
T.A.M. is a co-founder of Myracle Therapeutics, is on the scientific advisory boards of Eikonizo Therapeutics and Revier Therapeutics, received funding from Italfarmaco for an unrelated project, and has a subcontract from Eikonizo Therapeutics for an SBIR grant from the National Institutes of Health (HL154959).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| D2O (99.9%) | Cambridge Isotopes | DLM-4-10 |
| Thapsigargin | SelleckChem | S7895 |
| mTeSR Plus | STEMCELL Technologies | 100–0276 |
| DMEM/F12 | Gibco | 11320033 |
| Fetal bovine serum | Gibco | A5209402 |
| Phosphate buffered saline | Cytiva | BSS-PBS-1X6 |
| RPMI-1640 | Gibco | 11875093 |
| B-27 Supplement, minus insulin | Gibco | A3695201 |
| B-27 Supplement | Gibco | 17504044 |
| Y-27632 | SelleckChem | S1049 |
| FGM-3 Cardiac Fibroblast Growth Medium-3 Bullet Kit | Lonza | CC-4526 |
| Mass spectrometry grade trypsin | Promega | V5280 |
| RIPA Lysis and Extraction Buffer | Thermo | 89901 |
| Halt Protease and Phosphatase Inhibitor Cocktail | Thermo | 78430 |
| Ammonium Bicarbonate | Fisher Scientific | AC393210010 |
| Dithiothreitol | Fisher Scientific | PIA39255 |
| Iodoacetamide | Fisher Scientific | PIA39721 |
| 0.1% Formic acid | Fisher Scientific | LS118-500 |
| LC-MS grade anhydrous acetonitrile | Thermo | A9561 |
| CHIR-99021 | STEMCELL Technologies | 72052 |
| Doxorubicin | SelleckChem | S1208 |
| Deposited data | ||
| Mass spectrometry data of calibration and heavy water labeling in normal and stressed human AC16 cells | This study, Currie et al. 82 | ProteomeXchange: PXD048321 |
| Mass spectrometry data of hiPSC under dynamic heavy water labeling | This study | ProteomeXchange: PXD060287 |
| Mass spectrometry data of hiPSC-CM secretome | This study | ProteomeXchange: PXD060295 |
| Mass spectrometry data of protein abundance profiles during hiPSC differentiation | Lau et al. 62 | ProteomeXchange: PXD013426 |
| Mass spectrometry data of AC16 cells labeled with dynamic SILAC | Currie et al. 40 | ProteomeXchange: PXD038054 |
| Experimental models: Cell lines | ||
| AC16 Human Cardiomyocyte | Millipore | SCC109 RRID: CVCL_4U18 |
| hiPSC mono-allelic mEGFP-tagged MYL7 WTC iPSC line (tag at C-term) | Allen Institute/Coriell | AICS-0052-003 RRID: CVCL_UD15 |
| Primary human cardiac fibroblasts | University of Colorado Anschutz Medical Campus | COMIRB Protocol #01-568 |
| Software and algorithms | ||
| R | The R Project for Statistical Computing | https://www.r-project.org |
| Bioconductor | Bioconductor Project | https://www.bioconductor.org |
| Comet (v.2022.01) | Eng et al. 83 | https://github.com/UWPR/Comet |
| UniProt | The UniProt Consortium 84 | https://uniprot.org |
| DeepLoc (v.2.0) | Thumuluri et al. 85 | https://services.healthtech.dtu.dk/services/DeepLoc-2.0/ |
| Percolator (v.3.0) | The et al. 86 | https://crux.ms/ |
| MSFragger | Yu et al. 87 | https://msfragger.nesvilab.org |
| Philosopher (v.4.8.1) | da Veiga Leprevost et al. 88 | https://philosopher.nesvilab.org/ |
| Riana (v.0.8.0) | This study, Hammond et al. 73 and Currie et al. 82 |
https://github.com/ed-lau/riana https://doi.org/10.5281/zenodo.15643986 |
| Degronopedia | Szulc et al. 51 | https://degronopedia.com/ |
| Isotopomer visualizing webapp (v.0.1) | This study |
https://heart.shinyapps.io/D2O_Isotope/ https://doi.org/10.5281/zenodo.15652344 |
Experimental model and study participant details
Study approval and participants
All experiments utilizing human tissue were performed in accordance with COMIRB Protocol #01–568 at the University of Colorado Anschutz Medical Campus. Donor information was de-identified.
Human AC16 cell culture
Human AC16 cells (Millipore) were cultured in monolayer on tissue culture plates with DMEM/F12 medium (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) at 37°C, 5% CO2.
Human induced pluripotent stem cell culture
AICS-0052-003 hiPSC (mono-allelic C-terminus mEGFP-tagged MYL7 WTC-11; Allen Institute Cell Collection) line was acquired from Coriell Institute and seeded onto Geltrex (Gibco) coated 6 well plates and maintained in mTesR Plus basal media with Supplement (STEMCELL) media at 37°C, 5% CO2.
Human primary cardiac fibroblast culture
As previously described,89 primary human CFs were isolated from left ventricular tissue obtained from non-failing donor hearts not suitable for transplant by enzymatic digestion (100 mg collagenase type 2 [Worthington Biochemical Corporation]; 1 mg trypsin [Worthington TRL3]; and 15 mg bovine serum albumin [Sigma A5611] reconstituted in 40 mL of DMEM. Fibroblasts were maintained in FGM-3 Cardiac Fibroblast Growth Medium 3 BulletKit media (Lonza CC4526) and cultured to Passage 3. Fibroblasts were plated in 6-well plates at 80,000 cells/well and cultured for 2 days in FGM-3 medium at 37°C, 5% CO2 in low serum Fibroblast Basal Medium (CC-3131).
Method details
Human AC16 cell labeling calibration standards
The AC16 calibration standards data were initially generated for our parallel work to evaluate the effect of isotopomer selection on fractional synthesis calculation,37 and here utilized for an independent study goal. AC16 cells were cultured as described above. For labeling, the basal medium was diluted with either 6% vol/vol D2O (heavy labeled population) or 6% H2O (control population) at 37°C, 5% CO2. The cells were maintained in this medium for 3 passages, each passage with a split ratio of 1:8. This growth was estimated to constitute approximately 9 doublings of the cell populations. The cells were harvested by trypsinization, pelleted, washed once with phosphate buffered saline, and pelleted again before snap freezing in liquid nitrogen and storing at −80°C. At the time of processing each pellet was resuspended in 1 mL of RIPA buffer (Thermo Scientific) supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). Proteins were extracted with sonication in a Bioruptor Pico (Diagenode) with settings 10 × 30 s on 30 s off at 4°C. Insoluble debris was pelleted and removed from all samples by centrifugation at 14,000 ×g, 5 min.
Protein concentration of all samples was measured with the Rapid Gold BCA (Pierce) assay. Cell lysates from the D2O and H2O media populations were then combined in a labeling series expressed as the proportion of protein that was labeled with heavy water: 0, 0.125, 0.25, 0.375, 0.5, 0.625, 0.75, 0.875 and 1. The samples were trypsin digested using a modified version of the filter-aided sample preparation approach as previously described. A total of 50 μg protein per sample in 250 μL 8 M urea was loaded onto Pierce Protein Concentrators PES, 10K MWCO (Thermo Scientific) pre-washed with 100 mM ammonium bicarbonate (ABC). The samples were again washed with 8 M urea to denature proteins and remove SDS. The samples were washed with 300 μL 100 mM ABC twice. The samples were then reduced and alkylated with final concentrations 5 mM dithiothreitol (DTT) and 18 mM iodoacetamide (IAA) for 30 min at 37°C in the dark. DTT and IAA were removed with centrifugation and the samples were washed 3× with 100 mM ABC. Samples were digested atop the filters overnight at 37°C with mass spectrometry grade trypsin (Promega) at a ratio of 1:50 enzyme:protein. The following day samples were cleaned with Pierce C18 spin columns (Thermo Scientific) according to the manufacturer’s protocol. Eluted peptides were dried under vacuum and redissolved resuspended in 0.1% (vol/vol) formic acid.
