Abstract
Although different neurodegenerative diseases are defined by distinct pathological proteins, they share many common features including protein aggregation. Despite this commonality, most current therapeutic approaches in the field, such as anti-aggregate antibodies, are focused on individual diseases or single neuropathologies with only limited success. The endolysosomal proteins progranulin and TMEM106B were both initially associated with frontotemporal lobar degeneration but have subsequently also been linked to other neurodegenerative diseases. Thus, these proteins are predicted to participate in common pathogenic pathways shared across various neurodegenerative diseases. Importantly, recent discoveries of TMEM106B amyloid fibrils in varied neurodegenerative diseases and glycosphingolipid regulation by progranulin and TMEM106B further support their central roles in cross-disease neurodegenerative mechanisms. This review summarizes recent advances in progranulin and TMEM106B function within the endolysosomal system and neurodegenerative diseases. It describes preclinical models and therapeutic approaches for progranulin- and TMEM106B-associated diseases. We also discuss future direction leading to novel alternative therapies targeting shared mechanisms in neurodegenerative diseases.
Keywords: Progranulin, TMEM106B, Frontotemporal Lobar degeneration, Alzheimer’s disease, Parkinson’s disease, Aging, Endolysosome, Glycosphingolipid, Amyloid fibrils, GBA1
Background
Neurodegenerative diseases such as Alzheimer’s disease (AD), Parkinson’s disease (PD), frontotemporal lobar degeneration (FTLD), and amyotrophic lateral sclerosis (ALS), share many pathological features, including neuronal and synaptic loss, neuroinflammation, and accumulation of protein aggregates in the brain [1]. In addition, recent studies using human postmortem brains have shown that in addition to a primary proteinopathy, concomitant proteinopathies are frequently observed in patients with neurodegenerative diseases and influence their clinical course [2–4]. Despite these facts, current research and therapeutic approaches in the field have been focused on single protein neuropathologies, such as Aβ, tau, and -synuclein, with only limited success.
The endolysosomal system is essential for maintaining cellular proteostasis and its dysfunction is thought to be involved in accumulation of pathological proteins in various neurodegenerative diseases. Human genetic studies have identified numerous genes in the endolysosomal system associated with AD, PD, FTLD, and ALS, highlighting an importance of the endolysosomal system in a wide range of neurodegenerative diseases [5, 6]. The endolysosomal proteins progranulin (PGRN) and TMEM106B were initially implicated in FTLD with TAR DNA-binding protein 43 (TDP-43) inclusions (FTLD-TDP) [7–9]. Importantly, TMEM106B variants were also reported to contribute to genetic risk for FTLD-TDP in individuals with GRN mutations [7], suggesting a potential functional interaction between PGRN and TMEM106B in the endolysosomal system. Subsequently, these two proteins were also linked to many other neurodegenerative diseases including AD and PD [10, 11]. Thus, PGRN and TMEM106B may play a central role in common pathological mechanisms shared across various neurodegenerative diseases, enhancing their potential impact as therapeutic targets with broad application. In this review, we provide an overview of PGRN and TMEM106B in neurodegenerative diseases and the endolysosomal system. We describe recent discoveries of amyloid fibrils of C-terminal fragments of TMEM106B and lipid regulation by PGRN and TMEM106B. Advances in preclinical disease models as well as therapeutic targeting of PGRN- and TMEM106B-associated neurodegenerative diseases are reviewed. Finally, we consider future directions for PGRN and TMEM106B research designed to develop novel therapies targeting common mechanisms of multiple neurodegenerative diseases.
PGRN-associated neurodegenerative diseases
Heterozygous mutations in the GRN gene that result in PGRN haploinsufficiency cause FTLD [8, 9], the second most common cause of dementia in people under the age of 65 [12]. FTLD affects the frontal and temporal lobes of the brain, resulting in progressive changes in behavior, personality, and/or language [12, 13]. The two major pathological subtypes of FTLD are FTLD-tau characterized by tau pathology and FTLD-TDP associated with TDP-43 pathology, although 5–10% of patients may have inclusions immunoreactive for FUS, EWS, and TAF15 (FTLD-FET) [13]. GRN mutations account for approximately 5–25% of familial FTLD and are the second most common cause of inherited FTLD-TDP, after a CCCCGG hexanucleotide expansion in the non-coding region of C9orf72 [13]. The penetrance of GRN mutations is incomplete and reportedly age dependent with only 50% of carriers being affected by age 60 and 90% by age 70 [14]. Neuropathological analysis has shown that GRN mutations are exclusively associated with FTLD-TDP type A pathology, which is characterized by abundant cytoplasmic inclusions and short thick dystrophic neurites in the superficial cortical layers [12]. A significant increase in microgliosis has also been reported in the brain of FTLD patients with GRN mutations (FTLD-GRN) [15, 16]. In addition, recent single-nucleus RNA sequencing studies have revealed neurovascular dysfunction and astrocytic pathology in the brain of FTLD-GRN [17, 18]. At least 140 different loss-of-function mutations have been found in FTLD patients, and most of them lead to premature termination codons (PTCs) either directly due to nonsense mutations or indirectly through frameshift by splice-site mutations or small deletions and insertions. The net result is destruction of mutant GRN mRNA by nonsense-mediated mRNA decay with a consequent 50% loss of functional PGRN [19]. In addition to mutations leading to PTCs, a number of missense mutations have been also reported. Although for most of the missense cases, their contribution to the pathogenesis is not well defined, the W7R and A9D mutations in the signal sequence have been shown to affect mRNA stability, sorting, and/or secretion of PGRN [20, 21].
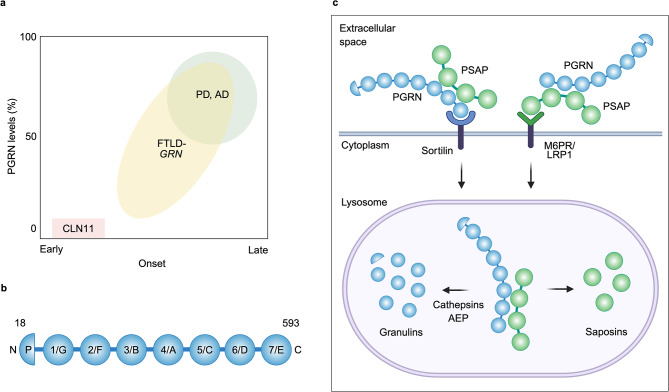
While PGRN haploinsufficiency causes FTLD-TDP, complete loss of function of PGRN due to homozygous GRN mutations leads to neuronal ceroid lipofuscinosis (CLN11), a rare lysosomal storage disorder characterized by excessive accumulation of lipofuscins in the lysosome [22]. Clinical features of CLN11 include cerebellar ataxia, epilepsy, visual loss, and progressive cognitive decline with the age of onset of ~ 15 years [22–27]. Importantly, a recent study however has reported six patients carrying homozygous GRN mutations with divergent phenotypes and variable ages of onset, including some presenting classical CLN11 symptoms at early age of onset and others showing a distinct delayed phenotype of frontotemporal dementia after 40 or 50 years, suggesting that CLN11 and FTLD-TDP are extreme phenotypes of a common spectrum of disorders caused by biallelic GRN mutations [28] (Fig. 1a).
Fig. 1.
Schematic illustration of structure and cellular uptake of PGRN. a, PGRN-associated neurodegenerative diseases. Haploinsufficiency (~50% loss) of PGRN causes FTLD, the second most common cause of dementia in people under the age of 65, while complete loss of PGRN causes the lysosomal storage disorder CLN11 with the age of onset of ~15 years, although rare homozygous GRN mutations were reported to cause FTLD with the age of onset of ~50 years. In addition, several GRN variants increase risk for AD and PD, which usually develop after age 65 and 60, respectively. A GRN variant is reported to cause 10-20% reduction in PGRN levels. Furthermore, heterozygous GRN mutations have been found in a substantial number of AD and PD patients. b, Schematic representation of PGRN. Human PGRN is a highly glycosylated protein composed of 7.5 cysteine-rich granulin domains (A to G and P) that are connected by short linker regions. P represents paragranulin. Amino acids 1 to 17 are the signal sequence. Granulins numbered 1 through 7 are based on the UniProtKB database: P28799. c, Schematic illustration of lysosomal delivery of PGRN and PSAP via their receptors. Extracellular PGRN binds to cell surface sortilin receptor through its C-terminal tail and is delivered to lysosomes, where PGRN is processed into granulins by cathepsins and AEP. PGRN also binds to PSAP extracellularly through the linker region between saposins B and C and can be delivered to lysosomes via the PSAP receptors M6PR and LRP1. The PGRN-PSAP complex may also be important to facilitate lysosomal delivery of extracellular PSAP via sortilin. In the lysosome, PSAP is processed into saposins, which are involved in glycosphingolipid degradation. Figure was created with BioRender.com.
Previous genetic studies including genome-wide association studies (GWAS) have suggested that common GRN variants are associated with increased risk for AD and PD [29–34]. In addition, GRN mutations have been found in a substantial number of AD and PD patients [14, 35–46] (Fig. 1a). A recent study has also shown an association of GRN mutations with Lewy body dementia (LBD) [47]. AD is pathologically characterized by accumulation of extracellular amyloid plaques composed of amyloid-β (Aβ) peptides and intracellular neurofibrillary tangles formed from hyperphosphorylated tau protein in the brain [48]. PGRN is known to be up-regulated in dystrophic neurites and microglia near amyloid plaques, while no or very weak PGRN immunoreactivity has been detected in neurofibrillary tangles in AD [49–51]. However, in human subjects of the Alzheimer’s Disease Neuroimaging Initiative (ADNI), the GRN rs5848 AD risk variant, which causes at least 10–20% reduction of PGRN protein levels, has no significant effects on florbetapir PET amyloid imaging or cerebrospinal fluid (CSF) Aβ levels, whereas it is associated with increased CSF tau levels [52], suggesting a potential effect of PGRN deficiency on tau pathology, rather than amyloid pathology. Importantly, accumulation of tau and α-synuclein, in addition to TDP-43 inclusions, is also found in the brain of several GRN mutation carriers [39, 45, 53–56]. In addition to AD and PD/LBD, GRN variation has also been implicated in Gaucher disease [57], FTLD-TDP and/or motor neuron disease with C9orf72 repeat expansions [58], ALS [59], and limbic-predominant age-related TDP-43 encephalopathy neuropathological change (LATE-NC) [60].
TMEM106B-associated neurological diseases
The TMEM106B gene was initially identified in 2010 by a GWAS as a genetic risk factor for FTLD-TDP [7]. For the top single-nucleotide polymorphism (SNP) rs1990622 (P = 1.08 × 10–11) (Fig. 2a), the minor C-allele conferred protection with an odds ratio of 0.61 (minor allele frequency 32.1% in cases and 43.6% in controls). Importantly, this study also reported that the disease-modulating effect was especially pronounced in FTLD patients with GRN mutations [7]. Subsequent studies have confirmed that TMEM106B is a strong modifier for FTLD-TDP caused by GRN mutations [61–66]. Remarkably, it has been consistently reported that homozygotes for TMEM106B protective minor alleles are rarely found in symptomatic FTLD patients with GRN mutations [61, 62, 64–66]. These results suggest that homozygosity for the TMEM106B minor alleles offers strong protection against developing FTLD in GRN mutation carriers and argue that TMEM106B functionally interacts with PGRN in the endolysosomal system to affect pathogenesis of FTLD-TDP. A few initial studies showed that the TMEM106B protective minor alleles were associated with increased plasma PGRN levels [62, 63], suggesting a potential protective mechanism. However, the increase does not appear to be robust enough to explain the strong protective effect of the minor alleles on FTLD. In addition, studies have failed to replicate the plasma PGRN effect of the TMEM106B variation [67, 68]. Therefore, it is likely that TMEM106B also participates in other disease mechanisms associated with PGRN.
Fig. 2.
Schematic illustration of structure and amyloid formation of TMEM106B. a, TMEM106B genomic region on chromosome 7p21. Exons (1-9) are indicated by blue boxes and the coding regions are labeled in dark blue. The major SNPs associated with neurodegenerative diseases are shown with protective/minor and risk/major alleles. Multiple SNPs near and in the TMEM106B gene, including rs1990620, rs1990622, rs3173615, rs6966915, and rs1020004, are in strong LD, constituting two common TMEM106B haplotypes, one associated with increased disease risk, and the other with a protective effect. AluYb8 insertion is found in 3’ UTR of the TMEM106B risk haplotype. b, Schematic representation of TMEM106B. The 274-amino acid human TMEM106B consists of an N-terminal intrinsically disordered cytoplasmic region, a single-pass transmembrane region, and a C-terminal luminal region with five glycosylation sites (N145, N151, N164, N183, and N256). T185S and D252N amino acid substitutions associated with FTLD and HLD are indicated. c, Amyloid fibril formation of CTF of TMEM106B. TMEM106B forms homodimers and undergoes shedding and C-terminal trimming by unknown lysosomal enzyme(s). It is currently unknown whether dimerization affects the processing of TMEM106B and what protease(s) mediate the processing to form the fibrils. A precise cleavage between resides 119 and 120 appears to be required for the amyloid fibril formation because Ser120 is buried in the fibril core, leaving no space for the other N-terminal residues. Upon fibrillization, the luminal region of TMEM106B undergoes a conformational change from a structure with a ubiquitous 7-bladed β sandwich fold (PDB: 8B7D) to ones with a five-layered fold consisting of 17-19 β-strands (PDB: 7QVC). The fibril formation is age-dependent and may be promoted by the TMEM106B risk haplotype, GRN mutation, or under LATE-NC condition. Note that, for the sake of simplicity, glycosylation is omitted from the illustration but the glycosylation sites within the fibril core (N145, N151, N164, and N183) are reported to be fully glycosylated. d, In TMEM106B fibrils, three major filament folds (I-III) and two doublet polymorphisms (1,2) have been reported. Unlike other amyloid proteins, no clear relationships between the filament folds and diseases were found. Folds I-III and doublets 1-2 are extracted from PDB: 7QVC, 7QWG, 7QWM, 7QVF, and 7SAS, respectively.Figure was created with BioRender.com.
