Abstract
Tripartite motif-containing 3 (TRIM 3), as a vital member of TRIM family, has been receiving significant attention in cancer research. This research aims to detect the effect and relevant molecular functions of TRIM3 in melanoma cells.GEPIA website and immunohistochemical analysis were performed to investigate TRIM3 level in melanoma samples. The growth ability, the mobility, and the apoptosis of M14 and A375 cells were determined by biological experiments. qPCR and western blot assays were performed to evaluate the expression of key genes. The interaction between TRIM3 and YAP1 was verified by immunoprecipitation and ubiquitination assays.A reduction of TRIM3 was observed in melanoma samples, and the loss of TRIM3 strengthened the proliferation and mobility of M14 and A375 cells and diminished apoptosis, and vice versa. TRIM3 expression only affected the protein level of YAP1, while YAP1 mRNA was not changed. Then, we demonstrated that TRIM3 directly interacted with YAP1, and TRIM3 reduced YAP1 stability by inducing ubiquitination modification. Finally, rescue assays showed that si-YAP1 treatment alleviated the effects of si-TRIM3 on melanoma cells.This study indicated that depletion of TRIM3 strengthened the proliferation and mobility of M14 and A375 cells, suppressed cell apoptosis, but these phenomena were counteracted by YAP1 down-regulation.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-11317-y.
Keywords: Melanoma, TRIM3, Ubiquitination, YAP1
Subject terms: Cancer, Genetics, Oncology, Cancer
Introduction
Melanoma, as a kind of malignant tumor, mostly occurs in the skin, and the incidence of melanoma has increased in the world, which has led to public health problems1. Primary extradermal melanoma can occur in the eyes, gastrointestinal tract, mucosa, meninges, genitourinary system, and lymph nodes. Surgical resection, chemotherapy, immunotherapy and radiation therapy as the conventional methods of melanoma treatment, greatly alleviate the symptoms of melanoma patients2. However, the high metastasis and recurrence rate of melanoma lead to relatively worse prognosis. Thus, revealing the relevant molecular functions of melanoma development and metastasis is imperative.
Tripartite motif (TRIM) is involved in diverse important biological behaviors, incorporating the modulation of cell growth, apoptosis, and mobility3. TRIM3 (tripartite motif-containing protein 3) is a crucial member of the TRIM family, known for its E3 ubiquitin ligase activity in regulating protein stability and function through the ubiquitin-proteasome system4. As a typical TRIM protein, TRIM3 contains a RING finger domain, B-box motifs, and a coiled-coil region, which collectively enable its interaction with E2 ubiquitin-conjugating enzymes and substrate proteins. The RING domain is particularly essential for its ligase function, as it facilitates the transfer of ubiquitin from E2 enzymes to specific target proteins, leading to their ubiquitination and subsequent degradation or functional modulation4. TRIM 3 is widely expressed in several cell types, and studies have been conducted on aspects such as innate immunity, autophagy, apoptosis and carcinogenesis, by selectively ubiquitinating key regulatory proteins5. For instance, studies have shown that TRIM3 targets SLC7A11/xCT through its NHL domain, leading to SCL7A11 K11-linked ubiquitination at K37, which promotes SLC7A11 proteasome-mediated degradation in non-small cell lung cancer (NSCLC)6. Additionally, TRIM3 has been implicated in the regulation of the cell cycle by modulating the stability of cyclin-dependent kinase inhibitors, further underscoring its role in maintaining cellular homeostasis7. Some reports presented a reduction of TRIM3 expression and confirmed its underlying function as a tumor inhibitor in several cancers, including colorectal8, gastric9, liver10 and cervical cancers11. Whereas, the expression and relevant molecular functions of TRIM3 in melanoma have not been reported.
The Hippo pathway, first discovered in Drosophila, is a primary developmental pathway that governs organ size and later found to be conserved in mammals12. Yes1-associated transcriptional regulator (YAP1) is the primary downstream effector of Hippo pathway13. Both animal and cell studies have confirmed the function of YAP1 in promoting the proliferation of cancer cells and tumor tissues, as well as cell migration, metastasis and resistance to therapeutic drugs14,15. Moreover, a previous study has indicated that YAP1 and its downstream target genes was associated with cancer angiogenesis in melanoma16. TRIM3, functioning as an E3 ubiquitin ligase, may directly or indirectly regulate YAP1 activity through ubiquitination modifications. Our preliminary studies have observed altered YAP1 expression upon TRIM3 depletion, providing initial evidence of their functional interplay. Notably, the biological functions of TRIM3 in melanoma remain unreported, and its connection with YAP1 signaling represents completely unexplored territory. Focusing on the TRIM3-YAP1 axis not only offers novel insights into TRIM3’s tumor-promoting mechanisms but also establishes a theoretical foundation for developing YAP1-targeted therapeutic strategies against melanoma.