The samples were analyzed on a Thermo Q-Exactive HF quadrupole-Orbitrap mass spectrometer coupled to a nanoflow Easy-nLC UPLC with the Thermo EasySpray electrospray ionization source. Peptides were separated with a PepMap RSLC C18 column 75 μm × 15 cm, 3 μm particle size (Thermo Scientific) with a 90-min gradient from 0 to 100% pH 2 solvent B (0.1% formic acid in 80% vol/vol LC-MS grade acetonitrile). The mass spectrometer was operated in data-dependent acquisition (DDA) mode with scans between m/z 200 and 1650 acquired at a mass resolution of 60,000. The maximum injection time was 20 ms, and the automatic gain control was set to 3e6. MS2 scans of the 15 most intense precursor ions with charge states of 2+ to 5+ were acquired with an isolation window of 2 m/z units, maximum injection time 110 ms, and automatic gain control of 2e5. Fragmentation of the peptides was by stepped normalized collision-induced dissociation energy (NCE) of 25–27. Dynamic exclusion of m/z values was used with an exclusion time of 30 s. The DDA mass spectrometry data were searched against UniProt Swiss-Prot database84 retrieved using Philosopher v.4.8.188 on 2023-06-27 with added contaminants using Comet v.2022.0183 including the following parameters: decoy_search = 1; peptide_mass_tolerance: 10.00 ppm; num_enzyme_termini = 1; isotope error: 0/1/2/3; fragment_bin_tol = 0.02; fragment_bin_offset = 0.0. Search results were post-processed using Percolator (crux-4.1 distribution)86 with the following options: --decoy-prefix DECOY_; --overwrite T; --maxiter 10. Peptide identifications at FDR adjusted q value of greater than 0.01 were excluded. Isotopomer intensity of the m0, m1, m2, m3, m4, and m5 peaks was extracted using Riana v.0.8.0 integrate73 with the following settings: -q (q-value) 0.01, -r (retention time in minutes) 0.25 -m (mass tolerance in ppm) 15.
Fractional synthesis calibration cells for hiPSC and hiPSC-CM
New calibration standards for hiPSC and hiPSC-CM were generated in this study. Briefly, AICS-0052-003 hiPSC were cultured and passaged as described above. The basal medium but not supplement was diluted with either 6% D2O (heavy labeled population) or 6% H2O (control population), and the cells were cultured at 37°C, 5% CO2 with daily media changes. At 80% confluency, cells were passaged at 1:6 using EDTA before resuspension in media supplemented with 10 μM Y-27632 (SelleckChem), for a total of 4 passages (equivalent to ≥10 doublings). The cells were then harvested by EDTA, processed, and analyzed by mass spectrometry as above.
To produce calibration cells for hiPSC-CM, fully labeled or unlabeled hiPSC were replated into Geltrex coated 12 well plates at a density of 4.5 × 105 cells/well and daily media changes continued until the cells reached 80% confluency, day 0 of cardiac differentiation. The hiPSC were differentiated into hiPSC-CM using a small molecule-based GSK-3β inhibition/Wnt inhibition protocol.90 Briefly, on day 0, cell media was replaced with 2 mL/well RPMI supplemented with B-27 minus insulin (Gibco) and 6 μM CHIR99021 (STEMCELL); on day 2, the media was changed to 2 mL/well RPMI+B-27 minus insulin. On day 3, the media was changed to 2 mL/well RPMI+B27 without insulin supplemented with 5 μM IWR-1-Endo (STEMCELL). On day 7, the media was changed to 2 mL RPMI+B27 with insulin. Differentiation was confirmed via visualization of morphology, spontaneous contraction of cells, and imaging of the GFP tagged MYL7/MLC-2a. Media was refreshed every other day with RPMI+B27 with insulin until day 14. Attached hiPSC-CMs were washed twice with PBS before incubating with 0.5 mL per well of TrypLE Express (1x, Thermo Fisher) for 11 min. An additional 1.5 mL PBS was added to dilute the TrypLE and the cells were triturated until fully detached. Cells were pelleted by centrifugation at 300 ×g for 3 min, washed with PBS before repelleting, and stored at −80°C until protein extraction. The samples were then processed and analyzed by mass spectrometry as above.
Prediction of deuterium labeling sites in cell culture
Because D2O labeling typically uses enrichment of 10% or lower excess deuterium, the resulting isotope pattern of labeled proteins is complex and overlaps with the unlabeled protein. Moreover, whereas in SILAC labeled amino acids are pre-synthesized with fixed mass shifts, upon D2O labeling, each amino acid residue has a characteristic number of deuterium-accessible labeling sites based on their biochemistry, hence the extent of mass shift a peptide exhibits upon labeling is sequence-dependent. Analysis therefore requires knowing the number of deuterium-accessible labeling sites in each amino acid, Saa. This information may be learned from asymptotically labeled peptides or nested fitting over many peptides, but this is not always practical to achieve (e.g., in short-term labeling or labeling of only few time-points) and may accrue additional greater fitting error, therefore calling for a general method to calculate the fractional synthesis of any peptide sequence irrespective of label duration.
To recover the fraction of newly synthesized protein θ (also referred to as f in the literature. We avoid the use of f due to potential confusion with other fractional or functional notations), isotope incorporation is analyzed as the change in the ratio between the monoisotopic peak over the complete isotope profile (A0) across labeling time-points (or in the case of the calibration experiment, across known experimental proportions of ground-truth θ). As fractional synthesis 0 ≤ θ ≤ 1 increases, A0(θ) decreases linearly from the initial position, toward the asymptote as determined by the excess deuterium enrichment (p) and number of accessible labeling sites of the peptides, Spep (sometimes denoted as n or NEH in the literature; Spep is used here to avoid confusion with number of replicates or the number of amino acids per peptide.)
The initial A0 (i.e., unlabeled, A0(θ = 0)) of a peptide can be calculated from natural isotopic distribution. The fully labeled A0(θ = 1) of the peptide is calculated from the naturally occurring A0, the total number of deuterium exchange sites on the peptide Spep, and the deuterium relative isotope abundance p where p = 0.06 in the 6% D2O experiments:
| (Equation 1) |
This could be refined by considering naturally occurring deuterium,91 but the background deuterium level (0.0001157098) is negligible compared to p and is ignored here.
In the calibration standard cells, Spep can be calculated from Equation 1 by considering the proportional abundance of the m0 peak, which contains no heavy isotopes in any atom center. This proportion decreases linearly as the proportion of labeled protein scales from 0 to 100%.
The number of deuterium accessible labeling sites in each amino acid, Saa, in human cell culture can be predicted by two machine learning strategies. In the first method, a linear regression model is used to find Saa from Spep:
| (Equation 2) |
Peptide series from the calibration standards that were identified at Percolator FDR 1% and quantified in all 9 mixture proportions were included. The peptides were then further filtered to include those with a linear fit (R2 > 0.9) of mixture proportion θ against A0. Training is done using the LinearRegression model in scikit-learn,92 with the settings fit_intercept = False, positive = True. following 80/20 train-test split with 1337 peptides in the training set and 335 in the test set.