Beyond FTLD-GRN, TMEM106B variation has also been linked to FTLD-TDP with C9orf72 repeat expansions [69, 70] and many other neurodegenerative diseases including AD [32, 71, 72], chronic traumatic encephalopathy [73], and LATE-NC [60]. In addition, a genetic study has found that TMEM106B is a genetic modifier of cognitive decline in PD [74] and of cognitive and motor functions in ALS [75, 76]. Furthermore, a dominant mutation (D252N) in TMEM106B was found in a patient with hypomyelinating leukodystrophy (HLD), a group of genetic disorders that affect the development of the myelin sheath in the brain [77], suggesting a role of TMEM106B in myelination.
Multiple SNPs near and in the TMEM106B gene, including the SNPs associated with neurodegenerative diseases described above, such as rs1990622, rs6966915, and rs1020004, are in strong linkage disequilibrium (LD), constituting two common TMEM106B haplotypes, one associated with increased disease risk, and the other with a protective effect [10, 78] (Fig. 2a). It is therefore difficult to determine the functional variant(s) on the haplotype that are responsible for modulating the disease risk. Within the haplotype block, rs3173615 is the only coding SNP, where threonine at amino acid position 185 on the risk haplotype is changed to serine on the protective haplotype (T185S). To date, cell culture and mouse studies to examine the effects of the amino acid substitution have yielded inconsistent results. We will discuss the T185S variant below in the sections “Endolysosomal functions of TMEM106B” and “Mouse models of PGRN- and/or TMEM106B-associated neurological diseases”. Another study has proposed that the noncoding rs1990620 risk variant increases TMEM106B mRNA expression by preferentially recruiting chromatin-organizing protein CCCTC-binding factor (CTCF) and thereby altering chromatin architecture [79]. However, while an independent study has confirmed an increase in TMEM106B mRNA in the frontal cortex of individuals with the risk haplotype [7], others have found no significant change [63, 80, 81]. Alternatively, recent studies have identified an AluYb8 element insertion in 3’UTR of the risk haplotype that may act as a functional variant [68, 82] (Fig. 2a). By using publicly available databases as well as a novel large dataset, one of the studies has also shown that the risk haplotype is associated with increased TMEM106B protein levels, but not with TMEM106B mRNA expression [68]. Consistent with this result, a proteomic study has reported an age-dependent increase in hippocampal TMEM106B protein levels specifically in the risk variant carriers [83]. Thus, it is possible that the AluYb8 insertion in the risk haplotype causes an increase in TMEM106B protein levels to affect the development and presentation of neurodegenerative diseases.
PGRN and TMEM106B in brain aging
Aging is a major risk factor for most neurodegenerative diseases [84]. Interestingly, human genetic studies have also linked PGRN and TMEM106B to brain aging. A GWAS study identified both the GRN and TMEM106B genes as genetic modifiers of biological aging in the human cerebral cortex [85]. In addition, GRN and TMEM106B variants were shown to interact genetically in the regulation of the biological aging [85]. In this study, TMEM106B variants were also found to be associated with inflammation and cognitive deficits even without known brain diseases [85]. Consistent with these results, another study has shown that the TMEM106B rs1990621 protective variant is associated with increased neuronal cell proportion specifically in elderly individuals, independent of disease status [86]. Moreover, a recent study finds that the GRN and TMEM106B AD protective alleles are strongly enriched in cognitively healthy centenarians [87]. These studies highlight the importance of PGRN and TMEM106B in maintained cognitive and brain health during aging.
The critical role of PGRN and TMEM106B in brain aging is also supported by mouse genetic studies, where deletion of either Grn or Tmem106b gene is reported to cause accelerated accumulation of lipofuscin in the brain [88, 89]. The accumulation of lipofuscin within postmitotic cells like neurons is considered a hallmark of aging because the rate of lipofuscin accumulation positively correlates with that of aging, although it is also associated with some pathological conditions such as lysosomal storage disorders [90, 91]. Importantly, studies have also shown that deleting both Grn and Tmem106b genes in mice further exacerbates lipofuscin accumulation in the brain [92, 93].
The precise mechanism by which GRN and TMEM106B variants affect brain aging and whether the relevant pathways also contribute to an increased risk for neurodegenerative diseases are currently unclear. However, given that one of the nine hallmarks of aging is loss of proteostasis [84, 94], it is likely that endolysosomal function of PGRN and TMEM106B plays an important role in brain aging.
Endolysosomal functions of PGRN
PGRN, encoded by the GRN gene in humans, is a 593 amino-acid highly glycosylated protein composed of N-terminal signal sequence followed by 7.5 cysteine-rich granulin domains that are connected by short linker regions [95, 96] (Fig. 1b). PGRN is mainly expressed in neurons and microglia in the brain [97–101], localized to the lysosome, and also secreted into the extracellular space. PGRN expression is regulated by the transcriptional factor EB (TFEB), a major regulator of lysosomal biogenesis and autophagy [102, 103]. There are two TFEB binding sites upstream of the GRN coding sequence [102]. PGRN secreted into the extracellular space is delivered to lysosomes by sortilin- or prosaposin (PSAP)-dependent endocytosis in neurons [104–106] (Fig. 1c). In addition, a recent study suggests the existence of additional sortilin and PSAP independent pathways mediating lysosomal trafficking of PGRN in microglia [105].
PGRN can be processed into mature granulins both extracellularly and intracellularly [95]. However, blocking lysosomal delivery of PGRN by knocking out sortilin significantly increases PGRN and decreases granulin levels in mouse brain lysates, suggesting that the PGRN processing occurs predominantly in the lysosome in the mouse brain [104, 105] (Fig. 1c). Several cathepsins and asparagine endopeptidase (AEP) have been proposed to mediate the lysosomal processing of PGRN into granulins [107–110]. Both PGRN and granulins are thought to play a critical role in lysosomal function [95, 96], although the exact functions of individual granulins versus PGRN remain unclear.
PGRN/granulins have been reported to bind to several endolysosomal proteins (Table 1). As described above, PGRN binds to sortilin and PSAP for its lysosomal delivery [104, 106] (Fig. 1c). Sortilin is a type I transmembrane protein of the vacuolar protein sorting 10 (VPS10) family localized to the cell surface, secretory, and endocytic compartments [111]. Sortilin functions as a cell surface PGRN receptor and regulates plasma, serum, and brain PGRN levels [104, 112]. A significant increase in PGRN levels has been reported in the brain and serum of sortilin knockout (KO) mice [104, 105]. PGRN binds to the β-propeller domain of sortilin through its C-terminal tail. It has been shown that deletion of the last three residues of PGRN (QLL) abolishes PGRN binding to sortilin [113]. PSAP is the precursor of saposin peptides that is, like PGRN, secreted into the extracellular space but is also localized or delivered to the lysosome via the PSAP receptors cation-independent mannose 6-phosphate receptor (M6PR) and low-density lipoprotein receptor-related protein 1 (LRP1) [106]. PGRN binds to PSAP through the linker region between saposins B and C [114], and can be delivered to the lysosome via M6PR and LRP1, independent of sortilin [106]. On the other hand, the PGRN-PSAP complex may also be important to facilitate neuronal uptake and lysosomal delivery of extracellular PSAP via sortilin. In the lysosome, PSAP can be processed into saposin peptides (A, B, C, and D), which are known to be involved in lysosomal glycosphingolipid degradation [115].
Table 1.
Endolysosomal PGRN or TMEM106B-binding proteins with known functional interaction
| Binding proteins | PGRN or TMEM106B | Procedures | Functions | References |
|---|---|---|---|---|
| Cathepsin D | PGRN | Co-IP (cell culture) | Stabilize and activate cathepsin D | [118–121] |
| Cathepsin D | TMEM106B | Brain IP-MS, co-IP (cell culture) | Stabilize cathepsin D | [132, 138] |
| Sortilin | PGRN | In vitro SPR, co-IP (cell culture), cell-based binding assay | Accelerate lysosomal delivery | [104, 113] |
| PSAP | PGRN | Co-IP (cell culture) | Accelerate lysosomal delivery | [106, 114] |
| GCase | PGRN | In vitro SPR, co-IP (cell culture) | Stabilize and activate GCase | [49, 206–208] |
| HexA | PGRN | Co-IP (cell culture) | Increase HexA activity | [219] |
| Rab2 | PGRN | Co-IP (cell culture) | Mediate autophagosome-lysosome fusion | [260] |
| CD68 | PGRN | Co-IP (cell culture), cell-based binding assay | Reciprocally stabilize their proteins | [261] |
| MAP6 | TMEM106B | Brain IP-MS, co-IP (Brain and cell culture) | Control dendritic trafficking of lysosomes in neurons | [146] |
| CHMP2B | TMEM106B | Co-IP (cell culture) | Regulate autophagic flux | [150] |
| V-ATPase AP1 | TMEM106B | Co-IP (cell culture) | Stabilize V-ATPase and lysosomal pH | [132, 137] |
| GALC | TMEM106B | Brain IP-MS, co-IP (cell culture) | Regulate GALC activity | [138] |
Abbreviations: SPR, surface plasmon resonance; IP-MS, immunoprecipitation-mass spectrometry
The other well-characterized PGRN/granulin-binding proteins in the endolysosomal system are β-glucocerebrosidase (GCase) and cathepsin D. We will discuss the role of PGRN-GCase interaction below in the section of “PGRN and TMEM106B in brain lipid metabolism”. Cathepsin D is a lysosomal aspartyl protease that has been linked to several neurodegenerative diseases, including AD and PD [116]. Like PGRN, homozygous mutations in the cathepsin D gene (CTSD) cause neuronal ceroid lipofuscinosis (CLN10) [117]. Several groups have independently reported that PGRN or several granulins can bind to cathepsin D [118–121], although the functional consequence of this interaction is still controversial. In vitro, recombinant PGRN and granulins have been shown to stimulate maturation of pro-cathepsin D and/or increase cathepsin D activity [118–120, 122]. Consistent with the in vitro results, reduced cathepsin D activity has been seen in fibroblasts and induced pluripotent stem cell (iPSC)-derived neurons generated from FTLD-GRN patients [119, 123]. However, in the mouse brain, PGRN deficiency has been consistently shown to cause an age-dependent increase in both pro- and mature-cathepsin D levels and cathepsin D activity [118, 124–128],, although some studies have reported that cathepsin D activity is decreased in PGRN KO brains when it is normalized to mature cathepsin D protein levels [118, 128]. Another study has shown that, at 2 months of age, cathepsin D activity is not changed in the brain, but decreased in the liver and spleen of PGRN KO mice [121]. Thus, the net effect of PGRN on cathepsin D function remains uncertain.
Endolysosomal functions of TMEM106B
TMEM106B is a 274 amino-acid highly glycosylated type II transmembrane protein localized to the endolysosomal system [10, 78] (Fig. 2b). TMEM106B is ubiquitously expressed in multiple tissues and in both neurons and glia in the central nervous system [129–133]. TMEM106B is known to form homodimers, which can be detected either by co-immunoprecipitation (co-IP) experiments [134, 135] or by SDS-PAGE under non-reducing conditions [136–138], although the functional importance of the dimers remains unknown. N-terminal cytoplasmic region of TMEM106B is intrinsically disordered without well-defined three-dimensional structure [139], but TMEM106B may undergo N-myristoylation [140–142]. In addition, a study has shown that the extended dileucine-based motif, ENQLVALI, in the cytoplasmic region is a potential lysosomal sorting signal of TMEM106B [143]. In contrast to the disordered N-terminal region, the crystal structure of C-terminal luminal region has revealed a compact fibronectin type III domain with a ubiquitous 7-bladed β sandwich fold, which is closely related to immunoglobulin-like domains [144]. The C-terminal luminal region contains five N-glycosylation sites with the consensus sequence motif N-X-S/T at N145, N151, N164, N183, and N256 [145]. A cell culture study using the glycosylation site mutants has shown that endoglycosidase H-resistant complex glycosylation occurs at N183 and N256 and the complex glycosylation is required for proper transport of TMEM106B to late endosomal/lysosomal compartments [145].
Given its endolysosomal localization, the role of TMEM106B in lysosomal functions has been investigated in numerous cell culture studies. Several studies have consistently shown that overexpression of TMEM106B induces enlargement of lysosomal compartments and formation of LAMP1-positive large cytoplasmic vacuolar structures, while its knockdown results in reduction in the size of lysosomes in immortalized cells and primary neurons [134–136, 143]. TMEM106B levels also affects motility of lysosomes in primary neurons. In DIV20 mouse cortical neurons, overexpression and knockdown of TMEM106B is shown to decrease and increase total lysosomal movements in dendrites, respectively [134]. Other studies have shown that TMEM106B knockdown or KO increases the number of moving lysosomes and retrograde transport of lysosomes in dendrites of DIV9 primary rat hippocampal neurons [146] and in axons of DIV7 primary mouse motoneurons [89]. Maintaining acidic pH (4.2–5.3) is essential for regulating many functions of lysosomes [147]. However, in primary mouse cortical neurons, TMEM106B deficiency has been shown to cause impairment in lysosomal acidification [137], although TMEM106B overexpression has mixed results with some showing an impairment in lysosomal acidification in HeLa cells [136, 143] and others reporting enhanced acidification in HEK293T cells and murine and human lung cancer cell lines [132, 148]. It remains to be determined whether the mixed results stem from the different cell lines used or the different level of overexpression. Although TMEM106B is expressed in both neurons and glia, there are only a few cell culture studies investigating its role in glia. One study using the Oli-Neu oligodendrocyte cell line has suggested that TMEM106B regulates lysosomal function and positioning and PLP trafficking in oligodendrocytes [132]. Another study has shown that TMEM106B deficiency decreases TREM2 levels and cell viability and increases inflammatory responses in cultured microglia [133].