Herein, we explored function of TRIM3 in melanoma cell lines, and detected the relationship between TRIM3 and YAP1 in melanoma.
Materials and methods
Immunohistochemical analysis
Tissue microarray (TMA) including 48 melanoma samples and 15 para-carcinoma samples was purchased from Zhongke Guanghua (Xi’an, China). TMA was dewaxed with xylene and rehydrated with gradient alcohol. Then, TMA was hatched with the corresponding antibodies and DAB/AEC developed color for 10 min.
Cell culture and cell transfection
Human melanoma cell lines M14 and A375 were purchased from Shanghai Anwei Biotechnology Co., Ltd. (Shanghai, China) and maintained in dulbecco’s modified eagle medium (DMEM, KeyGEN BioTECH, KGM12800N, Nanjing, China) medium followed by 10% fetal bovine serum (FBS, A31608, Invitrogen, USA) and 1% antibiotic in an incubator at 37 °C and 5% CO2.
For TRIM3 overexpression, pcDNA3.1-TRIM3 was used to artificially increase TRIM3 expression. For TRIM3 depletion, TRIM3-targeting small interference RNA (siRNA) was synthesized to knock down TRIM3 expression (si-TRIM3-1, 5’-GCTGTGAACAACAAGAATGAA-3’; si-TRIM3-2, 5’-GACATAATTGTGGCAGACTAT-3’). For YAP1 expression, YAP1-targeting siRNA was synthesized to knock down YAP1 expression (si-YAP1, 5’- GCCACCAAGCTAGATAAAGAA-3’).
Quantitative real-time polymerase chain reaction (qPCR)
Total RNA was extracted with the assistance of Trizol (CWBIO, CWO581S, Taizhou, China), and qPCR was operated with the assistance SYBR Green Real-time PCR Master Mix kit (CWBIO, CW0957M) based on the supplier’s direction and run on ABI Prism 7500 detection system. The relative expression of TRIM3 and YAP1 were analyzed by 2−ΔΔct method, and normalized to GAPDH. The primer sequences were listed as below:
TRIM3, F: 5’-GCAGACAGCAACAACCAGTG-3’, R: 5’-CCCCAGTCAGCCACAATGAT-3’; YAP1, F: 5’-CAACTCCAACCAGCAGCAAC-3’, R: 5’-TCCTGCAGACTTGGCATCAG-3’; GAPDH, F: 5’- GATTTGGTCGTATTGGGCGC-3’, R: 5’- AGTGATGGCATGGACTGTGG-3’.
Western blot
The whole proteins were separated from M14 and A375 cells by using lysis buffer and radioimmunoprecipitation assay (RIPA, CWBIO, CW2333S) buffer including 0.1% proteinase inhibitor. Then, the separation and transfer of proteins were carried out according to the previous methods. After incubating with the corresponding antibodies, enhanced chemiluminescent (ECL, PTG, B500024, USA) was used to develop the protein bands. With GAPDH as an internal control, the relative densitometry values were estimated by using Image J software and processed with GraphPad Prism 8.0. The primary antibodies were listed as follows: TRIM3 (abcam, ab111840, USA), Bcl-2 (abcam, ab182858, USA), BAX (abcam, ab32503, USA), caspase 3 (abcam, ab32351, USA), YAP1 (abcam, ab205270, USA), and GAPDH (abcam, ab9485, USA).
Cell proliferation assay
Cell counting kit-8 (CCK-8) assay and colony formation assay were utilized to investigate the growth ability of M14 and A375 cells. For CCK-8 assay, 3000 treated cells were cultivated in 96-well plates, and the optical density (OD) value was read every 24 h after adding CCK-8 solution (MCE, HY-K0301, USA) per well. The growth curve was drawn with GraphPad Prism 8.0. For colony formation assay, cells were gathered, cultivated onto a 60-mm dish at a number of 500 cells, and continued to be cultured for 2 weeks. After being fixed with paraformaldehyde (5 min) and stained with crystal violet (20 min), cells were imaged and counted.
Cell invasion assay
Transwell chambers (24- well-precoated with Matrigel, Millipore, USA) were utilized to evaluate the invasion capacity of M14 and A375 cells. In brief, cells were cultivated in the top chamber. Subsequently, the complete DMEM medium was filled into the bottom chamber. Cultured after 24 h, cells that invaded to the bottom surface of the chamber were stained with 0.1% crystal violet.
Wound healing assay
The migratory properties of M14 and A375 cells were analyzed by wound healing assay. First, M14 and A375 cells were cultivated in six-well plates with the density of 5 × 105 cells per well, and maintained with proper treatment. After reaching to 90% confluence, cells were wounded by a 200 µL sterile pipette tip in per well to made a scratch model. Cell healing images were captured at indicated time.