In the second strategy (direct prediction of isotopomer profiles), the empirical isotopic ratios as measured in the 25.0%, 50.0%, 75.0%, and 100.0% proportion of fully labeled cell lysates were used as targets of training. The full isotopomer profile of an unlabeled peptide is calculated using the isotope fine structure calculation algorithm IsoSpec2 (ref. 38) to resolve exact isotopologues of a peptide given its chemical composition. The isotope envelope of a labeled peptide is then approximated by performing the calculation with a modified elementary composition table to include a new element H∗ (exchangeable hydrogen) with 1 – p probability of having mass 1.007825, and p probability of having mass 2.014102. A train-test split of test size 0.2 was performed, resulting in 7,200 samples (peptide–proportion combination) in the train sets and 1,800 samples in the test sets. We then used the differential evolution (DE) algorithm in scipy.optimize93 to perform global optimization varying the labeling site of each amino acid residue as input to IsoSpec2 and then minimizing the median absolute scaled errors (m.a.s.e., calculated as median absolute error divided by median absolute deviation) between the IsoSpec2 predicted isotopic cluster and the actual empirical ratios of m0/m1, m0/m2, m1/m2, and m1/m3 in each of the four mixture proportion experiments. The differential evolution parameters are tol: 1e-5, disp: True, polish: False, maxiter: 2500, seed: 42. Lower and upper bounds of labeling sites were set between 0.08 and the number of hydrogen atoms per amino acid residue, respectively.
Acid hydrolysis of AC16 lysate and direct infusion mass spectrometry
To experimentally validate the predicted labeling sites in cell culture, non-labeled (i.e., 0% D2O) and fully labeled (i.e., nine doublings under 6% D2O) AC16 cell lysates (∼200 μg) were desalted and cleaned up using Zeba desalting columns (Thermo) with 7 kDa molecular weight cutoffs (MWCO). The samples were then acid hydrolyzed with 1 mL of 6 M of HCl at 110°C for 48 h in a vacuum hydrolysis tube filled with N2 to prevent oxidation. The hydrolysate was removed under a steady stream of N2 in a water bath at 90°C and then dried using a SpeedVac evaporator (Thermo). The dried samples were reconstituted with 300 μL of methanol/water (50:50) with 1% vol/vol formic acid, and directly infused with a syringe into a Q-Exactive HF Orbitrap mass spectrometer through a Nanospray Flex ion source. The following mass spectrometry settings were used to acquire MS1 signal for the hydrolyzed amino acids: ESI voltage of +3 kV, injection flow rate of 3 μL/min, mass range of m/z 50–250, scan time of 1 min, FT resolution of 30,000, 60,000, and 120,000, capillary temperature of 275°C, AGC Target of 1e6, max ion time of 50 ms, micro scan of 1 count.
AC16 and hiPSC culture for dynamic D2O labeling experiments
For the baseline and stressed AC16 cells, AC16 cells were grown in monolayer as above. While in log phase growth, cells were introduced to 6% D2O with or without 1 μM thapsigargin for 16 to 24 h. For hiPSC, cells were labeled in 6% D2O in mTeSR Plus and then collected at 9 time-points at 0, 1, 2, 3, 4, 6, 8, 12, and 24 h after the start of labeling. The cells were then harvested, and processed for DDA mass spectrometry analysis as described above.
For hiPSC time-course labeling, AICS-0052-003 hiPSC were passaged as above. Once the cells reach 50% confluency, mTeSR Plus media diluted with 6% D2O (heavy labeled population) or 6% H2O (control population) was introduced to the cells for the duration of each time interval at 0, 1, 2, 3, 4, 6, 8, 12 and 24 h as described in the text. At the end of each time interval, the labeling medium was aspirated, the cells were washed with PBS, and then collected with dissociation media (0.5 mL EDTA in PBS), and snap-frozen immediately. Proteins were then extracted using RIPA buffer with BioRuptor sonication as described above, then digested and analyzed with DDA mass spectrometry.
Directed differentiation of hiPSC for dynamic D2O labeling experiments
For hiPSC differentiation toward mesoderm/mesendoderm specification, cells are placed under CHIR-99021 to induce prolonged GSK3β inhibition and Wnt activation. ACIS-52 hiPSC were plated onto a 6-well plate coated with geltrex in mTesR Plus basal media with Supplement (STEMCELL) media and 10 μM Y-27632 (SelleckChem). Media changes were performed every day with mTesR Plus basal media with Supplement (STEMCELL) media. At 80% confluency, cells were split with TrypLE Select Enzyme (10X, Thermo Fisher) onto geltrex coated 12-well plates at 0.2 × 106 cells/well in mTesR Plus basal media with Supplement (STEMCELL) media and 10 μM Y-27632 (SelleckChem). Media changes were performed every day with mTesR Plus basal media with Supplement (STEMCELL) media. At 80% confluency, the medium was removed and replaced with 2 mL/well RPMI-1640 supplemented with B-27 minus insulin (Gibco), 6 μM CHIR99021 (STEMCELL), and 6% vol/vol D2O (Cambridge Isotope), except for time-point 0. At each time-point of 0, 1, 2, 3, 4, 6, 8, 12 and 24 h, media was aspirated from cells and cells were collected with 100 μL TrypLE Select Enzyme (10X, Thermo Fisher) for 2 min, then quenched with 500 μL RPMI-1640. For each replicate in each condition, 2 wells were combined, pelleted at 200 ×g for 5 min, rinsed 1× with 1 mL PBS, pelleted once more at 200 ×g for 5 min and immediately flash frozen. Protein extraction, digestion and peptide desalting were performed using the same protocol described above.
The digested peptides were resuspended in 0.1% formic acid (vol/vol) and separated using a Vanquish Neo UHPLC system (Thermo Fisher Scientific) on an Easy-Spray analytical column (ES900 C18, 75μm × 150 mm, 3 μm) with a 60-min gradient. Solvent A was 0.1% formic acid in LCMS-grade water and solvent B was 0.1% Formic acid in 80% LCMS-grade acetonitrile. The LC-system was coupled to an Orbitrap Exploris 480 (Thermo Fisher Scientific) operating in positive mode with the spray voltage at 2 kV and ion transfer tube temperature of 275°C. Full-mass spectra were acquired from 350 to 1650 m/z at 60,000 resolution with an AGC target at 300% and maximum ion injection time set to auto. The mass spectrometer was operated in DDA mode, selecting precursors within a charge state of 2–6 and minimum intensity of 3 x 102. The top 20 precursors were subsequently selected with a 2 m/z isolation window for HCD fragmentation with NCE set at 30%. The resolution was set to 15,000 with an AGC target at 200%, maximum ion injection time of 26 ms and dynamic exclusion of 45 s. The data were then analyzed as above.
For directed differentiation toward cardiac progenitors, data from our previous work62 were retrieved from ProteomeXchange under PXD013426. Briefly, hiPSC from three donor lines were directed to differentiate using a small molecule based GSK3β inhibition–Wnt inhibition protocol, first toward mesoderm (day 0 to day 2 post differentiation) and then toward cardiac progenitor stage (day 2 to day 6), and protein expression was sampled daily.
Protein turnover kinetics analysis of mass spectrometry data
Isotopomer intensity was extracted using Riana v.0.8.0 to extract the intensity over time of the m0, m1, m2, m3, m4, and m5 peaks as described above. For each peptide, Spep is calculated from the predicted Saa values (Table S1) using Equation 2. The plateau enrichment of a peptide can then be predicted using a modified version of Equation 1, that is, for . The fractional synthesis of a peptide at time t can then be calculated as:
| (Equation 3) |
The time-series of fractional synthesis at one or more experimental time-points was then fitted to a simple exponential kinetic model (Equation 4) to obtain the best-fit turnover rate constant (k) to explain the time-series, using the optim function in R.
| (Equation 4) |
We note that because the determined D2O labeling sites enable the asymptote of mass shifts to be calculated, the label incorporation kinetics of a single protein can be defined by a single variable, namely the apparent turnover rate constant (k) of the protein. This allows rise-to-plateau kinetics to be analyzed from as few as one time-point, provided the sampling time-point is within the informative region of the curve (i.e., an appreciate amount of isotope has been incorporated, and the curve has not plateaued).76,94 In other words, the approach is compatible with both single-point sampling and time-course designs (with multiple data-points across multiple values of t). Although single-point sampling approaches are challenged by the high variability of derived turnover rates, it is useful for evaluating whether the isotope envelope is quantified accurately to reflect expected median half-life of proteins. For the AC16 single time-point labeling analysis, no goodness-of-fit can be acquired. Therefore the best-fit turnover rates of the top 6 peptides ranked by intensity were used to calculate the median as the protein-level turnover rate, and proteins quantified with at least 3 peptides and with a relative median absolute deviation (calculated as median absolute deviation divided by median of the turnover rates) of 33% or less were accepted.