Because several studies demonstrated an association of the TMEM106B protective variants with increased human plasma PGRN levels [62, 63], the effect of TMEM106B expression on PGRN levels has been assessed in cell culture studies. Although it is consistently shown that TMEM106B knockdown has no significant effects on PGRN mRNA and protein levels [135, 145, 146], overexpression of TMEM106B has been reported to increase extracellular and/or intracellular PGRN levels [61, 135, 136] and reduce PGRN processing into granulins [108] in certain cell lines. Overexpression of TMEM106B has been reported to induce nuclear translocation of TFEB and expression of TFEB-regulated lysosomal genes in HEK293 cells, primary cortical neurons, and lung cancer cells [134, 148]. However, an increase in PGRN mRNA by TMEM106B overexpression has not been reported in other studies [135, 145]. It remains unclear whether the results from these cell culture studies are responsible for the human observations since the TMEM106B protective variants may reduce TMEM106B protein levels [7, 10, 68, 78, 79, 83].
Several studies have explored TMEM106B-binding proteins (Table 1). A yeast two-hybrid (Y2H) screen using cytoplasmic or luminal fragment of TMEM106B for bait and a prey library derived from human adult brain has identified TMEM106C, the endocytic adaptor proteins, AP2M1 and CLTC, and the vacuolar protein sorting proteins, VPS11 and VPS13D as N-terminal TMEM106B-binding proteins, although the functional importance of these interactions remains to be determined [134]. In the Y2H screen, no proteins have been found to bind to C-terminal TMEM106B [134]. An immunoprecipitation (IP) and liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis using P15 rat brain has identified microtubule-associated protein 6 (MAP6) as a TMEM106B-binding protein [146]. The TMEM106B-MAP6 interaction has been confirmed by co-IP experiments and shown to be crucial for controlling dendritic trafficking of lysosomes in primary rat neurons [146]. Another IP and LC-MS/MS analysis using WT and TMEM106B KO mouse brains has identified 22 potential lysosomal TMEM106B-binding proteins, including vacuolar-ATPase (V-ATPase), cathepsin D, and galactosylceramidase (GALC) [138]. Co-IP experiments have confirmed the TMEM106B-V-ATPase, especially V-ATPase accessary protein 1 (AP1) [132, 137], TMEM106B-cathepsin D [132, 138], and TMEM106B-GALC interactions [138]. V-ATPase is an ATP-driven proton pump involved in acidification of intracellular compartments [149]. TMEM106B deficiency has been shown to cause down-regulation of several V-ATPase V0 subunits and AP1 in the mouse brain and impair lysosomal acidification in primary mouse neurons [137]. The TMEM106B-cathespin D interaction may be important for maintaining proper cathepsin D levels [132]. We will discuss the role of TMEM106B-GALC interaction in the section of “PGRN and TMEM106B in lipid metabolism” below. A co-IP experiment using HEK293T cells has shown association between TMEM106B and charged multivesicular body protein 2B (CHMP2B), a subunit of the endosomal sorting complex required for transport-III (ESCRT-III) [150]. A mutation in CHMP2B has been reported to causes an autosomal dominant form of frontotemporal dementia [151]. TMEM106B-CHMP2B interaction may affect the ESCRT-mediated endosomal and autophagy pathways [150].
As aforementioned, T185S variant and D252N mutation of TMEM106B have been associated with FTLD and HLD, respectively. Multiple cell culture studies have assessed the effects of these amino acid substitutions on TMEM106B functions. First, the T185S variant and D252N mutation are located near the complex glycosylation sites N183 and N256. However, no significant effects of these amino acid substitutions on glycosylation have been reported so far [61, 132]. Studies have also shown that T185S variation has no significant effects on homodimerization of TMEM106B [135], on TMEM106B-GALC interaction [138], or on TMEM106B-induced lysosomal enlargement [135], nuclear translocation of TFEB [134], or increases of PGRN [61, 135]. An overexpression study has shown that T185S variation destabilizes TMEM106B protein [61], but this is not supported by other cell culture and mouse studies [135, 150, 152, 153]. Another study has reported that T185S variation significantly reduces association of TMEM106B with wild-type and mutant CHMP2BIntron5 and abolishes exacerbating effects of TMEM106B on CHMP2BIntron5-induced endosomal/autophagic defects and neurotoxicity [150]. For HLD-associated D252N mutation, studies have shown no significant effects of the mutation on the dimerization [132], TMEM106B-V-ATPase AP1 interaction [132], TMEM106B-cathepsin D interaction [132], and TMEM106B-GALC interaction [138]. However, D252N mutation has been reported to abolish TMEM106B-induced lysosomal enlargement and acidification [132]. In addition, a study using patient-derived fibroblasts has shown that heterozygous D252N mutation causes lysosomal dysfunction [154].
Amyloid fibrils of a C-terminal fragment (CTF) of TMEM106B
Recent studies using cryo-electron microscopy (cryo-EM) discovered amyloid fibrils composed of TMEM106B CTF corresponding to residues 120–254 from the brains of aged people ( > ~ 45 years) without known neurological disease as well as subjects with FTLD-TDP and a variety of other neurodegenerative diseases including AD, PD, LBD, progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), and ALS [155–161]. Immunohistochemical analysis using antibodies against the luminal domain of TMEM106B has shown that TMEM106B aggregates are found in both neurons and glia and are most abundant in brain astrocytes [129, 162, 163]. In addition, greater TMEM106B pathology has been observed in FTLD-GRN and LATE-NC cases [160, 163]. Immunoblot analysis has shown that the TMEM106B risk haplotype is associated with increased sarkosyl-insoluble 29 kDa CTF of TMEM106B [83, 164, 165], while decreasing dimeric TMEM106B in the RIPA-soluble fraction [164].
Although TMEM106B reportedly undergoes physiological processing [166, 167], the exact mechanism and proteases involved in the C-terminal cleavage of TMEM106B to form the fibrils are currently unclear. It appears that a precise processing of TMEM106B between resides 119 and 120 is required for the amyloid fibril formation because cryo-EM structure showed that Ser120 is buried in the fibril core 120–254, leaving no space for the other N-terminal residues [158]. In addition, there may be a second cleavage at some residue after Gly254 since antibodies against the extreme C-terminus (the residues 263–274, 253–274, and/or 259–274) of TMEM106B did not detect TMEM106B inclusions by immunostaining [129, 162] or sarkosyl-insoluble 29 kDa CTF by immunoblot [167] (Fig. 2c). A recent study has shown that the C-terminal trimming also occurs under physiological conditions [167]. Three major filament folds and two doublet polymorphisms have been described, although unlike other amyloid proteins such as tau, a-synuclein, and TDP-43, no relationship between the filament fold and disease was observed [156, 158] (Fig. 2d). So far, the TMEM106B fibrils have been identified only from human tissues and have not been found from any mouse tissues [129] or in cell culture systems. It is notable that a recent study has found amyloid fibrils of TMEM106B CTF in Biondi bodies, filamentous amyloid inclusions in ependymal cells of the choroid plexuses in aged human brains [168]. Biondi bodies have not been found in nonhuman mammals except for chimpanzees [169]. The result raises the possibility that TMEM106B fibrils may be formed only in certain primates.
It also remains unclear whether the TMEM106B fibrils play a role in brain aging and/or pathophysiology of neurodegenerative diseases. Recently, TMEM106B CTFs were shown to aggregate and cause neuronal loss and behavioral deficits when expressed in neurons of C. elegans [170]. However, the experimental system included N-terminal dendra2-tagged CTF, which likely forms aggregates with a structure distinct from ones observed in human brains. Therefore, the results need to be cautiously interpreted.
Mouse models of PGRN- and/or TMEM106B-associated neurological diseases
While PGRN haploinsufficiency causes FTLD-TDP, Grn heterozygous null (Grn+/−) mice develop no obvious disease-associated pathologies, such as TDP-43 inclusions, in the brain. However, some studies using a large cohort of mice have reported subtle but significant lysosomal dysregulation and behavioral abnormalities such as social deficits in Grn+/− mice, which may be relevant to FTLD-GRN [98, 171, 172]. In addition, a recent study has shown that Grn+/− mice have reduced dendritic spine head diameter in the prefrontal cortex [173].
While Grn homozygous null (Grn−/−) mice are viable and fertile, they exhibit several features of FTLD-GRN and CLN11 including lysosomal dysregulation, lipofuscinosis, neuroinflammation, synaptic loss, retinal ganglion cell (RGC) loss, and obsessive compulsive disorder (OCD)-like behavior as well as social deficits [16, 88, 137, 174–177] and therefore have been widely used as a model of FTLD-GRN and CLN11. However, Grn−/− mice fail to develop neurodegeneration or TDP-43 pathology in the cerebral cortex, although several studies have described selective loss of excitatory neurons and cytoplasmic TDP-43-positive aggregates in the thalamus of aged Grn−/− mice [175, 176, 178]. GrnR493X knockin mice harboring one of the most common human GRN mutations, a premature termination codon (PTC) at arginine 493 (R493X), have also been generated [179]. Homozygous GrnR493X knockin mice have significantly reduced Grn mRNA levels due to nonsense-mediated mRNA decay (NMD) and do not express detectable PGRN protein, resulting in a phenocopy of Grn−/− mice [179–181]. Thus, GrnR493X knockin mice may be a useful model to test therapeutic approaches targeting PTC or NMD (Table 2).
Table 2.
Mouse models of PGRN- and/or TMEM106B-associated neurological diseases
| Mouse models | Diseases | Behavioral phenotypes | Histological phenotypes | Biochemical phenotypes | References |
|---|---|---|---|---|---|
| Grn+/− mice | FTLD | Social deficits | Lysosomal dysfunction | Lysosomal dysfunction | [98, 171, 172] |
| Grn−/− mice | FTLD, CLN11 | Social deficits, OCD-like behavior | Lysosomal dysfunction, lipofuscinosis, neuroinflammation, complement activation, RGC loss, inhibitory synaptic and excitatory neuronal loss and Increased cytoplasmic TDP-43 in the thalamus | Lysosomal enzyme dysregulation, decreased BMP, increased GlcCer/Sph and gangliosides | [16, 88, 137, 174–178] |
| GrnR493X/R493X mice | FTLD, CLN11 | Social deficits, OCD-like behavior | Lysosomal dysfunction, lipofuscinosis, neuroinflammation, inhibitory synaptic and excitatory neuronal loss in the thalamus | Lysosomal enzyme dysregulation, decreased BMP, increased GlcSph and gangliosides | [179–181] |
| Tmem106b−/− mice | FTLD, HLD | Late-onset motor deficits | Minor myelination deficits, vacuole accumulation, late-onset cerebellar gliosis and Purkinje cell loss | Lysosomal enzyme dysregulation | [89, 132, 133, 187–190] |
| Grn−/− Tmem106b Tg micea | FTLD | No data available | Exacerbated lysosomal abnormalities and lipofuscinosis | No data available | [191] |
| Grn+/− Tmem106b+/− mice | FTLD | No amelioration of Grn+/− phenotypes | No data available | No amelioration of Grn+/− phenotypes | [192] |
| Grn−/− Tmem106b+/− mice | FTLD | No data available | No amelioration of Grn−/− phenotypes | No amelioration of Grn−/− phenotypes | [193] |
| Grn−/− Tmem106bH/H miceb | FTLD | Improved disinhibition, but late-onset motor deficits | Attenuated lysosomal enzyme dysfunction, reduced RGC loss, no amelioration of microglial activation and lipofuscin | Attenuated lysosomal enzyme dysregulation | [137, 194, 195] |
| Grn−/− Tmem106b−/− mice | FTLD | Early-onset motor deficits | Exacerbation of Grn−/− phenotypes, neurodegeneration and TDP-43 pathology in the brainstem and spinal cord. | Exacerbation of Grn−/− phenotypes, more insoluble p-TDP-43 in the brainstem and spinal cord | [92, 93, 193, 195] |
| Grn−/− Tmem106bT186S/T186S mice | FTLD | No data available | No amelioration of Grn−/− phenotypes | No amelioration of Grn−/− phenotypes | [153] |
| APPswe/PS1ΔE9 Grn−/− mice | AD | Improved learning deficit | Reduced diffuse Aβ plaque burden, more activated microglia near plaques, more complement deposition | Reduced insoluble Aβ | [52] |
| 5XFAD Grn−/− mice | AD | No obvious behavioral changes in the open-field and Y-maze | Reduced Aβ pathology in male, more activated microglia near plaques, increased expression of lysosomal proteins | Reduced APP and Aβ in young male, increased expression of lysosomal proteins | [182] |
| Tg2576 Grn+/− mice | AD | No data available | Reduced Aβ deposition | Reduced insoluble Aβ | [183] |
| J20 Grnflox/flox LysM-Cre | AD | exacerbated learning and memory deficits | Increased hippocampal Aβ deposition | No data available | [184] |
| JNPL3 Grn+/− mice | Tauopathy | No data available | More p-tau inclusions | Increased soluble and insoluble p-tau | [185] |
| PS19 Grn−/− mice | Tauopathy | Exacerbated disinhibition, but improved memory deficits | More GlcCer-positive tau and α-synuclein inclusions, but attenuated hippocampal atrophy | No change in insoluble tau, higher GlcCer | [49] |
| PS19 Tmem106b−/− mice | Tauopathy | Hyperactivity, exacerbated motor and cognitive deficits | Exacerbated tau pathology, neuroinflammation, and neurodegeneration | Increased soluble and insoluble p-tau | [152, 198] |
| PS19 Tmem106bT186S/T186S mice | Tauopathy | Improved cognitive decline | Attenuated hippocampal atrophy, no changes in glial and tau pathology | No change in p-tau | [152] |
a Tmem106b Tg, transgenic mice with elevated TMEM106B levels
b Tmem106bH/H, mice with homozygous Tmem106b hypomorphic alleles
To examine the role of neuronal versus microglial PGRN, several conditional PGRN KO mice have been generated. Studies using Grnflox/flox mice crossed with CaMKII-Cre, Nestin-Cre, or LyzM-Cre have demonstrated that neither neuronal nor microglial loss of PGRN is sufficient to induce lipofuscinosis and neuroinflammation in mice [97–99]. In addition, mice with depletion of both neuronal and microglial PGRN have also failed to develop lipofuscinosis and gliosis [101]. Therefore, these phenotypes appear to require essentially complete loss of PGRN in mice. Notably, two neuronal PGRN deficient mouse lines, CaMKII-Cre Grnflox/flox mice and Nestin-Cre Grnflox/flox mice, have been shown to develop social deficits, highlighting an important role for neuron-derived PGRN in social function [98]. In addition, microglial PGRN deficient mouse lines, LyzM-Cre Grnflox/flox mice and tamoxifen-treated Cx3Cr1-CreERT2 Grnflox/flox mice, exhibit excessive grooming, an OCD-like behavior, which is also observed in constitutive Grn−/− mice [100, 101].