Flow cytometry
Apoptosis was detected by utilizing AnnexinV-fluorescein isothiocyanate (FITC)/propidiumiodide (PI) kit (Beyotime, C1062S). In brief, the treated cells were centrifuged at 1500 rpm for 5 min, washed, and suspended in PBS. Then, the cells were collected and resuspended in Annexin V-FITC/PI solution for 10 min at 37 °C away from the light. Lastly, the cells were resuspended with binding buffer and measured by flow cytometric analysis and the Flowjo software.
Immunoprecipitation
Based on the supplier’s direction, cell lysates were incubated with immobilized antibody beads and the corresponding primary antibodies at 4 °C overnight (Millipore, USA). The degree of protein-protein interaction was determined by western blot analysis of immune-precipitated supernatants.
Protein stability assay
A total of 1 × 105 cells (M14 and A375) were cultivated and exposed to si-TRIM3. Then, the cells after cycloheximide (CHX) treatment were treated at indicated times. Lastly, the samples were subjected to western blot to calculate YAP1 level.
Analysis of YAP1 ubiquitination
To detect the ubiquitination of YAP1 in M14 and A375 cells, the cells were exposed to 2 µg YAP1 plasmid and 2 µg Flag-TRIM3 or Flag-PCMV into M14 or A375 cells. After 48 h of cultivation, cells were exposed to 10 µM MG132, then the ubiquitination level of YAP1 was determined by immunoprecipitation with an anti-YAP1 antibody, and then western blot analysis was carried out with an anti-HA antibody.
Statistical analysis
All experimental data were exhibited as mean ± standard deviation. Student’s test was used for the differences between the two groups, and one-way analysis of variance (ANOVA) followed by Bonferroni post hoc test were used for the differences among three or more groups. IHC analysis was conducted using the Chi-square test for 4 × 4 tables. When p < 0.0.5, the difference was considered statistically significant.
Results
TRIM3 was under-expressed in melanoma samples and suppressed the growth and mobility of melanoma cells.
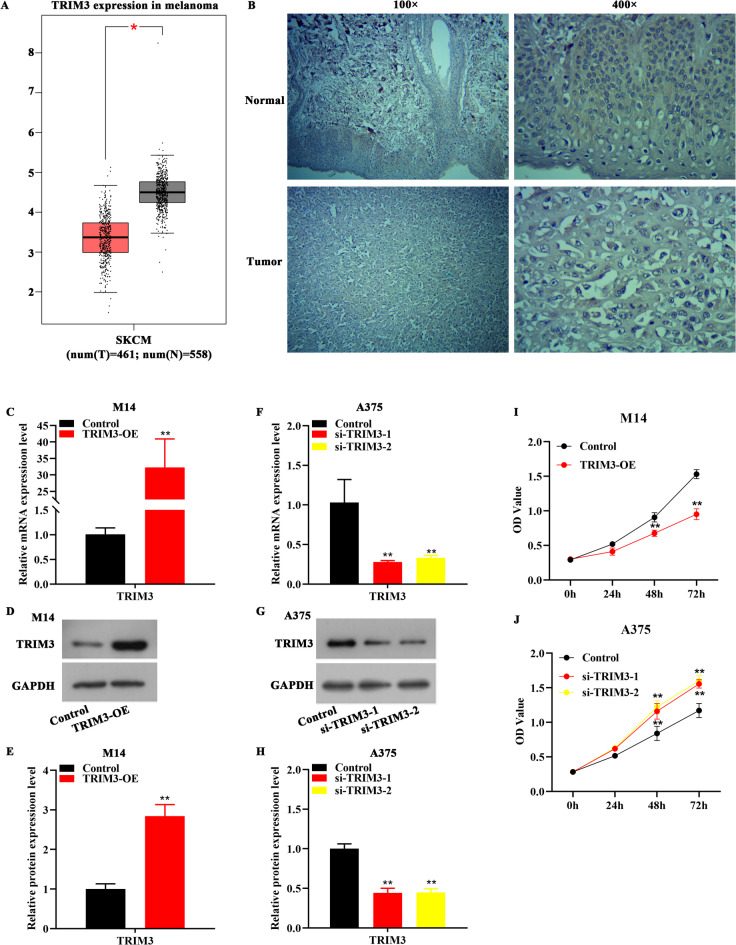
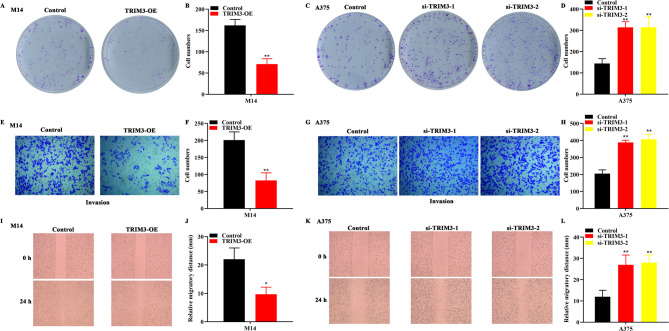
Gene Expression Profiling Interactive Analysis (GEPIA) website was utilized to detect TRIM3 level in melanoma, and the data showed that TRIM3 expression was significantly decreased in melanoma samples (Fig. 1A, tumor = 461, normal = 558). As exhibited in Fig. 1B; Table 1, TRIM3 was lower expressed in melanoma tissues in comparison with normal tissues, which was analyzed by immunohistochemistry. It was found that TRIM3 expression was independent of gender and age, but was related to I-II of pathological grading (Table 2). To further detect the function of TRIM3 in melanoma, we artificially overexpressed and knocked down TRIM3 in melanoma cells in vitro. Results from Fig. 1C-E displayed that the TRIM3 expression in M14 cells were elevated after TRIM3 overexpression (TRIM3-OE). Also, after si-TRIM3-1 or si-TRIM3-2 