Compared to single-point designs, time-course experiments enable a greater dynamic range of turnover rates to be measured, including fast turnover proteins that may have plateaued prior to sampling time. Moreover, the use of non-linear least square curve-fitting allows data filtering based on goodness-of-fit thresholds to remove problematic peptides that do not conform to the kinetic models (e.g., from low abundance peptides) (see below). For time-course labeling, goodness-of-fit is determined by the residual sum of squares (RSS) and total sum of squares (TSS) of the kinetic model curve-fitting:
| (Equation 5) |
A peptide-level R2 ≥ 0.8 is accepted as described in the text. All fractional synthesis data for all constituent peptides that passed this filter are then collected for a single protein-level fitting, where the sums of square weighted to a normalized log intensity of the peptide are minimized by the optim function in R. A protein-level weighted R2 ≥ 0.8 is accepted as described in the text.
Note that the reported turnover rate constants (k) represent apparent rates of isotope incorporation and do not distinguish the contributions from cell division rates, although this can be readily corrected in the kinetic model by subtracting the doubling rate from the rate constant.
Secretome analysis in D2O labeled hiPSC-CM
AICS-0052-003 hiPSC (mono-allelic C-terminus mEGFP-tagged MYL7 WTC-11; Allen Institute Cell Collection) were expanded and differentiated into hiPSC-CM as above. Media was refreshed every other day with RPMI+B27 with insulin until day 41, at which time the media was supplemented with 6% vol/vol D2O (Cambridge Isotope), and 1 μM doxorubicin (SelleckChem) or vehicle (DMSO). 24 h later, the media was collected and replaced with media supplemented with 6% vol/vol D2O. The collected conditioned media was centrifuged for 5 min at 300 ×g and 5 min at 14,000 ×g. Spun media was stored at −80°C. Another 24 h later (48 h of labeling total), the media was collected again and centrifuged as described. At the same time, the intracellular samples were collected and reduced, alkylated, and digested using an on-filter digestion protocol as described above.
To analyze secretome content, 300 μL of spun conditioned media per sample was depleted using the Seer Proteograph XT workflow (Seer, Inc.) utilizing two distinct nanoparticle (NP) mixtures (NPA, NPB). NP protein coronas were reduced, alkylated, and digested with Trypsin/Lys-C to generate tryptic peptides for LC-MS analysis. Digested peptides were desalted, eluted in a high-organic buffer into a deep-well collection plate, and quantified. In hiPSC-CM experiments, cleaned peptides were reconstituted in 0.1% formic acid to a final concentration of 0.0325 μg/μL and 0.116 μg/μL from NPA and NPB, respectively. 3 μL of peptides per sample were separated with online low-pH reversed-phase LC (PepMap C18 column, 3-μm particle, 100 Å pore; 75 μm × 15 cm; Thermo Fisher Scientific) via the EASYnLC 1200 system coupled to the Easy-Spray ion source (Thermo Fisher Scientific) at 300 nL/min with a 90-min gradient: 0–75 min: 0 to 30% B; 75–80 min: 30 to 70% B; 80–85 min: 70%–100% B; 85–90 min: 100% B (solvent A: 0.1% v/v formic acid; solvent B: 0.1% formic acid in 80% v/v acetonitrile). Mass spectra were acquired on a Thermo Scientific Q-Exactive HF Orbitrap mass spectrometer with the following settings: polarity, positive; data independent acquisition (DIA); MS resolution, 120,000; maximum ion injection time, 230 ms; MS automatic gain control (AGC) target, 3e6; normalized collision energy (NCE), 30; MS2 resolution, 30,000; MS2 maximum ion injection time, 45 ms; isolation window, 11 m/z; and MS2 AGC target, 1e6. DIA mass spectrometry data was searched using MSFragger v.3.887 using typical DIA settings against UniProt Swiss-Prot database84 retrieved using Philosopher v.4.8.188 on 2023-06-27 with added contaminants. The search results were post-processed using MSBooster v.1.1.695 followed by Percolator v.3. Peptide identification was accepted at 1% Percolator FDR. Mass isotopomer intensities were quantified using Riana and protein turnover rate constants calculated as described above.
Human primary fibroblast protein turnover kinetics and secretome analysis
Human primary fibroblasts were derived and cultured as described above, and subsequently cultured for 48 h in low serum Fibroblast Basal Medium (CC-3131) containing 6% D2O. This conditioned media was collected and processed with Seer Proteograph XT as above. Cleaned peptides from both NP mixtures were reconstituted in 0.1% formic acid to a final concentration of 0.1025 μg/μL. Resuspension concentrations were selected based on available mass of the digested peptides. In parallel, the intracellular samples were harvested and digested as described above. The samples were then analyzed using DIA mass spectrometry as above. Because primary cells are replicatively limited, they can only be cultured for a finite number of passages prior to senescence. We therefore did not pursue a fully labeled calibration standard, and instead opted to apply the deuterium accessible site values from hiPSC-CM toward hCF data analysis.
Additional data analysis
Subcellular localizations were predicted using DeepLoc v2.085 or UniProt84 annotations. Over-representation analysis and GSEA were performed using the ReactomePA96 and fgsea packages in R and using StringDB.97 Skewness of protein turnover rate distributions of secreted protein is calculated as Pearson’s moment coefficient of skewness, and the bootstrap significance is calculated as the proportion of samples with more skewed distributions among 1e6 random samples with replacement of the entire dataset, with sample sizes matched to the annotations being compared.
For degron set enrichment analysis, we retrieved 1,810 degron motifs from Degronopedia (2024-02-01 release).51 The degron motifs were curated on the Degronopedia server from multiple sources and included N-terminal, C-terminal, and internal degron motifs, that correspond to known E3 ubiquitin ligase targets as well as short linear motifs in disordered regions.51 The retrieved degron motifs are in regular expression, e.g., “R[A-Z]{2}G$” for the human C-terminus RxxG degron, which allowed us to loop through each motif and search for instances of motif matches within all quantified proteins in the.fasta protein database, using the str_count() function in the stringr package in R. We then filtered for 105 degron motifs that matched to 10 or more quantified proteins. Each degron motif, and the proteins with quantified turnover rates that it is found in, are then written in an MSigDB GMT format text file. Finally, we use the GMT file and the sorted protein list in descending turnover rates to perform GSEA, using the fgsea package in R, which looks for significant enrichment of each set of proteins associated with a degron in the ranked protein list. An FDR adjusted permutation p value of 0.1 is considered significant, and the degron and the associated leading edge proteins driving the enrichment score were retrieved.