To investigate the role of PGRN in AD and the mechanism by which GRN variants and mutations increase AD risk, numerous AD mouse models have been analyzed on PGRN-deficient background (Table 2). Studies using APPswe/PS1∆E9 and 5XFAD mouse models of amyloid pathology have shown that constitutive loss of PGRN has no exacerbating effects on Aβ pathology [52, 182], which is consistent with human ADNI biomarker data showing no effects of the GRN rs5848 AD variant on amyloid pathology [52]. In fact, a reduction of Aβ pathology has been reported in those mice with PGRN loss. In addition, a study has shown that PGRN haploinsufficiency reduces amyloid pathology in female Tg2576 APP transgenic mice although only a limited number of mice were examined [183]. In contrast, selective reduction of microglial PGRN by crossing with LysM-Cre mice has been shown to significantly increase Aβ pathology in J20 APP transgenic Grnflox/flox mice [184]. It remains unclear whether the contrasting results stem from differences in PGRN manipulation or simply from different AD mouse models utilized in those studies. The role of PGRN in tau pathology, another hallmark of AD, has also been investigated using several mouse models of tauopathy. Interestingly, not only complete loss but also haploinsufficiency of PGRN has been shown to increase tau pathology in P301L (JNPL3) and P301S (PS19) mouse models of tauopathy [49, 185], although tau-mediated neurodegeneration was paradoxically attenuated in PGRN-deficient PS19 tauopathy mice [49]. PGRN deficiency has been reported to have no significant effects on tau spreading induced by AD-derived tau fibrils in the mouse brain [186].
TMEM106B homozygous KO (Tmem106b−/−) mice are viable and fertile and exhibit no overt behavioral deficits during developmental and adult stages, but show minor myelination deficits in histopathological analysis [132, 187], consistent with human evidence that a de novo mutation in TMEM106B causes HLD [77]. The histological deficits include decreases in MBP, PLP, MOG, or OLIG2 immunostaining and Black Gold II stain in the corpus callosum [132, 133, 187]. Transcriptomic analysis has also shown global changes in myelin pathway in Tmem106b−/− brains [187]. In addition, Tmem106b−/− mice are more susceptible to cuprizone-induced demyelination and have a reduced capacity to remyelinate the corpus callosum [133, 187]. In addition to myelination deficits, accumulation of LAMP1-positive vacuoles is observed at the axon initial segment of motoneurons of Tmem106b−/− brains [89]. Aged Tmem106b−/− mice have been reported to develop motor deficits associated with cerebellar gliosis and Purkinje cell loss [188, 189] (Table 2). It is notable that increased Purkinje cell loss is also observed in human subjects with a TMEM106B disease risk allele [190]. To examine a role of TMEM106B in microglia in vivo, microglial TMEM106B deficient mice have been generated by treating Cx3Cr1-CreERT2 Tmem106bflox/flox mice with tamoxifen [133]. Microglia-specific loss of TMEM106B resulted in decreased microglial proliferation and activation in response to LPS [133]. Although TMEM106B is expressed in both neurons and glia [130–133], no other cell type-specific deletions of Tmem106b have been investigated so far.
Given the role of TMEM106B variants in FTLD-GRN [7, 61–66], several TMEM106B transgenic and KO mice have been crossbred with PGRN-deficient mice (Table 2). The TMEM106B protective/risk variants are predicted to reduce/increase TMEM106B protein without a complete loss [7, 10, 68, 78, 79, 83]. Thus, assessing the effects of partial reduction/increase of TMEM106B levels in Grn+/− or Grn−/− mice may be the most relevant to understanding human variants. Regarding this point, one study generated transgenic mice expressing human TMEM106B under the neuronal specific CaMKII promoter [191]. In these TMEM106B transgenic mice, total TMEM106B levels were not altered in the brain and were only significantly increased on Grn−/− background at 17–20 months of age. However, the elevated TMEM106B has been shown to exacerbate accumulation of lipofuscin and enlarged lysosomes in Grn−/− brains [191]. In contrast, TMEM106B haploinsufficiency has reportedly no significant effects on phenotypes of Grn+/− [192] and Grn−/− mice [193]. Although Tmem106bT186S knockin mice harboring the disease protective T185S variant have recently also been generated and crossed with Grn−/− mice, no protective effects of the T185S variant have been found in Grn−/− mice [153]. By immunoblot analysis, no significant changes in TMEM106B protein levels or apparent molecular weight have been observed in the brain of Tmem106bT186S knockin mice [152, 153], which is inconsistent with the results from the cell culture study described earlier [61]. The first TMEM106B-deficient mouse line reported [137] was generated by a gene trap strategy and found to be a hypomorph expressing 5–10% residual endogenous full-length TMEM106B [194, 195]. Interestingly, this hypomorphic mutation in Tmem106b ameliorates lysosomal and behavioral phenotypes of adult but not aged Grn−/− mice [137, 194, 195].
Several TMEM106B complete null mouse lines have also been crossbred with PGRN-deficient mice [92, 93]. Importantly, in contrast to the hypomorphic mutation described above, complete loss of TMEM106B on Grn−/− background leads to severe early motor dysfunction in mice. The double KO (DKO) mice also exhibit robust lysosomal and FTLD-GRN-like pathologies including neurodegeneration and TDP-43 inclusions in the brainstem and spinal cord [92, 93, 193]. Therefore, this DKO line has been proposed as a mouse model of FTLD-GRN and ALS/FTLD [196, 197], since single Grn−/− mice do not show robust neuronal loss or TDP-43 pathology as described above. However, it is worth noting that the TMEM106B risk variants do not cause complete loss of TMEM106B protein and previous studies have suggested that the risk variants may rather increase TMEM106B levels [7, 10, 68, 78, 79, 83]. Therefore, use of the DKO line as a model of FTLD-GRN with TMEM106B risk variants must be interpreted cautiously. Taken together with the hypophorphic allele results, the data suggest that TMEM106B levels have biphasic effects, reducing Grn−/− pathology with 90% reduction but exacerbating Grn−/− pathology with 100% TMEM106B reduction.
The role of TMEM106B in tauopathy has been also explored in mouse models. Recent studies have shown that complete loss of TMEM106B exacerbates tau pathology and tau-mediated neurodegeneration in the PS19 tauopathy mouse model [152, 198], while the T185S protective variant protected against cognitive deficits and neurodegeneration without affecting tau pathology [152] (Table 2). These results may be consistent with human genetic studies showing a link between TMEM106B variants and increased AD risk [32, 71, 72], although the underlying mechanism remains unknown. Effects of TMEM106B deficiency on tau spreading has been investigated in an AD-tau injection mouse model. However, similar to the results with Grn−/− mice, no significant effects of TMEM106B loss on tau spreading have been found [186]. To date, effects of TMEM106B deficiency on Aβ pathology and Aβ-associated deficits has not been reported for preclinical models.
Most recently, a novel transgenic mouse model expressing human TMEM106B under the CAG promotor was generated [199]. In contrast to the prior model described above [191], the transgenic mice successfully overexpress human TMEM106B, with a 4- to 8-fold increase in the protein levels in hemizygotes and homozygotes, respectively, as compared to the levels of endogenous mouse TMEM106B. Interestingly, down-regulation of immediate early genes, altered synaptic signaling, an anxiety-like behavior, and mild neuronal loss have been observed in these transgenic mice, suggesting that increased TMEM106B levels negatively impact brain health [199]. The transgenic mice have not been crossbred with PGRN-deficient mice or other mouse models of neurodegenerative diseases yet.
Human iPSC models of FTLD-GRN and CLN11
The differential effects of PGRN haploinsufficiency on human and mouse brains and difficulty of reproducing effects of the TMEM106B risk haplotype in mouse models may suggest the existence of human-specific pathogenic mechanisms in FTLD-GRN, and experimental systems using human patient-derived iPSC lines may be useful to explore such mechanisms. However, there are currently only a few published studies investigating in vitro models using iPSCs with heterozygous GRN mutations. So far, none of these studies show clear TDP-43 inclusions by immunostaining. Two studies using iPSC lines generated from FTLD-GRN patients carrying heterozygous S116X nonsense mutation and A9D missense mutation have observed PGRN haploinsufficiency and cytoplasmic mis-localization of TDP-43 in iPSC-derived neurons [119, 200]. The iPSC-derived neurons with GRN A9D mutation have also shown accumulation of insoluble TDP-43 and lipofuscin and reduced cathepsin D activity as mentioned earlier [119]. Another study using an iPSC line generated from FTLD patients carrying heterozygous GRNIVS1+5G >C mutation did not detect TDP-43 aggregation, but has observed insufficient differentiation into cortical neurons, but not motor neurons, which can be rescued by genetic correction or WNT inhibitor [201]. There are currently no studies examining iPSC lines generated from TMEM106B protective versus risk variants.
Although potentially less relevant to FTLD-GRN, several previous studies have established in vitro models using iPSC lines with homozygous GRN mutation. A recent study has reported that iPSC-derived cortical neurons carrying CLN11-causing GRN Thr272fsX10 homozygous mutation display TDP-43 mis-localization and cleavage as well as lipofuscin accumulation [202]. Another study using iPSC-derived neuron/astrocyte co-cultures has shown that GRNR493X/R493X astrocytes significantly delay spiking activity in developing neurons [203]. In addition, GRN homozygous null (GRN−/−) human mature brain organoids composed of iPSC-derived cortical neurons and astrocytes have been shown to recapitulate TDP-43 mis-localization, hyperphosphorylation, and mis-splicing of STMN2 transcripts [204], which is an indicator of TDP-43 pathology [205]. Finally, similar to Grn−/− Tmem106b−/− DKO mice, a study has observed a significant increase in TDP-43 cleavage and phosphorylation, and in cathepsin B, D, and L activities of co-cultures composed of WT iPSC-derived neurons and GRN−/− TMEM106B−/− DKO iPSC-derived microglia, although the effects of single gene KO were not evaluated [197].
PGRN and TMEM106B in brain lipid metabolism
Several groups have independently shown that PGRN physically interacts with glucocerebrosidase (GCase), a lysosomal enzyme that hydrolyzes the glycosphingolipids glucosylceramide (GlcCer) and glucosylsphingosine (GlcSph) [49, 206–208]. Biallelic mutations in the GBA1 gene, encoding GCase, result in the lysosomal storage disorder Gaucher disease, while heterozygous mutations have been identified as a common risk factor for PD [209]. PGRN deficiency attenuates GCase activity in the mouse brain [49, 206, 208, 210] and tissues [206]. Reduced GCase activity has also been reported in the brain of FTLD-GRN patients [208] as well as iPSC-derived GRN mutant neurons [211]. Lipidomic analysis confirms an increase in GlcCer or GlcSph levels in the brain of Grn−/− mice [49, 124, 210, 212] and Grn−/− Tmem106b−/− DKO mice [197]. An increase in GlcSph levels is also observed in the plasma of FTLD-GRN patients [210, 213]. Together, these studies suggest that PGRN binds GCase and potentiates the enzymatic activity to limit GlcCer levels in the brain (Fig. 3). PGRN binding to GCase may also stabilize lysosomal localization of GCase [49, 57]. Besides the direct binding to GCase, PGRN has also been suggested to increase GCase activity indirectly by affecting co-factors such as saposin C [211] or bis(monoacylglycero)phosphate (BMP) [210] as described below. It is important to note that a previous study has shown association of GRN variants with Gaucher disease [57], demonstrating a genetic interaction between GRN and GBA1. Consistent with the human genetic study, PGRN deficiency has been shown to exacerbate Gaucher disease-associated phenotypes of Gba1D409V/D409V knockin mice [214].
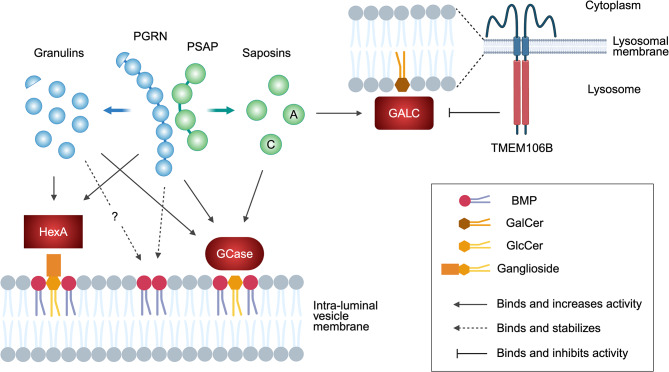
Fig. 3.
Glycosphingolipid regulation by PGRN and TMEM106B. PGRN and/or granulins bind to GCase and HexA and promote their activity to limit accumulation of GlcCer (and GlcSph) and gangliosides in lysosomes. PGRN also binds and stabilizes anionic phospholipid BMP, which stimulates GCase activity and ganglioside degradation at the intraluminal vesicles of lysosomes. Granulins may also bind and stabilize BMP, although it has not been proven. TMEM106B binds to GALC and regulates its activity to maintain GalCer levels in lysosomes. PSAP is cleaved into saposins A to D in lysosomes and saposins A and C are known to stimulate GALC and GCase activity, respectively. Figure was created with BioRender.com
Recent lipidomic studies from multiple groups have found a significant and robust decreases in BMP levels in the brain of Grn−/− mice [49, 124, 210, 212, 215–217] and Grn−/− Tmem106b−/− DKO mice [197]. BMP is an anionic phospholipid enriched in the endolysosomal compartment and its intralumenal vesicles [218]. A study has shown that adding liposomes containing BMP restores attenuated GCase activity in PGRN-deficient mouse brain lysates, suggesting that BMP deficiency may underlie GCase activity defects in Grn−/− mice [210]. A decrease in BMP levels has also been observed in CSF and the brains of FTLD-TDP patients with and without GRN mutations [210, 215], while an increase in BMP has been detected in urine samples of FTLD-GRN patients treated with AAV expressing PGRN [216]. The mechanism underlying this positive correlation between PGRN and BMP levels is still not fully understood. In vitro, His-tagged PGRN binds to liposomes containing BMP [210, 215], suggesting a direct effect of PGRN on BMP stability and/or turnover (Fig. 3). However, one study has found that only a minor fraction of untagged PGRN binds to liposomes containing BMP, and a control protein, His-tagged Cas9, also binds to the liposomes [215], suggesting that the His-tag might be responsible. Further investigation is required to elucidate the exact mechanism.