treatment, TRIM3 expression was reduced (Fig. 1F-H). Subsequently, a series of biological experiments were performed to evaluate the growth and mobility of melanoma cells. The result from Fig. 1I displayed that TRIM3 overexpression diminished the OD values of M14 cells, whereas knocked down of TRIM3 increased the OD values of A375 cells (Fig. 1J). Data from Fig. 2A-B displayed that overexpression of TRIM3 decreased the number of M14 cell clones, whereas depletion of TRIM3 increased the number of A375 cell clones (Fig. 2C-D). Moreover, the invasion number of M14 cells was reduced after TRIM3 up-regulation (Fig. 2E-F), whereas silencing TRIM3 increased the invasion number of A375 cells (Fig. 2G-H). The migratory distance of M14 cells was diminished after TRIM3 overexpression (Fig. 2I-J), whereas down-regulation of TRIM3 increased the migratory distance of A375 cells (Fig. 2K-L).
Fig. 1.
TRIM3 was under-expressed in melanoma samples and suppressed the proliferation of melanoma cells. (A) Data from GEPIA website showed low expression of TRIM3 in melanoma samples, p < 0.5. (B) Immunohistochemical analysis showed that TRIM3 expression was decreased in melanoma samples. C-E. TRIM3 expression was increased in M14 cells after TRIM3-overexpression (TRIM3-OE) transfection, **p < 0.01 vs. control. F-H. si-TRIM3-1 or si-TRIM3-2 transfection reduced TRIM3 expression in A375 cells, **p < 0.01 vs. control. I. TRIM3-OE transfection reduced the OD values of M14 cells, **p < 0.01 vs. control. J. si-TRIM3-1 or si-TRIM3-2 transfection increased the OD values of A375 cells, **p < 0.01 vs. control.
Table 1.
TRIM3 expression in melanoma compared with para-carcinoma tissue.
| Group | n | TRIM3 expression | P | |
|---|---|---|---|---|
| Low (n%) | High (n%) | |||
| Melanoma | 48 | 23 (47.9) | 25 (52.1) | 0.037* |
| para-carcinoma | 15 | 2 (13.3) | 13 (86.7) | |
Table 2.
TRIM3 expression associated with the clinicopathological parameters in Melanoma.
| Clinicopathological parameters | n | TRIM3 Low (n%) | TRIM3 High (n%) | P |
|---|---|---|---|---|
| Gender | ||||
| Male | 23 | 13 (56.5) | 10 (43.5) | 0.392 |
| Female | 25 | 10 (40.0) | 15 (60.0) | |
| Age (years) | ||||
| ≤50 | 22 | 10 (45.5) | 12 (54.5) | 0.981 |
| >50 | 26 | 13 (50.0) | 13 (50.0) | |
| Pathological grading | ||||
| I-II | 27 | 10 (37.0) | 17 (63.0) | 0.035* |
| III-IV | 5 | 5 (100.0) | 0 (0.0) | |
Fig. 2.
TRIM3 inhibited the clonogenesis and motility of melanoma cells. (A-B). TRIM3 overexpression decreased the clone numbers of M14 cells. C-D. si-TRIM3-1 or si-TRIM3-2 transfection elevated the clone numbers of A375 cells. (E-H). The invasion number of M14 cells was decreased after TRIM3 overexpression, while the invasion number of A375 cells was elevated after TRIM3 knocked down. I-L. The migrated distance of M14 cells was reduced after TRIM3 overexpression, whereas depletion of TRIM3 increased the migrated distance of A375 cells. **p < 0.01 vs. control.