Protein energetic cost is calculated using the energy cost per amino acid (ECPA) method with human/heterotroph H11 cost values.50 Protein copy number per cell is calculated using the proteomic ruler98 method from the MS1 label free quantity of peptides in labeled and unlabeled samples, integrated from m0 to m5 using Riana:
| (Equation 6) |
To verify the calculations, the derived protein copy numbers are compared with prior per-protein values in mouse embryonic fibroblasts 14 and histone proportion per cell. 98 The hiPSC calculations indicate 4.5% [4.1–4.8%] of histone mass as a total protein mass, roughly comparable to prior estimates in cancer cells (∼2–4%). The total protein mass and cell mass predicted from the proteomic ruler are also in range with values predicted from geometry. The proteomic ruler produces an estimate of hiPSC cell mass of 826 [736–916] ng (95% c.i.). For estimation from cell size, we assume hiPSC to be spherical cells between 10 and 12 μm in diameter. Dry mass is assumed to be 30% of cellular mass and protein mass 60% of total dry mass based on literature estimates.99 Assuming a cellular density of 1.06 g/cm3 yields cell mass of 0.555–0.959 ng, which is roughly in line with the proteomic ruler numbers.
Quantification and statistical analysis
Isotopomer intensity was extracted using Riana v.0.8.0 to extract the intensity over time of the m0, m1, m2, m3, m4, and m5 peaks as described above. Kinetics curve-fitting is performed using the optim function in R. Individual time-point samples contribute to the curve independently and are considered replicates. Peptide-level and protein-level R2 ≥ 0.8 are considered well fitted. For differential protein abundance analysis, the linear model and empirical bayes method in limma (Bioconductor v.3.21)100 is used and an FDR-adjusted p value of 0.01 is considered significant. For degron/gene set enrichment analyses, an FDR adjusted permutation p value of 0.1 is considered significant, and the degron/gene sets and the associated leading edge proteins driving the enrichment score are reported. Full details for proteomic quantification and calibration are reported in sections above.
Published: July 10, 2025
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2025.101104.
Contributor Information
Maggie P.Y. Lam, Email: maggie.lam@cuanschutz.edu.
Edward Lau, Email: edward.lau@cuanschutz.edu.
Supplemental information
References
- 1.Rayon T., Stamataki D., Perez-Carrasco R., Garcia-Perez L., Barrington C., Melchionda M., Exelby K., Lazaro J., Tybulewicz V.L.J., Fisher E.M.C., Briscoe J. Species-specific pace of development is associated with differences in protein stability. Science. 2020;369 doi: 10.1126/science.aba7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Francavilla C., Rigbolt K.T.G., Emdal K.B., Carraro G., Vernet E., Bekker-Jensen D.B., Streicher W., Wikström M., Sundström M., Bellusci S., et al. Functional Proteomics Defines the Molecular Switch Underlying FGF Receptor Trafficking and Cellular Outputs. Mol. Cell. 2013;51:707–722. doi: 10.1016/j.molcel.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Hanna J., Meides A., Zhang D.P., Finley D. A Ubiquitin Stress Response Induces Altered Proteasome Composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 4.Teixeira L.K., Reed S.I. Ubiquitin ligases and cell cycle control. Annu. Rev. Biochem. 2013;82:387–414. doi: 10.1146/annurev-biochem-060410-105307. [DOI] [PubMed] [Google Scholar]
- 5.Werner A., Manford A.G., Rape M. Ubiquitin-Dependent Regulation of Stem Cell Biology. Trends Cell Biol. 2017;27:568–579. doi: 10.1016/j.tcb.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eden E., Geva-Zatorsky N., Issaeva I., Cohen A., Dekel E., Danon T., Cohen L., Mayo A., Alon U. Proteome Half-Life Dynamics in Living Human Cells. Science. 2011;331:764–768. doi: 10.1126/science.1199784. [DOI] [PubMed] [Google Scholar]
- 7.Welle K.A., Zhang T., Hryhorenko J.R., Shen S., Qu J., Ghaemmaghami S. Time-resolved Analysis of Proteome Dynamics by Tandem Mass Tags and Stable Isotope Labeling in Cell Culture (TMT-SILAC) Hyperplexing. Mol. Cell. Proteomics. 2016;15:3551–3563. doi: 10.1074/mcp.M116.063230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varshavsky A. N-degron pathways. Proc. Natl. Acad. Sci. 2024;121 doi: 10.1073/pnas.2408697121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowan A.D., Ciulli A. Driving E3 Ligase Substrate Specificity for Targeted Protein Degradation: Lessons from Nature and the Laboratory. Annu. Rev. Biochem. 2022;91:295–319. doi: 10.1146/annurev-biochem-032620-104421. [DOI] [PubMed] [Google Scholar]
- 10.Lee J.M., Hammarén H.M., Savitski M.M., Baek S.H. Control of protein stability by post-translational modifications. Nat. Commun. 2023;14:201. doi: 10.1038/s41467-023-35795-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larance M., Ahmad Y., Kirkwood K.J., Ly T., Lamond A.I. Global Subcellular Characterization of Protein Degradation Using Quantitative Proteomics. Mol. Cell. Proteomics. 2013;12:638–650. doi: 10.1074/mcp.M112.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Cai Z., Vaites L.P., Shen N., Mitchell D.C., Huttlin E.L., Paulo J.A., Harry B.L., Gygi S.P. Proteome-wide mapping of short-lived proteins in human cells. Mol. Cell. 2021;81:4722–4735.e5. doi: 10.1016/j.molcel.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bulut-Karslioglu A., Macrae T.A., Oses-Prieto J.A., Covarrubias S., Percharde M., Ku G., Diaz A., McManus M.T., Burlingame A.L., Ramalho-Santos M. The Transcriptionally Permissive Chromatin State of Embryonic Stem Cells Is Acutely Tuned to Translational Output. Cell Stem Cell. 2018;22:369–383.e8. doi: 10.1016/j.stem.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 15.Boisvert F.-M., Ahmad Y., Gierliński M., Charrière F., Lamont D., Scott M., Barton G., Lamond A.I. A Quantitative Spatial Proteomics Analysis of Proteome Turnover in Human Cells. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathieson T., Franken H., Kosinski J., Kurzawa N., Zinn N., Sweetman G., Poeckel D., Ratnu V.S., Schramm M., Becher I., et al. Systematic analysis of protein turnover in primary cells. Nat. Commun. 2018;9:689. doi: 10.1038/s41467-018-03106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doherty M.K., Hammond D.E., Clague M.J., Gaskell S.J., Beynon R.J. Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome Res. 2009;8:104–112. doi: 10.1021/pr800641v. [DOI] [PubMed] [Google Scholar]
- 18.Woodside K.H. Effects of cycloheximide on protein degradation and gluconeogenesis in the perfused rat liver. Biochim. Biophys. Acta. 1976;421:70–79. doi: 10.1016/0304-4165(76)90170-7. [DOI] [PubMed] [Google Scholar]
- 19.Cheatham A.M., Sharma N.R., Satpute-Krishnan P. Competition for calnexin binding regulates secretion and turnover of misfolded GPI-anchored proteins. J. Cell Biol. 2023;222 doi: 10.1083/jcb.202108160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herath K., Bhat G., Miller P.L., Wang S.-P., Kulick A., Andrews-Kelly G., Johnson C., Rohm R.J., Lassman M.E., Previs S.F., et al. Equilibration of 2H labeling between body water and free amino acids: Enabling studies of proteome synthesis. Anal. Biochem. 2011;415:197–199. doi: 10.1016/j.ab.2011.04.031. [DOI] [PubMed] [Google Scholar]