Accumulation of gangliosides, which are sialic acid-containing glycosphingolipids, is also reported in the brain of Grn−/− mice [212, 215, 219], Grn−/− Tmem106b−/− DKO mice [197], and FTLD-GRN patients [215]. Similar to GCase activity defects described above, a study has shown that ganglioside accumulation can be rescued by supplementation of BMP in PGRN-deficient HeLa cells [215]. Therefore, the ganglioside accumulation may also be a consequence of BMP deficiency caused by PGRN loss, which is consistent with a previous study demonstrating that BMP stimulates ganglioside degradation [220]. However, there is also a published study demonstrating that PGRN binds and increases the enzymatic activity of β-hexosaminidase A (HexA), a lysosomal enzyme that breaks down GM2 ganglioside [219] (Fig. 3).
In addition to specific lipid classes such as GlcCer and BMP, PGRN may regulate formation of lipid droplets in the cell. Lipid droplets are lipid-storing organelles composed of a neutral lipid core, primarily consisting of triacylglycerols (TAGs) and/or cholesterol esters, surrounded by a phospholipid monolayer [221] and known to accumulate in microglia in the aging brain [222]. Lipid-droplet-accumulating microglia were shown to be defective in phagocytosis and secrete proinflammatory cytokines [222, 223]. A CRISPR-Cas9 screen using BV2 mouse microglial cell line identified Grn as a genetic regulator of lipopolysaccharide-induced lipid droplet formation [222]. It was also reported that Grn−/− mice contain high numbers of lipid-droplet-rich microglia [222]. Another study has shown an increase in perilipin 2-positive lipid droplets in microglia of cuprizone-treated female Grn−/− mice [224]. In addition, PGRN haploinsufficiency has been shown to lead to aberrant lipid droplet formation in human monocyte-derived microglia-like cells [225]. The mechanism by which PGRN deficiency increases lipid droplets in microglia is currently unknown. Although an increase in polyunsaturated TAGs has been reported in Grn−/− mouse embryonic fibroblasts and in the liver of Grn−/− mice [226], it remains unclear whether TAGs are also increased in Grn−/− brains and whether the increase is a consequence or a cause of lipid droplet formation.
TMEM106B has also been implicated in lipid metabolism. A previous study has predicted that TMEM106B is a lipid transfer protein [227]. The luminal region of TMEM106B is found to be a member of late embryogenesis abundant-2 (LEA-2) domain superfamily, which has a long, conserved lipid-binding groove. Interestingly, two identified yeast LEA-2 homologues, Vac7 and Tag1, are localized to the degrative vacuole in yeast (equivalent to the lysosome in humans) and regulate PI(3,5)P2 generation and autophagy, respectively [227].
A recent targeted lipidomic analysis using TMEM106B-deficient mice has revealed that TMEM106B deficiency significantly decreases the glycosphingolipids galactosylceramide (GalCer) and its sulfated derivative sulfatide levels in the brain [138]. A decrease in several species of GalCer is also reported in the brain of Grn−/− Tmem106b−/− DKO mice, which was not rescued by AAV-mediated liver expression of brain-penetrant PGRN biologics [197]. GalCer and sulfatide are two major classes of myelin lipids, which together constitute ~ 27% of the myelin lipid [228]. Thus, the results are consistent with the finding that TMEM106B is a causal gene of HLD [77]. In humans, hexosylceramide and sulfatide levels are shown to be significantly lower in the TMEM106B disease risk variant carriers [83]. Mechanistically, TMEM106B physically interacts with galactosylceramidase (GALC), a lysosomal enzyme that hydrolyzes GalCer, via its luminal region (Fig. 3). GALC activity, but not protein levels, is significantly increased in TMEM106B-deficient mouse brains [138]. Whether alteration of GalCer and/or sulfatide levels is responsible for TMEM106B-associated neurological diseases such as HLD requires further investigation.
PGRN-boosting therapies
There are currently no approved therapies for FTLD-GRN and CLN11. As FTLD-GRN and CLN11 are caused by PGRN deficiency, an effective therapy for these diseases may be to increase PGRN protein levels by gene therapy or protein replacement therapy. Indeed, adeno-associated virus (AAV)-based gene therapy has been successfully used to restore PGRN levels and to improve pre-existing pathologies and deficits in Grn+/− and Grn−/− mice [98, 128, 216, 229]. In addition, AAV1/9-mediated PGRN expression has been reported to partially prevent brain pathologies and motor deficits of Grn−/− Tmem106b−/− DKO mice [196]. Interim results from a phase 1/2 clinical trial of AAV-based GRN gene therapy (PR006) have demonstrated that one-time injection of PR006 into the cisterna magna is generally safe and well tolerated and increases CSF PGRN levels [216]. In addition, two other AAV-based GRN gene therapies, PBFT02 and AVB-101, are currently in phase 1/2 clinical trials (NCT04747431 and NCT06064890, respectively) (Table 3). To avoid the logistics of central nervous system (CNS) injection and its potential safety concerns [230], recent preclinical studies have investigated therapeutic strategies using transferrin receptor (TfR) binding, brain-penetrant PGRN biologics, enabling TfR-mediated transcytosis across the blood-brain barrier (BBB) and enhanced CNS delivery [210, 231]. Peripheral administration and AAV-mediated liver expression of brain-penetrant PGRN biologics have been shown to rescue phenotypes of Grn−/− mice and Grn−/− Tmem106b−/− DKO mice, respectively [197, 210]. A brain-penetrant recombinant PGRN biologic (DNL593) is currently under investigation in a phase 1/2 clinical trial (NCT05262023) (Table 3).
Table 3.
PGRN-boosting therapies in clinical trials
| Modality | Name | Stage | CTGID | Mechanism of action | Route of administration | Result | References |
|---|---|---|---|---|---|---|---|
| Anti-sortilin antibody | Latozinemab (AL001) | Phase 3 | NCT04374136 | Block lysosomal delivery of PGRN to increase extracellular PGRN | Intravenous (IV) | Under investigation | [232, 236] |
| Anti-sortilin antibody | Latozinemab (AL001) | Phase 2 | NCT03987295 | Block lysosomal delivery of PGRN to increase extracellular PGRN | Intravenous (IV) | Under investigation | [232, 236] |
| Anti-sortilin antibody | Latozinemab (AL001) | Phase 1 | NCT03636204 | Block lysosomal delivery of PGRN to increase extracellular PGRN | Intravenous (IV) | Well tolerated, reduced WBC sortilin, and increased plasma and CSF PGRN in healthy volunteers and aFTD-GRN participants. | [232, 236] |
| Anti-sortilin antibody | AL101 | Phase 2 | NCT06079190 | Block lysosomal delivery of PGRN to increase extracellular PGRN | Intravenous (IV) | Under investigation | No published preclinical data |
| Anti-sortilin antibody | AL101 | Phase 1 | NCT04111666 | Block lysosomal delivery of PGRN to increase extracellular PGRN | Intravenous (IV) or subcutaneous (SC) | Generally safe and well tolerated, and increased plasma and CSF PGRN in healthy volunteers | No published preclinical data |
| GRN gene therapy |
PR006/ LY3884963 |
Phase 1/2 | NCT04408625 | AAV-mediated GRN gene delivery | Intracisterna magna (ICM) | increased CSF PGRN in all patients | [216] |
| GRN gene therapy | PBFT02 | Phase 1/2 | NCT04747431 | AAV-mediated GRN gene delivery | Intracisterna magna (ICM) | Under investigation | [229] |
| GRN gene therapy | AVB-101 | Phase 1/2 | NCT06064890 | AAV-mediated GRN gene delivery | Bilateral intrathalamic infusion | Under investigation | No published preclinical data |
| PGRN protein replacement therapy | DNL593 | Phase 1/2 | NCT05262023 | Brain-penetrant recombinant PGRN biologic | Intravenous (IV) | Under investigation | [210] |
| Alkalizing agent | Amiodarone | Completed (phase 2) | 2011-004571-37 | Upregulate GRN mRNA | Oral | No significant effects on serum PGRN and disease course | [240] |
| HDAC inhibitor | FRM-0334 | Completed (phase 2a) | NCT02149160 | Upregulate GRN mRNA | Oral | Safe and well tolerated, but no effects on plasma and CSF PGRN | [244] |
| Calcium channel blocker | Nimodipine | Completed (Phase 1) | NCT01835665 | Increase secreted PGRN | Oral | Safe and well tolerated, but no effects on plasma and CSF PGRN | [238] |
Abbreviations: WBC, white blood cell
For FTLD-GRN, several other approaches have also been explored to increase PGRN from the intact GRN allele and restore its protein levels in the brain. One such approach is targeting sortilin-PGRN interaction to inhibit cellular uptake of PGRN [232–235]. Latozinemab (also known as AL001), an anti-sortilin antibody that blocks sortilin-PGRN interactions and targets sortilin for degradation [232], has demonstrated favorable safety and an increase in plasma and CSF PGRN levels in FTLD-GRN participants in a phase 1 clinical trial [236], and is currently in a phase 3 clinical trial (NCT04374136) (Table 3). In addition, AL101, another anti-sortilin antibody, is currently in a phase 2 clinical trial for the treatment of neurodegenerative diseases, including AD and PD (NCT06079190).
Another approach to increase PGRN levels used in a recent study is focusing on microRNAs (miRs) that negatively regulates PGRN protein levels. Antisense oligonucleotides (ASOs) to target the binding site of miR-29b in the 3’ UTR of the human GRN mRNA have been shown to effectively increase PGRN translation in human iPSC-derived neurons and in a humanized GRN mouse model [237].
Small molecules that can increase PGRN levels have also been explored and some have been tested in clinical trials (Table 3). Nimodipine, an FDA-approved BBB-penetrant calcium channel blocker, increases PGRN levels in Grn+/− mice through unknown mechanisms, but did not alter plasma or CSF PGRN levels in a phase 1 clinical trial [238]. V-ATPase inhibitors and alkalizing reagents such as chloroquine and amiodarone have been shown to increase intracellular and secreted PGRN in cell culture via a translational mechanism independent of autophagy, lysosomal degradation, or endocytosis [239]. However, amiodarone administration had no significant effects on serum PGRN and disease course in a pilot phase 2 clinical trial with five FTLD-GRN patients [240]. Histone deacetylase (HDAC) inhibitors have been shown to upregulate PGRN transcription in cell culture systems, including iPSC-derived neurons [241–243] but failed to increase plasma and CSF PGRN levels in participants with GRN haploinsufficiency in a phase 2a clinical trial [244]. There are preclinical studies reporting that the disaccharide trehalose, BBB-penetrant benzoxazole derivatives, and bromodomain and extra-terminal domain (BET) inhibitors significantly increase PGRN mRNA and protein levels in iPSC-derived neurons [245–247] and in Grn+/− mouse brains [245, 246].
As many FTLD-GRN cases are caused by nonsense mutations, which result in PTCs in the GRN mRNA, another potential treatment strategy may be nonsense suppression therapy, which has been investigated in many other genetic diseases caused by nonsense mutations [248]. Two previous studies have shown that aminoglycosides-induced PTC readthrough upregulates PGRN expression in some but not all nonsense mutations tested in cell culture and preclinical models [249, 250]. It is however known that long-term administration of aminoglycosides causes off-target toxic side effects [248]. Therefore, further investigation will be needed to find safe, effective PTC readthrough agents. Inhibiting nonsense-mediated mRNA decay (NMD) may be an alternative approach. However, a study has reported that ASO-based inhibition of NMD of GrnR493X mutant mRNA and genetic deletion of the NMD factor Upf3b do not increase Grn mRNA levels in the GrnR493X mouse model although the ASOs successfully increases Grn mRNA levels in fibroblasts derived from GrnR493X knockin mice [251].
Beyond FTLD-GRN and CLN11, effects of boosting PGRN levels have also been assessed in several mouse models of AD and PD. Lentivirus-mediated PGRN overexpression lowered plaque load and prevents neuronal loss and memory deficits in 5XFAD mice [184]. Intrahippocampal injection of recombinant PGRN protein also reduces hippocampal Aβ deposition in 5XFAD mice [252]. Lentivirus-mediated PGRN overexpression was also effective in reducing plaque burden and synaptic atrophy in Tg2576 APP transgenic mice [253]. For PD, lentivirus-mediated GRN gene delivery has been reported to protect against 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced dopaminergic neuronal loss and locomotor deficits [254]. So far, effects of boosting PGRN levels on tau and α-synuclein pathologies have not been investigated.
Conclusion and future direction
PGRN boosting therapy is a promising therapeutic strategy for FTLD-GRN and CLN11, and also potentially for other neurodegenerative diseases. However, it should be noted that PGRN overexpression has long been known to be associated with many tumors [255, 256]. Safety and adverse effects of long-term treatment therefore will need to be assessed carefully (BOX 1). In terms of the sortilin antibody approach, it is also important to note that a recent study has shown that lysosomal PGRN in neurons and the granulins/PGRN ratio are significantly decreased in sortilin KO mouse brain despite increased total PGRN levels in the brain [105]. Therefore, potential effects of selective loss of neuronal PGRN and granulins in subject treated with therapeutic anti-sortilin antibodies will need to be considered. So far, little is known about the physiological or pathological role(s) of granulins versus PGRN in the brain. A recent preclinical study has shown that AAV-mediated expression of granulin 2/F or 4/A rescues several phenotypes of Grn−/− mice [212], revealing a protective role of granulins in the brain. Further studies are necessary to understand the exact functions of individual granulins (BOX 1).