TRIM3 promoted the apoptosis of melanoma cells
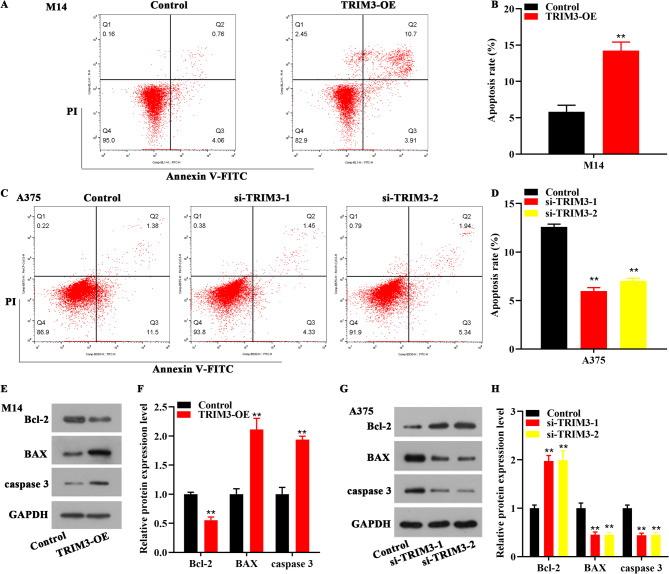
Furthermore, the function of TRIM3 on melanoma cells was determined by flow cytometry. As presented in Fig. 3A-D, TRIM3 overexpression elevated the apoptosis rate of M14 cells, however, TRIM3 depletion reduced the apoptosis of A375 cells. Additionally, the expression of apoptosis-related proteins was detected by western blot. Bcl-2, an anti-apoptotic protein, inhibited apoptosis by preventing the release of cytochrome c from mitochondria, thus blocking the caspase-cascade activation17. In contrast, Bax, a pro-apoptotic Bcl-2 family member, triggered the release of cytochrome c and other apoptogenic factors, initiating the caspase-mediated apoptotic pathway17. Caspase-3, a key executioner caspase, was activated by upstream caspases in response to apoptotic signals and then cleaved numerous intracellular substrates, leading to the characteristic morphological and biochemical changes of apoptosis, such as cell shrinkage, chromatin condensation, and DNA fragmentation17. TRIM3 overexpression reduced Bcl-2 protein level, and elevated the protein level of BAX and caspase-3 in M14 cells, which was reversed by si-TRIM3 transfection in A375 cells (Fig. 3E-H).
Fig. 3.
TRIM3 promoted the apoptosis of melanoma cells. (A-D). The apoptosis rate of M14 cells increased after TRIM3-overexpression, while depletion of TRIM3 reduced the apoptosis rate of A375 cells. (E-F). In M14 cells, TRIM3 overexpression reduced Bcl-2 expression, as well as increased the expression of BAX and caspase 3. (G-H). In A375 cells, depletion of TRIM3 increased Bcl-2 expression, and diminished the expression of BAX and caspase 3. **p < 0.01 vs. control.
TRIM3 directly interacted with YAP1 and induced YAP1 ubiquitination degradation
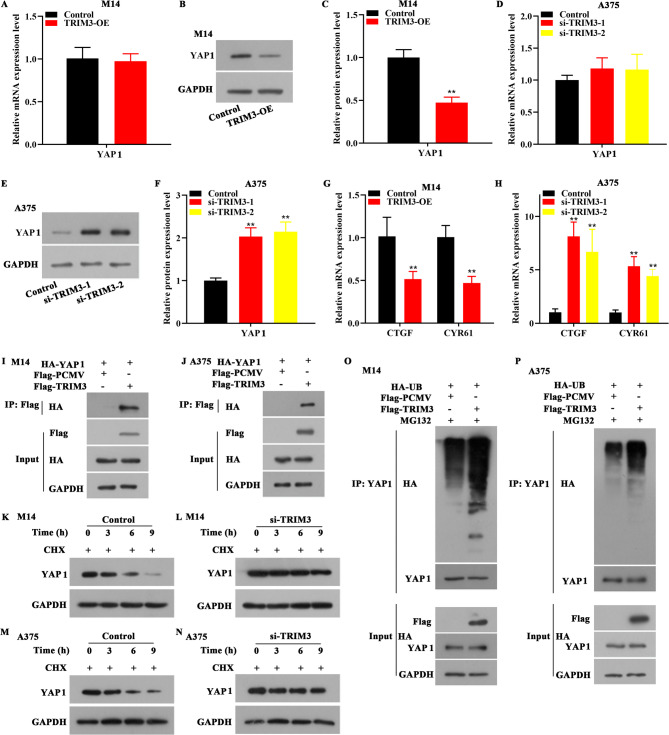
YAP1 plays a prominent role in tumor initiation, invasiveness, and therapeutic drug resistance18, and studies have demonstrated high expression of YAP1 in melanoma19. Herein, we discovered that YAP1 had no obvious change (mRNA level) after TRIM3 overexpression in M14 cells, whereas the protein level of YAP1 was reduced after TRIM3 overexpression in M14 cells (Fig. 4A-C). Moreover, in A375 cells, the YAP1 had no obvious change (mRNA level) after TRIM3 depletion, however, the protein level of YAP1 was increased (Fig. 4D-F). As downstream targets of YAP1, the levels of CTGF and CYR61 diminished after TRIM3 overexpression and elevated after TRIM3 knocked down (Fig. 4G-H). To further detect the relationship between TRIM3 and YAP1, immunoprecipitation assay was performed. The data from Fig. 4I-J showed that TRIM3 could bind with YAP1 protein in M14 and A375 cells. To further detect whether TRIM3 could regulate the stability of YAP1 protein, protein stability assay was carried out. The result indicated that the degradation rate of YAP1 slowed down after TRIM3 was knocked down in M14 and A375 cells, which indicated that TRIM3 negatively regulated YAP1 by proteasome mediated degradation (Fig. 4K-N). To detect whether TRIM3 induced ubiquitous modification of YAP1, the ubiquitination assay was performed. The data showed that TRIM3 could degrade YAP1 by ubiquitination in M14 and A375 cells (Fig. 4O-P).