- 21.Rachdaoui N., Austin L., Kramer E., Previs M.J., Anderson V.E., Kasumov T., Previs S.F. Measuring proteome dynamics in vivo: as easy as adding water? Mol. Cell. Proteomics. 2009;8:2653–2663. doi: 10.1074/mcp.M900026-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busch R., Kim Y.-K., Neese R.A., Schade-Serin V., Collins M., Awada M., Gardner J.L., Beysen C., Marino M.E., Misell L.M., Hellerstein M.K. Measurement of protein turnover rates by heavy water labeling of nonessential amino acids. Biochim. Biophys. Acta. 2006;1760:730–744. doi: 10.1016/j.bbagen.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 23.Fanara P., Banerjee J., Hueck R.V., Harper M.R., Awada M., Turner H., Husted K.H., Brandt R., Hellerstein M.K. Stabilization of Hyperdynamic Microtubules Is Neuroprotective in Amyotrophic Lateral Sclerosis. J. Biol. Chem. 2007;282:23465–23472. doi: 10.1074/jbc.M703434200. [DOI] [PubMed] [Google Scholar]
- 24.Fanara P., Wong P.-Y.A., Husted K.H., Liu S., Liu V.M., Kohlstaedt L.A., Riiff T., Protasio J.C., Boban D., Killion S., et al. Cerebrospinal fluid–based kinetic biomarkers of axonal transport in monitoring neurodegeneration. J. Clin. Investig. 2012;122:3159–3169. doi: 10.1172/JCI64575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drake J.C., Peelor F.F., Biela L.M., Watkins M.K., Miller R.A., Hamilton K.L., Miller B.F. Assessment of Mitochondrial Biogenesis and mTORC1 Signaling During Chronic Rapamycin Feeding in Male and Female Mice. J. Gerontol. Ser. A. 2013;68:1493–1501. doi: 10.1093/gerona/glt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Previs S.F., Fatica R., Chandramouli V., Alexander J.C., Brunengraber H., Landau B.R. Quantifying rates of protein synthesis in humans by use of 2H2O: application to patients with end-stage renal disease. Am. J. Physiol. Endocrinol. Metab. 2004;286:E665–E672. doi: 10.1152/ajpendo.00271.2003. [DOI] [PubMed] [Google Scholar]
- 27.Price J.C., Holmes W.E., Li K.W., Floreani N.A., Neese R.A., Turner S.M., Hellerstein M.K. Measurement of human plasma proteome dynamics with 2H2O and liquid chromatography tandem mass spectrometry. Anal. Biochem. 2012;420:73–83. doi: 10.1016/j.ab.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Wilkinson D.J., Franchi M.V., Brook M.S., Narici M.V., Williams J.P., Mitchell W.K., Szewczyk N.J., Greenhaff P.L., Atherton P.J., Smith K. A validation of the application of D2 O stable isotope tracer techniques for monitoring day-to-day changes in muscle protein subfraction synthesis in humans. Am. J. Physiol. Endocrinol. Metab. 2014;306:E571–E579. doi: 10.1152/ajpendo.00650.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller B.F., Wolff C.A., Peelor F.F., Shipman P.D., Hamilton K.L. Modeling the contribution of individual proteins to mixed skeletal muscle protein synthetic rates over increasing periods of label incorporation. J. Appl. Physiol. 2015;118:655–661. doi: 10.1152/japplphysiol.00987.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dufner D.A., Bederman I.R., Brunengraber D.Z., Rachdaoui N., Ismail-Beigi F., Siegfried B.A., Kimball S.R., Previs S.F. Using 2 H 2 O to study the influence of feeding on protein synthesis: effect of isotope equilibration in vivo vs. in cell culture. Am. J. Physiol. Endocrinol. Metab. 2005;288:E1277–E1283. doi: 10.1152/ajpendo.00580.2004. [DOI] [PubMed] [Google Scholar]
- 31.De Riva A., Deery M.J., McDonald S., Lund T., Busch R. Measurement of protein synthesis using heavy water labeling and peptide mass spectrometry: Discrimination between major histocompatibility complex allotypes. Anal. Biochem. 2010;403:1–12. doi: 10.1016/j.ab.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller B.F., Reid J.J., Price J.C., Lin H.-J.L., Atherton P.J., Smith K. CORP: The use of deuterated water for the measurement of protein synthesis. J. Appl. Physiol. 2020;128:1163–1176. doi: 10.1152/japplphysiol.00855.2019. [DOI] [PubMed] [Google Scholar]
- 33.Bruns D.R., McNair B.D., Peelor F.F., Borowik A.K., Pranay A., Yusifov A., Miller B.F. Skeletal and cardiac muscle have different protein turnover responses in a model of right heart failure. GeroScience. 2023;45:2545–2557. doi: 10.1007/s11357-023-00777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolff C.A., Reid J.J., Musci R.V., Linden M.A., Konopka A.R., Peelor F.F., Miller B.F., Hamilton K.L., Bruns D.R. Differential Effects of Rapamycin and Metformin in Combination With Rapamycin on Mechanisms of Proteostasis in Cultured Skeletal Myotubes. J. Gerontol. A Biol. Sci. Med. Sci. 2020;75:32–39. doi: 10.1093/gerona/glz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prevosto C., Usmani M.F., McDonald S., Gumienny A.M., Key T., Goodman R.S., Gaston J.S.H., Deery M.J., Busch R. Allele-Independent Turnover of Human Leukocyte Antigen (HLA) Class Ia Molecules. PLoS One. 2016;11 doi: 10.1371/journal.pone.0161011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Commerford S.L., Carsten A.L., Cronkite E.P. The distribution of tritium among the amino acids of proteins obtained from mice exposed to tritiated water. Radiat. Res. 1983;94:151–155. [PubMed] [Google Scholar]
- 37.Currie J., Ng D.C.M., Pandi B., Black A., Manda V., Pavelka J., Lam M.P.Y., Lau E. Improved determination of protein turnover rate with heavy water labeling by mass isotopomer ratio selection. bioRxiv. 2024 doi: 10.1101/2024.06.04.597043. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Łącki M.K., Valkenborg D., Startek M.P. IsoSpec2: Ultrafast Fine Structure Calculator. Anal. Chem. 2020;92:9472–9475. doi: 10.1021/acs.analchem.0c00959. [DOI] [PubMed] [Google Scholar]
- 39.Rutherfurd S.M., Gilani G.S. Amino Acid Analysis. Curr. Protoc. Protein Sci. 2009;Chapter 11:11.9.1–11.9.37. doi: 10.1002/0471140864.ps1109s58. [DOI] [PubMed] [Google Scholar]
- 40.Currie J., Manda V., Robinson S.K., Lai C., Agnihotri V., Hidalgo V., Ludwig R.W., Zhang K., Pavelka J., Wang Z.V., et al. Simultaneous proteome localization and turnover analysis reveals spatiotemporal features of protein homeostasis disruptions. Nat. Commun. 2024;15:2207. doi: 10.1038/s41467-024-46600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hua Y., Yoshimochi K., Li J., Takekita K., Shimotsuma M., Li L., Qu X., Zhang J., Sawa Y., Liu L., Miyagawa S. Development and evaluation of a novel xeno-free culture medium for human-induced pluripotent stem cells. Stem Cell Res. Ther. 2022;13:223. doi: 10.1186/s13287-022-02879-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau E., Cao Q., Ng D.C.M., Bleakley B.J., Dincer T.U., Bot B.M., Wang D., Liem D.A., Lam M.P.Y., Ge J., Ping P. A large dataset of protein dynamics in the mammalian heart proteome. Sci. Data. 2016;3 doi: 10.1038/sdata.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kluever V., Russo B., Mandad S., Kumar N.H., Alevra M., Ori A., Rizzoli S.O., Urlaub H., Schneider A., Fornasiero E.F. Protein lifetimes in aged brains reveal a proteostatic adaptation linking physiological aging to neurodegeneration. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abn4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hosios A.M., Hecht V.C., Danai L.V., Johnson M.O., Rathmell J.C., Steinhauser M.L., Manalis S.R., Vander Heiden M.G. Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev. Cell. 2016;36:540–549. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y., Beyer A., Aebersold R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 46.Dekel E., Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 47.Zecha J., Meng C., Zolg D.P., Samaras P., Wilhelm M., Kuster B. Peptide Level Turnover Measurements Enable the Study of Proteoform Dynamics. Mol. Cell. Proteomics. 