Box 1.
Key outstanding questions regarding PGRN and TMEM106B in the field
| What are functions of individual granulin peptides versus PGRN in the brain? |
| How do TMEM106B variants affect FTLD-GRN and other neurodegenerative diseases? |
| Do PGRN and TMEM106B exert their deleterious/protective effects predominantly in neurons versus glia? |
| What are the mechanisms of amyloid fibril formation of C-terminal fragments of TMEM106B? |
| Do the TMEM106B fibrils directly contribute to pathophysiology of neurodegenerative diseases? |
| Can we establish a preclinical model that fully reproduces effects of PGRN haploinsufficiency? |
| What are the mechanisms by which PGRN deficiency causes alteration in lipids such as BMP and glycosphingolipids? |
| Is the lipid alteration caused by PGRN or TMEM106B deficiency directly responsible for PGRN- or TMEM106B-associated neurological diseases? |
| Do PGRN-boosting therapies have side effects? |
TMEM106B has not been considered as a therapeutic target because preclinical studies have found that both complete loss and overexpression of TMEM106B cause lysosomal abnormalities in the brain. It is important to note however that, given their presence in multiple neurodegenerative diseases as well as in aged brains, if TMEM106B amyloid fibrils are found to have a pathological role, preventing formation of and/or removing these amyloid fibrils may be a therapeutic strategy for various neurodegenerative diseases. Thus, better understanding of the TMEM106B fibrils through relevant in vitro and in vivo models recapitulating formation of TMEM106B amyloid fibrils will be necessary (BOX 1). Importantly, there is currently no standardized criteria and methods for identification of the TMEM106B fibrils. While determining cryo-EM structure proves the presence of the TMEM106B fibrils, immunohistochemistry studies using antibodies against luminal region of TMEM106B could potentially detect abnormally overexpressed full-length TMEM106B or cleaved but non-fibrillar luminal region of TMEM106B. In addition, the molecular weight of the sarkosyl-insoluble CTF in immunoblot analysis varies between published studies [83, 156–158, 170]. More specific antibodies, probes and/or image analyses to detect the TMEM106B fibrils may help solving these issues.
The recent discovery of an involvement of PGRN and TMEM106B in lipid metabolism by directly regulating several lipid classes such as GlcCer, BMP, gangliosides, and GalCer is a significant advance. It is interesting to note that both PGRN and TMEM106B appear to be involved in glycosphingolipid metabolism (Fig. 3). PGRN regulation of GlcCer has been demonstrated to be critical for developing Gaucher disease [57] and potentially for synucleinopathy and tauopathy [49]. However, it remains unclear whether other lipids contribute to brain aging or the pathophysiology of PGRN- or TMEM106B-associated neurological diseases (BOX 1). In addition, lipid-droplet-accumulating Grn−/− microglia were recently shown to be defective in phagocytosis [222]. In contrast, several previous studies have reported that PGRN deficiency enhances phagocytosis and/or synaptic pruning in macrophages and microglia [16, 52, 257–259]. Resolving these issues and apparent contradictions may lead to development of novel alternative therapeutic approaches targeting common lipid pathways shared by various neurodegenerative diseases.
Finally, it is important to note that although commonly used Grn−/− and Tmem106b−/− mice and their crosses have been valuable tools to understand physiological functions of PGRN and TMEM106B and their general roles under pathological conditions, these mice with complete loss of PGRN or TMEM106B do not accurately model human diseases associated with PGRN and TMEM106B except for CLN11. Thus, to better understand disease mechanisms and to facilitate the development of novel therapies, it is necessary to establish more relevant preclinical models recapitulating effects of human GRN mutations or variants and the TMEM106B risk haplotype (BOX 1). Importantly, it remains unclear how the TMEM106B haplotype modifies a disease risk across various neurodegenerative diseases as well as brain aging (BOX 1). The TMEM106B risk haplotype has been suggested to increase TMEM106B protein levels [7, 61, 68, 79, 83]. However, this needs to be further verified experimentally by determining the functional variant(s) on the haplotype and elucidating the underlying mechanism. The verification is especially important because (1) inconsistent results have been obtained in its effect on mRNA levels [7, 63, 68, 79–81] and in the protein stability of the T185S protective variant [61, 135, 150, 152, 153], (2) cell culture and mouse studies have shown that both loss and overexpression of TMEM106B negatively affect some aspects of lysosomal function and exacerbate phenotypes of PGRN-deficient mice [89, 92, 93, 134–137, 143, 191, 195], and (3) a few studies have reported potential functional impacts of the T185S protective variant [150, 152]. It also remains ill-defined whether PGRN and TMEM106B exert their deleterious/protective effects predominantly in neurons versus glia and whether PGRN and TMEM106B interact intracellularly or intercellularly between different cell types to affect neurodegeneration and brain aging (BOX 1). For example, PGRN-deficient microglia may specifically influence neurons with increased TMEM106B levels, similar to the results shown in a previous study [178]. Further studies using co-culture, 3D culture, or xenotransplantation of human iPSC-derived neurons and glia generated from patients with or without GRN mutations and/or with the TMEM106B risk or protective haplotype may achieve the goal. Once the link between the risk haplotype and increased TMEM106B protein levels is confirmed, animal models with increased TMEM106B levels like the recent human TMEM106B transgenic mice [199] will be also useful.
Acknowledgements
Figures were created with BioRender.com.
Abbreviations
- AAV
Adeno-associated virus
- Aβ
Amyloid-β
- AD
Alzheimer’s disease
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- AEP
Asparagine endopeptidase
- ALS
Amyotrophic lateral sclerosis
- AP1
Accessary protein 1
- ASO
Antisense oligonucleotide
- BBB
Blood-brain barrier
- BMP
Bis(monoacylglycero)phosphate
- CBD
Corticobasal degeneration
- CHMP2B
Charged multivesicular body protein 2B
- CLN11
Neuronal ceroid lipofuscinosis type 11
- CNS
Central nervous system
- Co-IP
Co-immunoprecipitation
- Cryo-EM
Cryo-electron microscopy
- CSF
Cerebrospinal fluid
- CTCF
CCCTC-binding factor
- CTF
C-terminal fragment
- DKO
Double knockout
- ESCRT-III
Endosomal sorting complex required for transport-III
- FTLD
Frontotemporal lobar degeneration
- FTLD-TDP
FTLD with TDP-43 inclusions
- FTLD-GRN
FTLD with GRN mutations
- GALC
Galactosylceramidase
- GalCer
Galactosylceramide
- GCase
β-Glucocerebrosidase
- GlcCer
Glucosylceramide
- GlcSph
Glucosylsphingosine
- GWAS
Genome-wide association study
- HDAC
Histone deacetylase
- HexA
β-Hexosaminidase A
- HLD
Hypomyelinating leukodystrophy
- IP
Immunoprecipitation
- IP-MS
Immunoprecipitation-mass spectrometry
- iPSC
Induced pluripotent stem cell
- KO
Knockout
- LATE-NC
Limbic-predominant age-related TDP-43 encephalopathy neuropathological change
- LBD
Lewy body dementia
- LC-MS/MS
Liquid chromatography tandem mass spectrometry
- LD
Linkage disequilibrium
- LEA-2
Late embryogenesis abundant-2
- LRP1
Low-density lipoprotein receptor-related protein 1
- MAP6
Microtubule-associated protein 6
- miR
MicroRNA
- MPTP
1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- M6PR
Mannose 6-phosphate receptor
- NMD
Nonsense-mediated mRNA decay
- OCD
Obsessive compulsive disorder
- PD
Parkinson’s disease
- PGRN
Progranulin
- PSAP
Prosaposin
- PSP
Progressive supranuclear palsy
- PTC
Premature termination codon
- RGC
Retinal ganglion cell
- SNP
Single-nucleotide polymorphism
- SPR
Surface plasmon resonance
- TAGs
Triacylglycerols
- TDP-43
TAR DNA-binding protein 43
- TFEB
Transcriptional factor EB
- TfR
Transferrin receptor
- V-ATPase
Vacuolar-ATPase
- VPS10
Vacuolar protein sorting 10
- WBC
White blood cell
- Y2H
Yeast two-hybrid
Author contributions
H.T. and S.M.S. wrote the main manuscript text, and H.T. prepared all figures and tables. All authors reviewed the manuscript.
Funding
This work was supported by National Institute on Aging of the National Institutes of Health under grant number R01AG034924 and R01AG066165 to SMS.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wilson DM 3rd, Cookson MR, Van Den Bosch L, Zetterberg H, Holtzman DM, Dewachter I. Hallmarks of neurodegenerative diseases. Cell. 2023;186(4):693–714. [DOI] [PubMed] [Google Scholar]
- 2.Robinson JL, Lee EB, Xie SX, Rennert L, Suh E, Bredenberg C, Caswell C, Van Deerlin VM, Yan N, Yousef A, et al. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain. 2018;141(7):2181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kovacs GG. Are comorbidities compatible with a molecular pathological classification of neurodegenerative diseases? Curr Opin Neurol. 2019;32(2):279–91. [DOI] [PubMed] [Google Scholar]
- 4.Spires-Jones TL, Attems J, Thal DR. Interactions of pathological proteins in neurodegenerative diseases. Acta Neuropathol. 2017;134(2):187–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Telpoukhovskaia MA, Bahr BA, Chen X, Gan L. Endo-lysosomal dysfunction: a converging mechanism in neurodegenerative diseases. Curr Opin Neurobiol. 2018;48:52–8. [DOI] [PubMed] [Google Scholar]
- 6.Todd TW, Shao W, Zhang YJ, Petrucelli L. The endolysosomal pathway and ALS/FTD. Trends Neurosci. 2023;46(12):1025–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, et al. Common variants at 7p21 are associated with frontotemporal Lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42(3):234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–9. [DOI] [PubMed] [Google Scholar]
- 9.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442(7105):920–4. [DOI] [PubMed] [Google Scholar]
- 10.Feng T, Lacrampe A, Hu F. Physiological and pathological functions of TMEM106B: a gene associated with brain aging and multiple brain disorders. Acta Neuropathol. 2021;141(3):327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhinn H, Tatton N, McCaughey S, Kurnellas M, Rosenthal A. Progranulin as a therapeutic target in neurodegenerative diseases. Trends Pharmacol Sci. 2022;43(8):641–52. [DOI] [PubMed] [Google Scholar]
- 12.Van Mossevelde S, Engelborghs S, van der Zee J, Van Broeckhoven C. Genotype-phenotype links in frontotemporal Lobar degeneration. Nat Rev Neurol. 2018;14(6):363–78. [DOI] [PubMed] [Google Scholar]
- 13.Grossman M, Seeley WW, Boxer AL, Hillis AE, Knopman DS, Ljubenov PA, Miller B, Piguet O, Rademakers R, Whitwell JL, et al. Frontotemporal Lobar degeneration. Nat Rev Dis Primers. 2023;9(1):40. [DOI] [PubMed] [Google Scholar]
- 14.Rademakers R, Baker M, Gass J, Adamson J, Huey ED, Momeni P, Spina S, Coppola G, Karydas AM, Stewart H, et al. Phenotypic variability associated with progranulin haploinsufficiency in patients with the common 1477C–>T (Arg493X) mutation: an international initiative. Lancet Neurol. 2007;6(10):857–68. [DOI] [PubMed] [Google Scholar]
- 15.Sakae N, Roemer SF, Bieniek KF, Murray ME, Baker MC, Kasanuki K, Graff-Radford NR, Petrucelli L, Van Blitterswijk M, Rademakers R, et al. Microglia in frontotemporal Lobar degeneration with progranulin or C9ORF72 mutations. Ann Clin Transl Neurol. 2019;6(9):1782–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, Shang Y, Oldham MC, Martens LH, Gao F, et al. Progranulin deficiency promotes Circuit-Specific synaptic pruning by microglia via complement activation. Cell. 2016;165(4):921–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsan E, Velmeshev D, Ramsey A, Patel RK, Zhang J, Koontz M, Andrews MG, de Majo M, Mora C, Blumenfeld J et al. Astroglial toxicity promotes synaptic degeneration in the thalamocortical circuit in frontotemporal dementia with GRN mutations. J Clin Invest 2023, 133(6). [DOI] [PMC free article] [PubMed]
- 18.Gerrits E, Giannini LAA, Brouwer N, Melhem S, Seilhean D, Le Ber I, Brainbank Neuro CEBNN, Kamermans A, Kooij G, de Vries HE, et al. Neurovascular dysfunction in GRN-associated frontotemporal dementia identified by single-nucleus RNA sequencing of human cerebral cortex. Nat Neurosci. 2022;25(8):1034–48. [DOI] [PubMed] [Google Scholar]
- 19.Zhou X, Kukar T, Rademakers R. Lysosomal dysfunction and other pathomechanisms in FTLD: evidence from progranulin genetics and biology. Adv Exp Med Biol. 2021;1281:219–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shankaran SS, Capell A, Hruscha AT, Fellerer K, Neumann M, Schmid B, Haass C. Missense mutations in the progranulin gene linked to frontotemporal Lobar degeneration with ubiquitin-immunoreactive inclusions reduce progranulin production and secretion. J Biol Chem. 2008;283(3):1744–53. [DOI] [PubMed] [Google Scholar]