Fig. 4.
TRIM3 suppressed YAP1 by inducing ubiquitous degradation of YAP1. (A-C). In M14 cells, overexpression of TRIM3 inhibited the protein expression level of YAP1, while the mRNA expression level of YAP1 remained unchanged. (D-F). In A375 cells, TRIM3 depletion increased YAP1 expression at protein level, whereas the mRNA expression level of YAP1 remained unchanged. (G). In M14 cells, overexpression of TRIM3 reduced the mRNA levels of CTGF and CYR61. (H). In A375 cells, depletion of TRIM3 increased the mRNA level of CTGF and CYR61. (I-J). M14 and A375 cells were co-transfected with HA-YAP1 and Flag-TRIM3 plasmids, and being further cultured for 24 h before immunoprecipitation assay, and the binding between TRIM3 and YAP1 was detected by immunoprecipitation assay. (K-N). In M14 and A375 cells, depletion of TRIM3 reduced the degradation rate of YAP1 protein. (O-P). M14 and A375 cells were transfected with HA-UB plasmid, together with Flag-TRIM3 plasmid, the ubiquitous status of YAP1 was analyzed by immunoprecipitation. **p < 0.01 vs. control.
Knocked down of YAP1 reversed the effects of si-TRIM3 on A375 cells
To further detect the relationship between YAP1 and TRIM3, a rescue assay was performed. The data from Fig. 5A-B showed that si-TRIM3 transfection increased the number of A375 cell clones, whereas knocked down of YAP1 alleviated the beneficial role of si-TRIM3 on A375 cell clone numbers. The result from Fig. 5C-F exhibited that depletion of TRIM3 elevated the invasion number and migratory distance of A375 cells, which was counteracted after si-YAP1 treatment. The data from Fig. 5G-H showed that silencing TRIM3 reduced the apoptosis rate of A375 cells, which was rescued after YAP1 knocked down. The result from Fig. 5I exhibited that the OD value of A375 cells was increased after TRIM3 depletion, whereas si-YAP1 treatment inversed this phenomenon. Moreover, the elevated expression of CTGF and CYR61 induced by TRIM3 knocked down was also inhibited by si-YAP1 transfection (Fig. 5J).
Fig. 5.
Depletion of YAP1 alleviated the effects of si-TRIM3 on A375 cells. (A-B). Knocked down of TRIM3 increased the number of A375 cell clones, while transfection with si-YAP1 reversed this phenomenon. (C-D). Depletion of TRIM3 increased the number of A375 cell invasions, which was counteracted by si-YAP1 transfection. (E-F). Silencing TRIM3 elevated the migrated distance of A375 cells, whereas knocked down of YAP1 reversed this phenomenon. (G-H). The reduction in A375 apoptosis rate induced by TRIM3 knocked down was reversed by si-YAP1 transfection. (I). The increase in OD value of A375 cells caused by TRIM3 knocked down was counteracted by si-YAP1 transfection. (J). The up-regulation of CTGF and CYR61 expression in A375 cells induced by TRIM3 knocked down was reversed by si-YAP1 transfection. **p < 0.01 vs. control, #p < 0.05, ##p < 0.01 vs. si-TRIM3.