2018;17:974–992. doi: 10.1074/mcp.RA118.000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dill K.A., Ghosh K., Schmit J.D. Physical limits of cells and proteomes. Proc. Natl. Acad. Sci. USA. 2011;108:17876–17882. doi: 10.1073/pnas.1114477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aspromonte M.C., Nugnes M.V., Quaglia F., Bouharoua A., DisProt Consortium. Tosatto S.C.E., Piovesan D., Chasapi A., Fichó E., Balatti G.E. DisProt in 2024: improving function annotation of intrinsically disordered proteins. Nucleic Acids Res. 2024;52:D434–D441. doi: 10.1093/nar/gkad928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H., Wang Y., Li J., Chen H., He X., Zhang H., Liang H., Lu J. Biosynthetic energy cost for amino acids decreases in cancer evolution. Nat. Commun. 2018;9:4124. doi: 10.1038/s41467-018-06461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szulc N.A., Stefaniak F., Piechota M., Soszyńska A., Piórkowska G., Cappannini A., Bujnicki J.M., Maniaci C., Pokrzywa W. DEGRONOPEDIA: a web server for proteome-wide inspection of degrons. Nucleic Acids Res. 2024;52:W221–W232. doi: 10.1093/nar/gkae238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emenecker R.J., Griffith D., Holehouse A.S. Metapredict: a fast, accurate, and easy-to-use predictor of consensus disorder and structure. Biophys. J. 2021;120:4312–4319. doi: 10.1016/j.bpj.2021.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim T.-Y., Wang D., Kim A.K., Lau E., Lin A.J., Liem D.A., Zhang J., Zong N.C., Lam M.P.Y., Ping P. Metabolic labeling reveals proteome dynamics of mouse mitochondria. Mol. Cell. Proteomics. 2012;11:1586–1594. doi: 10.1074/mcp.M112.021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahdessian D., Cesnik A.J., Gnann C., Danielsson F., Stenström L., Arif M., Zhang C., Le T., Johansson F., Schutten R., et al. Spatiotemporal dissection of the cell cycle with single-cell proteogenomics. Nature. 2021;590:649–654. doi: 10.1038/s41586-021-03232-9. [DOI] [PubMed] [Google Scholar]
- 55.Kumar M., Michael S., Alvarado-Valverde J., Mészáros B., Sámano-Sánchez H., Zeke A., Dobson L., Lazar T., Örd M., Nagpal A., et al. The Eukaryotic Linear Motif resource: 2022 release. Nucleic Acids Res. 2022;50:D497–D508. doi: 10.1093/nar/gkab975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhuang M., Calabrese M.F., Liu J., Waddell M.B., Nourse A., Hammel M., Miller D.J., Walden H., Duda D.M., Seyedin S.N., et al. Structures of SPOP-Substrate Complexes: Insights into Molecular Architectures of BTB-Cul3 Ubiquitin Ligases. Mol. Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bouchard J.J., Otero J.H., Scott D.C., Szulc E., Martin E.W., Sabri N., Granata D., Marzahn M.R., Lindorff-Larsen K., Salvatella X., et al. Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol. Cell. 2018;72:19–36.e8. doi: 10.1016/j.molcel.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Theurillat J.-P.P., Udeshi N.D., Errington W.J., Svinkina T., Baca S.C., Pop M., Wild P.J., Blattner M., Groner A.C., Rubin M.A., et al. Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science. 2014;346:85–89. doi: 10.1126/science.1250255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blattner M., Liu D., Robinson B.D., Huang D., Poliakov A., Gao D., Nataraj S., Deonarine L.D., Augello M.A., Sailer V., et al. SPOP Mutation Drives Prostate Tumorigenesis In Vivo through Coordinate Regulation of PI3K/mTOR and AR Signaling. Cancer Cell. 2017;31:436–451. doi: 10.1016/j.ccell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang P., Wang D., Zhao Y., Ren S., Gao K., Ye Z., Wang S., Pan C.-W., Zhu Y., Yan Y., et al. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat. Med. 2017;23:1055–1062. doi: 10.1038/nm.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li G., Ci W., Karmakar S., Chen K., Dhar R., Fan Z., Guo Z., Zhang J., Ke Y., Wang L., et al. SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer Cell. 2014;25:455–468. doi: 10.1016/j.ccr.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lau E., Han Y., Williams D.R., Thomas C.T., Shrestha R., Wu J.C., Lam M.P.Y. Splice-Junction-Based Mapping of Alternative Isoforms in the Human Proteome. Cell Rep. 2019;29:3751–3765.e5. doi: 10.1016/j.celrep.2019.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knecht S., Eberl H.C., Kreisz N., Ugwu U.J., Starikova T., Kuster B., Wilhelm S. An Introduction to Analytical Challenges, Approaches, and Applications in Mass Spectrometry–Based Secretomics. Mol. Cell. Proteomics. 2023;22 doi: 10.1016/j.mcpro.2023.100636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirschner F., Arnold-Schild D., Leps C., Łącki M.K., Klein M., Chen Y., Ludt A., Marini F., Kücük C., Stein L., et al. Modulation of cellular transcriptome and proteome composition by azidohomoalanine—implications on click chemistry–based secretome analysis. J. Mol. Med. 2023;101:855–867. doi: 10.1007/s00109-023-02333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim K.E., Park I., Kim J., Kang M.-G., Choi W.G., Shin H., Kim J.-S., Rhee H.-W., Suh J.M. Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice. Nat. Commun. 2021;12:5204. doi: 10.1038/s41467-021-25546-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hammond D.E., Kumar J.D., Raymond L., Simpson D.M., Beynon R.J., Dockray G.J., Varro A. Stable Isotope Dynamic Labeling of Secretomes (SIDLS) Identifies Authentic Secretory Proteins Released by Cancer and Stromal Cells. Mol. Cell. Proteomics. 2018;17:1837–1849. doi: 10.1074/mcp.TIR117.000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roelofsen H., Dijkstra M., Weening D., De Vries M.P., Hoek A., Vonk R.J. Comparison of Isotope-labeled Amino Acid Incorporation Rates (CILAIR) Provides a Quantitative Method to Study Tissue Secretomes. Mol. Cell. Proteomics. 2009;8:316–324. doi: 10.1074/mcp.M800254-MCP200. [DOI] [PubMed] [Google Scholar]
- 68.Kristensen L.P., Chen L., Nielsen M.O., Qanie D.W., Kratchmarova I., Kassem M., Andersen J.S. Temporal Profiling and Pulsed SILAC Labeling Identify Novel Secreted Proteins During Ex Vivo Osteoblast Differentiation of Human Stromal Stem Cells. Mol. Cell. Proteomics. 2012;11:989–1007. doi: 10.1074/mcp.M111.012138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grancharova T., Gerbin K.A., Rosenberg A.B., Roco C.M., Arakaki J.E., DeLizo C.M., Dinh S.Q., Donovan-Maiye R.M., Hirano M., Nelson A.M., et al. A comprehensive analysis of gene expression changes in a high replicate and open-source dataset of differentiating hiPSC-derived cardiomyocytes. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-94732-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wolling H., Konze S.A., Höfer A., Erdmann J., Pich A., Zweigerdt R., Buettner F.F.R. Quantitative Secretomics Reveals Extrinsic Signals Involved in Human Pluripotent Stem Cell Cardiomyogenesis. Proteomics. 2018;18 doi: 10.1002/pmic.201800102. [DOI] [PubMed] [Google Scholar]