- 21.Pinarbasi ES, Karamyshev AL, Tikhonova EB, Wu IH, Hudson H, Thomas PJ. Pathogenic signal sequence mutations in progranulin disrupt SRP interactions required for mRNA stability. Cell Rep. 2018;23(10):2844–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, Morbin M, Rossi G, Pareyson D, Mole SE, Staropoli JF, et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet. 2012;90(6):1102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canafoglia L, Morbin M, Scaioli V, Pareyson D, D’Incerti L, Fugnanesi V, Tagliavini F, Berkovic SF, Franceschetti S. Recurrent generalized seizures, visual loss, and palinopsia as phenotypic features of neuronal ceroid lipofuscinosis due to progranulin gene mutation. Epilepsia. 2014;55(6):e56–59. [DOI] [PubMed] [Google Scholar]
- 24.Almeida MR, Macario MC, Ramos L, Baldeiras I, Ribeiro MH, Santana I. Portuguese family with the co-occurrence of frontotemporal Lobar degeneration and neuronal ceroid lipofuscinosis phenotypes due to progranulin gene mutation. Neurobiol Aging. 2016;41:200. e201-200 e205. [DOI] [PubMed] [Google Scholar]
- 25.Faber I, Prota JR, Martinez AR, Lopes-Cendes I, Franca MCJ. A new phenotype associated with homozygous GRN mutations: complicated spastic paraplegia. Eur J Neurol. 2017;24(1):e3–4. [DOI] [PubMed] [Google Scholar]
- 26.Kamate M, Detroja M, Hattiholi V. Neuronal ceroid lipofuscinosis type-11 in an adolescent. Brain Dev. 2019;41(6):542–5. [DOI] [PubMed] [Google Scholar]
- 27.Nobrega PR, Paiva ARB, Amorim Junior AD, Lima P, Cabral KSS, Barcelos IP, Pessoa ALS, Souza-Lima CFL, Castro MAA, Freua F, et al. Further description of the phenotypic spectrum of neuronal ceroid lipofuscinosis type 11. Genet Med. 2025;27(1):101291. [DOI] [PubMed] [Google Scholar]
- 28.Huin V, Barbier M, Bottani A, Lobrinus JA, Clot F, Lamari F, Chat L, Rucheton B, Fluchere F, Auvin S, et al. Homozygous GRN mutations: new phenotypes and new insights into pathological and molecular mechanisms. Brain. 2020;143(1):303–19. [DOI] [PubMed] [Google Scholar]
- 29.Lee MJ, Chen TF, Cheng TW, Chiu MJ. rs5848 variant of progranulin gene is a risk of alzheimer’s disease in the Taiwanese population. Neurodegener Dis. 2011;8(4):216–20. [DOI] [PubMed] [Google Scholar]
- 30.Sheng J, Su L, Xu Z, Chen G. Progranulin polymorphism rs5848 is associated with increased risk of alzheimer’s disease. Gene. 2014;542(2):141–5. [DOI] [PubMed] [Google Scholar]
- 31.Xu HM, Tan L, Wan Y, Tan MS, Zhang W, Zheng ZJ, Kong LL, Wang ZX, Jiang T, Tan L, et al. PGRN is associated with Late-Onset alzheimer’s disease: a Case-Control replication study and Meta-analysis. Mol Neurobiol. 2017;54(2):1187–95. [DOI] [PubMed] [Google Scholar]
- 32.Wightman DP, Jansen IE, Savage JE, Shadrin AA, Bahrami S, Holland D, Rongve A, Borte S, Winsvold BS, Drange OK, et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for alzheimer’s disease. Nat Genet. 2021;53(9):1276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A, et al. Identification of novel risk loci, causal insights, and heritable risk for parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18(12):1091–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellenguez C, Kucukali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, Naj AC, Campos-Martin R, Grenier-Boley B, Andrade V, et al. New insights into the genetic etiology of alzheimer’s disease and related dementias. Nat Genet. 2022;54(4):412–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brouwers N, Nuytemans K, van der Zee J, Gijselinck I, Engelborghs S, Theuns J, Kumar-Singh S, Pickut BA, Pals P, Dermaut B, et al. Alzheimer and Parkinson diagnoses in progranulin null mutation carriers in an extended founder family. Arch Neurol. 2007;64(10):1436–46. [DOI] [PubMed] [Google Scholar]
- 36.Brouwers N, Sleegers K, Engelborghs S, Maurer-Stroh S, Gijselinck I, van der Zee J, Pickut BA, Van den Broeck M, Mattheijssens M, Peeters K, et al. Genetic variability in progranulin contributes to risk for clinically diagnosed alzheimer disease. Neurology. 2008;71(9):656–64. [DOI] [PubMed] [Google Scholar]
- 37.Cortini F, Fenoglio C, Guidi I, Venturelli E, Pomati S, Marcone A, Scalabrini D, Villa C, Clerici F, Dalla Valle E, et al. Novel exon 1 progranulin gene variant in alzheimer’s disease. Eur J Neurol. 2008;15(10):1111–7. [DOI] [PubMed] [Google Scholar]
- 38.Kelley BJ, Haidar W, Boeve BF, Baker M, Shiung M, Knopman DS, Rademakers R, Hutton M, Adamson J, Kuntz KM, et al. Alzheimer disease-like phenotype associated with the c.154delA mutation in progranulin. Arch Neurol. 2010;67(2):171–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry DC, Lehmann M, Yokoyama JS, Karydas A, Lee JJ, Coppola G, Grinberg LT, Geschwind D, Seeley WW, Miller BL, et al. Progranulin mutations as risk factors for alzheimer disease. JAMA Neurol. 2013;70(6):774–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piaceri I, Imperiale D, Ghidoni E, Atzori C, Bagnoli S, Ferrari C, Ungari S, Ambrogio L, Sorbi S, Nacmias B. Novel GRN mutations in alzheimer’s disease and frontotemporal Lobar degeneration. J Alzheimers Dis. 2018;62(4):1683–9. [DOI] [PubMed] [Google Scholar]
- 41.Redaelli V, Rossi G, Maderna E, Kovacs GG, Piccoli E, Caroppo P, Cacciatore F, Spinello S, Grisoli M, Sozzi G, et al. Alzheimer neuropathology without frontotemporal Lobar degeneration hallmarks (TAR DNA-binding protein 43 inclusions) in missense progranulin mutation Cys139Arg. Brain Pathol. 2018;28(1):72–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonato G, Campagnolo M, Emmi A, Misenti V, Carrer T, Fogliano C, Salviati L, Carecchio M, Antonini A. Progranulin mutation manifesting as Parkinson disease: A case series from the PADUA-CESNE cohort. Mov Disord Clin Pract 2025. [DOI] [PMC free article] [PubMed]
- 43.Rovelet-Lecrux A, Deramecourt V, Legallic S, Maurage CA, Le Ber I, Brice A, Lambert JC, Frebourg T, Hannequin D, Pasquier F, et al. Deletion of the progranulin gene in patients with frontotemporal Lobar degeneration or Parkinson disease. Neurobiol Dis. 2008;31(1):41–5. [DOI] [PubMed] [Google Scholar]
- 44.Wauters E, Van Mossevelde S, Sleegers K, van der Zee J, Engelborghs S, Sieben A, Vandenberghe R, Philtjens S, Van den Broeck M, Peeters K, et al. Clinical variability and onset age modifiers in an extended Belgian GRN founder family. Neurobiol Aging. 2018;67:84–94. [DOI] [PubMed] [Google Scholar]
- 45.Kelley BJ, Haidar W, Boeve BF, Baker M, Graff-Radford NR, Krefft T, Frank AR, Jack CR Jr., Shiung M, Knopman DS, et al. Prominent phenotypic variability associated with mutations in progranulin. Neurobiol Aging. 2009;30(5):739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu C, Dong L, Wang J, Li J, Huang X, Lei D, Mao C, Chu S, Sha L, Xu Q, et al. GRN mutation spectrum and genotype-phenotype correlation in Chinese dementia patients: data from PUMCH dementia cohort. J Med Genet. 2024;61(6):543–8. [DOI] [PubMed] [Google Scholar]
- 47.Reho P, Koga S, Shah Z, Chia R, International LBDGC, American Genome C, Rademakers R, Dalgard CL, Boeve BF, Beach TG, et al. GRN mutations are associated with lewy body dementia. Mov Disord. 2022;37(9):1943–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL. Alzheimer’s disease. Nat Rev Dis Primers. 2015;1:15056. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi H, Bhagwagar S, Nies SH, Ye H, Han X, Chiasseu MT, Wang G, Mackenzie IR, Strittmatter SM. Reduced progranulin increases Tau and alpha-synuclein inclusions and alters mouse Tauopathy phenotypes via glucocerebrosidase. Nat Commun. 2024;15(1):1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendsaikhan A, Tooyama I, Bellier JP, Serrano GE, Sue LI, Lue LF, Beach TG, Walker DG. Characterization of lysosomal proteins progranulin and prosaposin and their interactions in alzheimer’s disease and aged brains: increased levels correlate with neuropathology. Acta Neuropathol Commun. 2019;7(1):215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pereson S, Wils H, Kleinberger G, McGowan E, Vandewoestyne M, Van Broeck B, Joris G, Cuijt I, Deforce D, Hutton M, et al. Progranulin expression correlates with dense-core amyloid plaque burden in alzheimer disease mouse models. J Pathol. 2009;219(2):173–81. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi H, Klein ZA, Bhagat SM, Kaufman AC, Kostylev MA, Ikezu T, Strittmatter SM. Alzheimer’s disease neuroimaging I: opposing effects of progranulin deficiency on amyloid and Tau pathologies via microglial TYROBP network. Acta Neuropathol. 2017;133(5):785–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hosokawa M, Kondo H, Serrano GE, Beach TG, Robinson AC, Mann DM, Akiyama H, Hasegawa M, Arai T. Accumulation of multiple neurodegenerative disease-related proteins in Familial frontotemporal Lobar degeneration associated with granulin mutation. Sci Rep. 2017;7(1):1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leverenz JB, Yu CE, Montine TJ, Steinbart E, Bekris LM, Zabetian C, Kwong LK, Lee VM, Schellenberg GD, Bird TD. A novel progranulin mutation associated with variable clinical presentation and tau, TDP43 and alpha-synuclein pathology. Brain. 2007;130(Pt 5):1360–74. [DOI] [PubMed] [Google Scholar]
- 55.Sieben A, Van Mossevelde S, Wauters E, Engelborghs S, van der Zee J, Van Langenhove T, Santens P, Praet M, Boon P, Miatton M, et al. Extended FTLD pedigree segregating a Belgian GRN-null mutation: neuropathological heterogeneity in one family. Alzheimers Res Ther. 2018;10(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeda T, Seilhean D, Le Ber I, Millecamps S, Sazdovitch V, Kitagawa K, Uchihara T, Duyckaerts C. Amygdala TDP-43 pathology in frontotemporal Lobar degeneration and motor neuron disease. J Neuropathol Exp Neurol. 2017;76(9):800–12. [DOI] [PubMed] [Google Scholar]
- 57.Jian J, Zhao S, Tian QY, Liu H, Zhao Y, Chen WC, Grunig G, Torres PA, Wang BC, Zeng B, et al. Association between progranulin and gaucher disease. EBioMedicine. 2016;11:127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Blitterswijk M, Mullen B, Wojtas A, Heckman MG, Diehl NN, Baker MC, DeJesus-Hernandez M, Brown PH, Murray ME, Hsiung GY, et al. Genetic modifiers in carriers of repeat expansions in the C9ORF72 gene. Mol Neurodegener. 2014;9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sleegers K, Brouwers N, Maurer-Stroh S, van Es MA, Van Damme P, van Vught PW, van der Zee J, Serneels S, De Pooter T, Van den Broeck M, et al. Progranulin genetic variability contributes to amyotrophic lateral sclerosis. Neurology. 2008;71(4):253–9. [DOI] [PubMed] [Google Scholar]
- 60.Nelson PT, Dickson DW, Trojanowski JQ, Jack CR, Boyle PA, Arfanakis K, Rademakers R, Alafuzoff I, Attems J, Brayne C, et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain. 2019;142(6):1503–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicholson AM, Finch NA, Wojtas A, Baker MC, Perkerson RB 3rd, Castanedes-Casey M, Rousseau L, Benussi L, Binetti G, Ghidoni R, et al. TMEM106B p.T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J Neurochem. 2013;126(6):781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, Crook R, Hunter T, Ghidoni R, Benussi L, et al. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76(5):467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cruchaga C, Graff C, Chiang HH, Wang J, Hinrichs AL, Spiegel N, Bertelsen S, Mayo K, Norton JB, Morris JC, et al. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol. 2011;68(5):581–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perneel J, Manoochehri M, Huey ED, Rademakers R, Goldman J. Case report: TMEM106B haplotype alters penetrance of GRN mutation in frontotemporal dementia family. Front Neurol. 2023;14:1160248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pottier C, Zhou X, Perkerson RB 3rd, Baker M, Jenkins GD, Serie DJ, Ghidoni R, Benussi L, Binetti G, Lopez de Munain A, et al. Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal Lobar degeneration and GRN mutations: a genome-wide association study. Lancet Neurol. 2018;17(6):548–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lattante S, Le Ber I, Galimberti D, Serpente M, Rivaud-Pechoux S, Camuzat A, Clot F, Fenoglio C, French research network on FTD, Ftd ALS. Defining the association of TMEM106B variants among frontotemporal Lobar degeneration patients with GRN mutations and C9orf72 repeat expansions. Neurobiol Aging. 2014;35(11):e26582651–5. [DOI] [PubMed] [Google Scholar]
- 67.Serpente M, Fenoglio C, Clerici F, Bonsi R, Arosio B, Cioffi SM, Rotondo E, Franceschi M, Martinelli Boneschi F, Mari D, et al. Transmembrane protein 106B gene (TMEM106B) variability and influence on progranulin plasma levels in patients with alzheimer’s disease. J Alzheimers Dis. 2015;43(3):757–61. [DOI] [PubMed] [Google Scholar]
- 68.Chemparathy A, Le Guen Y, Zeng Y, Gorzynski J, Jensen TD, Yang C, Kasireddy N, Talozzi L, Belloy M, Stewart I, et al. A 3’UTR insertion is a candidate causal variant at the TMEM106B locus associated with increased risk for FTLD-TDP. Neurol Genet. 2024;10(1):e200124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Blitterswijk M, Mullen B, Nicholson AM, Bieniek KF, Heckman MG, Baker MC, DeJesus-Hernandez M, Finch NA, Brown PH, Murray ME, et al. TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol. 2014;127(3):397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gallagher MD, Suh E, Grossman M, Elman L, McCluskey L, Van Swieten JC, Al-Sarraj S, Neumann M, Gelpi E, Ghetti B, et al. TMEM106B is a genetic modifier of frontotemporal Lobar degeneration with C9orf72 hexanucleotide repeat expansions. Acta Neuropathol. 2014;127(3):407–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bellenguez C, Kucukali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, Naj AC, Campos-Martin R, Grenier-Boley B, Andrade V et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet 2022. [DOI] [PMC free article] [PubMed]