Discussion
Melanoma is a malignant tumor that generates from melanocytes and is the most invasive type of skin cancer with worse prognosis20. Melanoma is a particularly challenging type of cancer as it is prone to metastasis, resulting in worse survival rates for advanced patients21. Melanoma is induced by the collection of abnormal gene expression that disrupts the regular effect of melanocytes and promotes their infinite multiplication22. In this research, we discovered that TRIM3 was down-regulated in melanoma, and silencing TRIM3 elevated the growth and mobility of melanoma cells, and suppressed the apoptosis of melanoma cells. Additionally, we also discovered that the disturbance of TRIM3 regulated YAP1 expression through the ubiquitination pathway. Moreover, knocked down of YAP1 alleviated the function of si-TRIM3 on melanoma cells.
The aberrant expression of TRIM3 was closely associated with the initiation and progression of diverse tumors, such as lung, colorectum, ovarian, etc6,8,23. When TRIM3 was highly expressed in these cancers, it exhibits promising anti-cancer effects. TRIM3 not only regulated the a series of signaling pathway that controlled tumor development, but also exerted tumor inhibitory effects by regulating cell cycle, EMT, and other mechanisms24. At present, there is limited research on TRIM3, and the anti-tumor effect of TRIM3 is not fully understood. Its relevant functions and signal transduction need to be further explored. Therefore, it is necessary to conduct further comprehensive and in-depth research to reveal the specific functions by which TRIM3 plays a role in these different tumors. Herein, our study found that TRIM3 level was inhibited in melanoma samples. Moreover, TRIM3 overexpression diminished the OD values, number of cell clones and cell invasion, and migratory distance of melanoma cells, but promoted cell apoptosis. Conversely, silencing TRIM3 strengthened the proliferation and mobility of melanoma cells, as well as suppressed apoptosis.
YAP1, as a transcription co-activator, has been demonstrated to be an important oncogene in many cancer types, which could modulate cell migration, tumor growth, cancer stemness and so on25. For example, the post-translational dysregulation of Yap1 was genetically regulated in liver cancer, which determines the fate of human disease and stem cell like behavior26, YAP1 increased PD-L1 level and inhibited T cell activation, resulting in immune escape in small cell lung cancer27, and YAP1 accelerated the proliferation and invasion of adenocarcinoma by activating the Axl28. Herein, our study discovered that the protein level of YAP1 was decreased when TRIM3 up-regulation, whereas TRIM3 knocked down increased YAP1 protein expression. However, the mRNA level of YAP1 had no significant change. TRIM3 is a ubiquitination associated protein that is related with cell growth, mobility, and metastasis in the process of tumor development. To further investigate the relationship between YAP1 and TRIM3, immunoprecipitation and ubiquitination assays were performed. Our data illustrated that high expression of TRIM3 led to ubiquitination degradation of YAP1, resulting in down-regulation of YAP1 protein expression. Moreover, our rescue assays demonstrated that silencing YAP1 alleviated the effects of si-TRIM3 on melanoma cells.
Although several achievements have been made in our research, some problems still need to be addressed. Among them, the most important point is that our results have only been obtained in vitro cell experiments and need to be further validated in animals. Secondly, the molecular mechanism downstream of YAP1 has not been revealed.
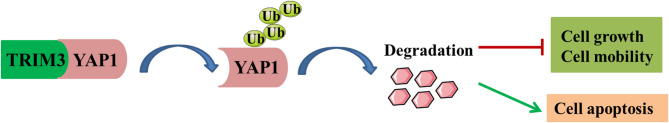
In summary, our data illustrated that low expression of TRIM3 accelerated the growth and motility of melanoma cells, and suppressed cell apoptosis rate, but these phenomena are counteracted by YAP1 knocked down. Therefore, the tumor inhibitory effect of TRIM3 in melanoma is partially achieved by degradation of YAP1 through the ubiquitination pathway (Fig. 6).
Fig. 6.
High TRIM3 expression leads to ubiquitination and degradation of YAP1, leading to downregulation of YAP1 protein expression, inhibition of cell proliferation and promotion of apoptosis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the patients, investigators, clinicians, technical personnel, and funding bodies who contributed to GEPIA(http://gepia2.cancer-pku.cn/#index)database, thereby making this study possible.
Author contributions
YW and JZ designed the study. YW、MS and JZ collected the data.YW was responsible for the experiment.All authors wrote the manuscript. All authors have read and approved the final manuscript.