- 71.Deberneh H.M., Abdelrahman D.R., Verma S.K., Linares J.J., Murton A.J., Russell W.K., Kuyumcu-Martinez M.N., Miller B.F., Sadygov R.G. Quantifying label enrichment from two mass isotopomers increases proteome coverage for in vivo protein turnover using heavy water metabolic labeling. Commun. Chem. 2023;6:72. doi: 10.1038/s42004-023-00873-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sadygov R.G., Avva J., Rahman M., Lee K., Ilchenko S., Kasumov T., Borzou A. d2ome, Software for in Vivo Protein Turnover Analysis Using Heavy Water Labeling and LC-MS, Reveals Alterations of Hepatic Proteome Dynamics in a Mouse Model of NAFLD. J. Proteome Res. 2018;17:3740–3748. doi: 10.1021/acs.jproteome.8b00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hammond D.E., Simpson D.M., Franco C., Wright Muelas M., Waters J., Ludwig R.W., Prescott M.C., Hurst J.L., Beynon R.J., Lau E. Harmonizing Labeling and Analytical Strategies to Obtain Protein Turnover Rates in Intact Adult Animals. Mol. Cell. Proteomics. 2022;21 doi: 10.1016/j.mcpro.2022.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Naylor B.C., Porter M.T., Wilson E., Herring A., Lofthouse S., Hannemann A., Piccolo S.R., Rockwood A.L., Price J.C. DeuteRater: a tool for quantifying peptide isotope precision and kinetic proteomics. Bioinformatics. 2017;33:1514–1520. doi: 10.1093/bioinformatics/btx009. [DOI] [PubMed] [Google Scholar]
- 75.Price J.C., Khambatta C.F., Li K.W., Bruss M.D., Shankaran M., Dalidd M., Floreani N.A., Roberts L.S., Turner S.M., Holmes W.E., Hellerstein M.K. The Effect of Long Term Calorie Restriction on in Vivo Hepatic Proteostatis: A Novel Combination of Dynamic and Quantitative Proteomics. Mol. Cell. Proteomics. 2012;11:1801–1814. doi: 10.1074/mcp.M112.021204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lam M.P.Y., Wang D., Lau E., Liem D.A., Kim A.K., Ng D.C.M., Liang X., Bleakley B.J., Liu C., Tabaraki J.D., et al. Protein kinetic signatures of the remodeling heart following isoproterenol stimulation. J. Clin. Investig. 2014;124:1734–1744. doi: 10.1172/JCI73787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deberneh H.M., Abdelrahman D.R., Verma S.K., Linares J.J., Murton A.J., Russell W.K., Kuyumcu-Martinez M.N., Miller B.F., Sadygov R.G. A large-scale LC-MS dataset of murine liver proteome from time course of heavy water metabolic labeling. Sci. Data. 2023;10:635. doi: 10.1038/s41597-023-02537-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Varum S., Rodrigues A.S., Moura M.B., Momcilovic O., Easley C.A., Ramalho-Santos J., Van Houten B., Schatten G. Energy Metabolism in Human Pluripotent Stem Cells and Their Differentiated Counterparts. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu R., Markowetz F., Unwin R.D., Leek J.T., Airoldi E.M., MacArthur B.D., Lachmann A., Rozov R., Ma’ayan A., Boyer L.A., et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin B., Suter D.M. An out-of-equilibrium definition of protein turnover. Bioessays. 2023;45 doi: 10.1002/bies.202200209. [DOI] [PubMed] [Google Scholar]
- 81.Kobak K.A., Lawrence M.M., Pharaoh G., Borowik A.K., Peelor F.F., Shipman P.D., Griffin T.M., Van Remmen H., Miller B.F. Determining the contributions of protein synthesis and breakdown to muscle atrophy requires non-steady-state equations. J. Cachexia Sarcopenia Muscle. 2021;12:1764–1775. doi: 10.1002/jcsm.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Currie J., Ng D.C.M., Pandi B., Black A., Manda V., Durham C., Pavelka J., Lam M.P.Y., Lau E. Improved Method to Determine Protein Turnover Rates with Heavy Water Labeling by Mass Isotopomer Ratio Selection. J. Proteome Res. 2025;24:1992–2005. doi: 10.1021/acs.jproteome.4c01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eng J.K., Hoopmann M.R., Jahan T.A., Egertson J.D., Noble W.S., MacCoss M.J. A deeper look into Comet--implementation and features. J. Am. Soc. Mass Spectrom. 2015;26:1865–1874. doi: 10.1007/s13361-015-1179-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.The U.P.C., Bateman A., Martin M.-J., Orchard S., Magrane M., Ahmad S., Alpi E., Bowler-Barnett E.H., Britto R., Bye-A-Jee H., et al. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023;51:D523–D531. doi: 10.1093/nar/gkac1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thumuluri V., Almagro Armenteros J.J., Johansen A.R., Nielsen H., Winther O. DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022;50:W228–W234. doi: 10.1093/nar/gkac278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.The M., MacCoss M.J., Noble W.S., Käll L. Fast and Accurate Protein False Discovery Rates on Large-Scale Proteomics Data Sets with Percolator 3.0. J. Am. Soc. Mass Spectrom. 2016;27:1719–1727. doi: 10.1007/s13361-016-1460-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu F., Teo G.C., Kong A.T., Fröhlich K., Li G.X., Demichev V., Nesvizhskii A.I. Analysis of DIA proteomics data using MSFragger-DIA and FragPipe computational platform. Nat. Commun. 2023;14:4154. doi: 10.1038/s41467-023-39869-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.da Veiga Leprevost F., Haynes S.E., Avtonomov D.M., Chang H.-Y., Shanmugam A.K., Mellacheruvu D., Kong A.T., Nesvizhskii A.I. Philosopher: a versatile toolkit for shotgun proteomics data analysis. Nat. Methods. 2020;17:869–870. doi: 10.1038/s41592-020-0912-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rubino M., Travers J.G., Headrick A.L., Enyart B.T., Lemieux M.E., Cavasin M.A., Schwisow J.A., Hardy E.J., Kaltenbacher K.J., Felisbino M.B., et al. Inhibition of Eicosanoid Degradation Mitigates Fibrosis of the Heart. Circ. Res. 2023;132:10–29. doi: 10.1161/CIRCRESAHA.122.321475. [DOI] [PubMed] [Google Scholar]
- 90.Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M., et al. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sadygov R.G. Using Heavy Mass Isotopomers for Protein Turnover in Heavy Water Metabolic Labeling. J. Proteome Res. 2021;20:2035–2041. doi: 10.1021/acs.jproteome.0c00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 93.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ilchenko S., Haddad A., Sadana P., Recchia F.A., Sadygov R.G., Kasumov T. Calculation of the Protein Turnover Rate Using the Number of Incorporated 2H Atoms and Proteomics Analysis of a Single Labeled Sample. Anal. Chem. 2019;91:14340–14351. doi: 10.1021/acs.analchem.9b02757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang K.L., Yu F., Teo G.C., Li K., Demichev V., Ralser M., Nesvizhskii A.I. MSBooster: improving peptide identification rates using deep learning-based features. Nat. Commun. 2023;14:4539. doi: 10.1038/s41467-023-40129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu G., He Q.-Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016;12:477–479. doi: 10.1039/C5MB00663E. [DOI] [PubMed] [Google Scholar]
- 97.Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S., et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wiśniewski J.R., Hein M.Y., Cox J., Mann M. A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteomics MCP. 2014;13:3497–3506. doi: 10.1074/mcp.M113.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feijó Delgado F., Cermak N., Hecht V.C., Son S., Li Y., Knudsen S.M., Olcum S., Higgins J.M., Chen J., Grover W.H., Manalis S.R. Intracellular Water Exchange for Measuring the Dry Mass, Water Mass and Changes in Chemical Composition of Living Cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0067590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw MS data are available on ProteomeXchange under accession numbers ProteomeXchange: PXD048321, PXD060287, PXD060295.
-
•
Riana is open-source and available on GitHub at http://github.com/ed-lau/riana. A visualization web application for isotopomer envelopment is also available at http://heart.shinyapps.io/D2O_Isotope/. Archival DOIs for the versions used in this paper are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.