Data availability
Data is provided within the manuscript or supplementary information files.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
We have obtained ethical approval for animal experiments and permission for personnel to participate.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ahmed, B., Qadir, M. I. & Ghafoor, S. Malignant melanoma: skin Cancer-Diagnosis, prevention, and treatment. Crit. Rev. Eukaryot. Gene Expr. 30 (4), 291–297 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Namikawa, K. & Yamazaki, N. Targeted therapy and immunotherapy for melanoma in Japan. Curr. Treat. Options Oncol.20 (1), 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esposito, D., Koliopoulos, M. G. & Rittinger, K. Structural determinants of TRIM protein function. Biochem. Soc. Trans.45 (1), 183–191 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Esposito, D. & Dudley-Fraser, J. Divergent self-association properties of paralogous proteins TRIM2 and TRIM3 regulate their E3 ligase activity. Nat. Commun.13 (1), 7583 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li, W. W. et al. Ubiquitination of TLR3 by TRIM3 signals its ESCRT-mediated trafficking to the endolysosomes for innate antiviral response. 117(38) , 23707–23716. (2020). [DOI] [PMC free article] [PubMed]
- 6.Wang, Z. et al. TRIM3 facilitates ferroptosis in non-small cell lung cancer through promoting SLC7A11/xCT K11-linked ubiquitination and degradation. Cell. Death Differ.31 (1), 53–64 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raheja, R. et al. The ability of TRIM3 to induce growth arrest depends on RING-dependent E3 ligase activity. Biochem. J.458 (3), 537–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuo, Q. et al. TRIM3 inhibits colorectal cancer cell migration and lipid droplet formation by promoting FABP4 degradation. Histol. Histopathol. 39 (2), 239–250 (2024). [DOI] [PubMed] [Google Scholar]
- 9.Farhadi, J. et al. Decreased expression of TRIM3 gene predicts a poor prognosis in gastric cancer. 53(1),179–186. (2022). [DOI] [PubMed]
- 10.Lu, K. et al. TRIM proteins in hepatocellular carcinoma. J. Biomed. Sci.29 (1), 69 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song, Y. et al. Tripartite motif-containing protein 3 plays a role of tumor inhibitor in cervical cancer. Biochem. Biophys. Res. Commun.498 (3), 686–692 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Wu, S. et al. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with Salvador and warts. Cell114 (4), 445–456 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Nishioka, N. et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell.16 (3), 398–410 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Cunningham, R. & Hansen, C. G. The Hippo pathway in cancer: YAP/TAZ and TEAD as therapeutic targets in cancer. 136(3), 197–222. (2022). [DOI] [PMC free article] [PubMed]
- 15.Song, Q. et al. YAP enhances autophagic flux to promote breast cancer cell survival in response to nutrient deprivation. PLoS One. 10 (3), e0120790 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen, Y. & Wang, X. STAT3-YAP/TAZ signaling in endothelial cells promotes tumor angiogenesis. 14(712), eabj8393. (2021). [DOI] [PubMed]
- 17.Xue, Q., Kang, R. & Klionsky, D. J. Copper Metabolism Cell. Death Autophagy Autophagy, 19(8), 2175–2195. (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szulzewsky, F., Holland, E. C. & Vasioukhin, V. YAP1 and its fusion proteins in cancer initiation, progression and therapeutic resistance. Dev. Biol.475, 205–221 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, R. et al. Combined BET and MEK Inhibition synergistically suppresses melanoma by targeting YAP1. Theranostics14 (2), 593–607 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomas, A., Leonardi-Bee, J. & Bath-Hextall, F. A systematic review of worldwide incidence of nonmelanoma skin cancer. Br. J. Dermatol.166 (5), 1069–1080 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Ferlay, J. et al. Cancer statistics for the year 2020: An overview. (2021). [DOI] [PubMed]
- 22.Shain, A. H. & Bastian, B. C. From melanocytes to melanomas. Nat. Rev. Cancer. 16 (6), 345–358 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Cong, Y. et al. Tripartite-motif 3 represses ovarian cancer progression by downregulating lactate dehydrogenase A and inhibiting AKT signaling. (2024). [DOI] [PubMed]
- 24.Teng, W. et al. Advances in the antitumor mechanisms of tripartite motif-containing protein 3. J. Cancer Res. Clin. Oncol.150 (2), 105 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jing, L. et al. CircSETD2 inhibits YAP1 by interaction with HuR during breast cancer progression. 24(1), 2246205. (2023). [DOI] [PMC free article] [PubMed]
- 26.Simile, M. M. et al. Post-translational deregulation of YAP1 is genetically controlled in rat liver cancer and determines the fate and stem-like behavior of the human disease. Oncotarget7 (31), 49194–49216 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen, P. et al. YAP1 expression is associated with survival and immunosuppression in small cell lung cancer. Cell. Death Dis.14 (9), 636 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui, Z. L. et al. YES-associated protein 1 promotes adenocarcinoma growth and metastasis through activation of the receptor tyrosine kinase Axl. Int. J. Immunopathol. Pharmacol.25 (4), 989–1001 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.