- 72.Hu Y, Sun JY, Zhang Y, Zhang H, Gao S, Wang T, Han Z, Wang L, Sun BL, Liu G. rs1990622 variant associates with alzheimer’s disease and regulates TMEM106B expression in human brain tissues. BMC Med. 2021;19(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cherry JD, Mez J, Crary JF, Tripodis Y, Alvarez VE, Mahar I, Huber BR, Alosco ML, Nicks R, Abdolmohammadi B, et al. Variation in TMEM106B in chronic traumatic encephalopathy. Acta Neuropathol Commun. 2018;6(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tropea TF, Mak J, Guo MH, Xie SX, Suh E, Rick J, Siderowf A, Weintraub D, Grossman M, Irwin D, et al. TMEM106B effect on cognition in Parkinson disease and frontotemporal dementia. Ann Neurol. 2019;85(6):801–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Manini A, Ratti A, Brusati A, Maranzano A, Fogh I, Peverelli S, Messina S, Gentilini D, Verde F, Poletti B et al. TMEM106B acts as a modifier of cognitive and motor functions in amyotrophic lateral sclerosis. Int J Mol Sci 2022, 23(16). [DOI] [PMC free article] [PubMed]
- 76.Vass R, Ashbridge E, Geser F, Hu WT, Grossman M, Clay-Falcone D, Elman L, McCluskey L, Lee VM, Van Deerlin VM, et al. Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121(3):373–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simons C, Dyment D, Bent SJ, Crawford J, D’Hooghe M, Kohlschutter A, Venkateswaran S, Helman G, Poll-The BT, Makowski CC, et al. A recurrent de Novo mutation in TMEM106B causes hypomyelinating leukodystrophy. Brain. 2017;140(12):3105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perneel J, Rademakers R. Identification of TMEM106B amyloid fibrils provides an updated view of TMEM106B biology in health and disease. Acta Neuropathol. 2022;144(5):807–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gallagher MD, Posavi M, Huang P, Unger TL, Berlyand Y, Gruenewald AL, Chesi A, Manduchi E, Wells AD, Grant SFA, et al. A Dementia-Associated risk variant near TMEM106B alters chromatin architecture and gene expression. Am J Hum Genet. 2017;101(5):643–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van der Zee J, Van Langenhove T, Kleinberger G, Sleegers K, Engelborghs S, Vandenberghe R, Santens P, Van den Broeck M, Joris G, Brys J, et al. TMEM106B is associated with frontotemporal Lobar degeneration in a clinically diagnosed patient cohort. Brain. 2011;134(Pt 3):808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu L, De Jager PL, Yang J, Trojanowski JQ, Bennett DA, Schneider JA. The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology. 2015;84(9):927–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodney A, Karanjeet K, Benzow K, Koob MD. A common Alu insertion in the 3’UTR of TMEM106B is associated with risk of dementia. Alzheimers Dement. 2024;20(7):5071–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee JY, Harney DJ, Teo JD, Kwok JB, Sutherland GT, Larance M, Don AS. The major TMEM106B dementia risk allele affects TMEM106B protein levels, fibril formation, and Myelin lipid homeostasis in the ageing human hippocampus. Mol Neurodegener. 2023;18(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hou Y, Dan X, Babbar M, Wei Y, Hasselbalch SG, Croteau DL, Bohr VA. Ageing as a risk factor for neurodegenerative disease. Nat Rev Neurol. 2019;15(10):565–81. [DOI] [PubMed] [Google Scholar]
- 85.Rhinn H, Abeliovich A. Differential aging analysis in human cerebral cortex identifies variants in TMEM106B and GRN that regulate aging phenotypes. Cell Syst. 2017;4(4):404–15. e405. [DOI] [PubMed] [Google Scholar]
- 86.Li Z, Farias FHG, Dube U, Del-Aguila JL, Mihindukulasuriya KA, Fernandez MV, Ibanez L, Budde JP, Wang F, Lake AM, et al. The TMEM106B FTLD-protective variant, rs1990621, is also associated with increased neuronal proportion. Acta Neuropathol. 2020;139(1):45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tesi N, van der Lee S, Hulsman M, van Schoor NM, Huisman M, Pijnenburg Y, van der Flier WM, Reinders M, Holstege H. Cognitively healthy centenarians are genetically protected against alzheimer’s disease. Alzheimers Dement. 2024;20(6):3864–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ahmed Z, Sheng H, Xu YF, Lin WL, Innes AE, Gass J, Yu X, Wuertzer CA, Hou H, Chiba S, et al. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am J Pathol. 2010;177(1):311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luningschror P, Werner G, Stroobants S, Kakuta S, Dombert B, Sinske D, Wanner R, Lullmann-Rauch R, Wefers B, Wurst W, et al. The FTLD risk factor TMEM106B regulates the transport of lysosomes at the axon initial segment of motoneurons. Cell Rep. 2020;30(10):3506–19. e3506. [DOI] [PubMed] [Google Scholar]
- 90.Brunk UT, Terman A. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med. 2002;33(5):611–9. [DOI] [PubMed] [Google Scholar]
- 91.Moreno-Garcia A, Kun A, Calero O, Medina M, Calero M. An overview of the role of Lipofuscin in Age-Related neurodegeneration. Front Neurosci. 2018;12:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feng T, Mai S, Roscoe JM, Sheng RR, Ullah M, Zhang J, Katz II, Yu H, Xiong W, Hu F. Loss of TMEM106B and PGRN leads to severe lysosomal abnormalities and neurodegeneration in mice. EMBO Rep. 2020;21(10):e50219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Werner G, Damme M, Schludi M, Gnorich J, Wind K, Fellerer K, Wefers B, Wurst W, Edbauer D, Brendel M, et al. Loss of TMEM106B potentiates lysosomal and FTLD-like pathology in progranulin-deficient mice. EMBO Rep. 2020;21(10):e50241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Paushter DH, Du H, Feng T, Hu F. The lysosomal function of progranulin, a guardian against neurodegeneration. Acta Neuropathol. 2018;136(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Simon MJ, Logan T, DeVos SL, Di Paolo G. Lysosomal functions of progranulin and implications for treatment of frontotemporal dementia. Trends Cell Biol. 2023;33(4):324–39. [DOI] [PubMed] [Google Scholar]
- 97.Petkau TL, Blanco J, Leavitt BR. Conditional loss of progranulin in neurons is not sufficient to cause neuronal ceroid lipofuscinosis-like neuropathology in mice. Neurobiol Dis. 2017;106:14–22. [DOI] [PubMed] [Google Scholar]
- 98.Arrant AE, Filiano AJ, Unger DE, Young AH, Roberson ED. Restoring neuronal progranulin reverses deficits in a mouse model of frontotemporal dementia. Brain. 2017;140(5):1447–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Petkau TL, Kosior N, de Asis K, Connolly C, Leavitt BR. Selective depletion of microglial progranulin in mice is not sufficient to cause neuronal ceroid lipofuscinosis or neuroinflammation. J Neuroinflammation. 2017;14(1):225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Krabbe G, Minami SS, Etchegaray JI, Taneja P, Djukic B, Davalos D, Le D, Lo I, Zhan L, Reichert MC, et al. Microglial NFkappaB-TNFalpha hyperactivation induces obsessive-compulsive behavior in mouse models of progranulin-deficient frontotemporal dementia. Proc Natl Acad Sci U S A. 2017;114(19):5029–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arrant AE, Filiano AJ, Patel AR, Hoffmann MQ, Boyle NR, Kashyap SN, Onyilo VC, Young AH, Roberson ED. Reduction of microglial progranulin does not exacerbate pathology or behavioral deficits in neuronal progranulin-insufficient mice. Neurobiol Dis. 2019;124:152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Belcastro V, Siciliano V, Gregoretti F, Mithbaokar P, Dharmalingam G, Berlingieri S, Iorio F, Oliva G, Polishchuck R, Brunetti-Pierri N, et al. Transcriptional gene network inference from a massive dataset elucidates transcriptome organization and gene function. Nucleic Acids Res. 2011;39(20):8677–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–7. [DOI] [PubMed] [Google Scholar]
- 104.Hu F, Padukkavidana T, Vaegter CB, Brady OA, Zheng Y, Mackenzie IR, Feldman HH, Nykjaer A, Strittmatter SM. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68(4):654–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Du H, Zhou X, Feng T, Hu F. Regulation of lysosomal trafficking of progranulin by sortilin and prosaposin. Brain Commun. 2022;4(1):fcab310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou X, Sun L, Bastos de Oliveira F, Qi X, Brown WJ, Smolka MB, Sun Y, Hu F. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J Cell Biol. 2015;210(6):991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee CW, Stankowski JN, Chew J, Cook CN, Lam YW, Almeida S, Carlomagno Y, Lau KF, Prudencio M, Gao FB, et al. The lysosomal protein cathepsin L is a progranulin protease. Mol Neurodegener. 2017;12(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Holler CJ, Taylor G, Deng Q, Kukar T. Intracellular proteolysis of progranulin generates stable, lysosomal granulins that are haploinsufficient in patients with frontotemporal dementia caused by GRN mutations. eNeuro 2017, 4(4). [DOI] [PMC free article] [PubMed]
- 109.Zhou X, Paushter DH, Feng T, Sun L, Reinheckel T, Hu F. Lysosomal processing of progranulin. Mol Neurodegener. 2017;12(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mohan S, Sampognaro PJ, Argouarch AR, Maynard JC, Welch M, Patwardhan A, Courtney EC, Zhang J, Mason A, Li KH, et al. Processing of progranulin into granulins involves multiple lysosomal proteases and is affected in frontotemporal Lobar degeneration. Mol Neurodegener. 2021;16(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Willnow TE, Petersen CM, Nykjaer A. VPS10P-domain receptors - regulators of neuronal viability and function. Nat Rev Neurosci. 2008;9(12):899–909. [DOI] [PubMed] [Google Scholar]
- 112.Carrasquillo MM, Nicholson AM, Finch N, Gibbs JR, Baker M, Rutherford NJ, Hunter TA, DeJesus-Hernandez M, Bisceglio GD, Mackenzie IR, et al. Genome-wide screen identifies rs646776 near sortilin as a regulator of progranulin levels in human plasma. Am J Hum Genet. 2010;87(6):890–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zheng Y, Brady OA, Meng PS, Mao Y, Hu F. C-terminus of progranulin interacts with the beta-propeller region of sortilin to regulate progranulin trafficking. PLoS ONE. 2011;6(6):e21023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou X, Sullivan PM, Sun L, Hu F. The interaction between progranulin and prosaposin is mediated by granulins and the linker region between Saposin B and C. J Neurochem. 2017;143(2):236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou X, Sun L, Bracko O, Choi JW, Jia Y, Nana AL, Brady OA, Hernandez JCC, Nishimura N, Seeley WW, et al. Impaired prosaposin lysosomal trafficking in frontotemporal Lobar degeneration due to progranulin mutations. Nat Commun. 2017;8:15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vidoni C, Follo C, Savino M, Melone MA, Isidoro C. The role of cathepsin D in the pathogenesis of human neurodegenerative disorders. Med Res Rev. 2016;36(5):845–70. [DOI] [PubMed] [Google Scholar]
- 117.Hersheson J, Burke D, Clayton R, Anderson G, Jacques TS, Mills P, Wood NW, Gissen P, Clayton P, Fearnley J, et al. Cathepsin D deficiency causes juvenile-onset ataxia and distinctive muscle pathology. Neurology. 2014;83(20):1873–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Beel S, Moisse M, Damme M, De Muynck L, Robberecht W, Van Den Bosch L, Saftig P, Van Damme P. Progranulin functions as a cathepsin D chaperone to stimulate axonal outgrowth in vivo. Hum Mol Genet. 2017;26(15):2850–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Valdez C, Wong YC, Schwake M, Bu G, Wszolek ZK, Krainc D. Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum Mol Genet. 2017;26(24):4861–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Butler VJ, Cortopassi WA, Argouarch AR, Ivry SL, Craik CS, Jacobson MP, Kao AW. Progranulin stimulates the in vitro maturation of Pro-Cathepsin D at acidic pH. J Mol Biol. 2019;431(5):1038–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou X, Paushter DH, Feng T, Pardon CM, Mendoza CS, Hu F. Regulation of cathepsin D activity by the FTLD protein progranulin. Acta Neuropathol. 2017;134(1):151–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Butler VJ, Cortopassi WA, Gururaj S, Wang AL, Pierce OM, Jacobson MP, Kao AW. Multi-Granulin domain peptides bind to Pro-Cathepsin D and stimulate its enzymatic activity more effectively than progranulin in vitro. Biochemistry. 2019;58(23):2670–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ward ME, Chen R, Huang HY, Ludwig C, Telpoukhovskaia M, Taubes A, Boudin H, Minami SS, Reichert M, Albrecht P et al. Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci Transl Med 2017, 9(385). [DOI] [PMC free article] [PubMed]
- 124.Reifschneider A, Robinson S, van Lengerich B, Gnorich J, Logan T, Heindl S, Vogt MA, Weidinger E, Riedl L, Wind K, et al. Loss of TREM2 rescues hyperactivation of microglia, but not lysosomal deficits and neurotoxicity in models of progranulin deficiency. EMBO J. 2022;41(4):e109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang M, Modeste E, Dammer E, Merino P, Taylor G, Duong DM, Deng Q, Holler CJ, Gearing M, Dickson D, et al. Network analysis of the progranulin-deficient mouse brain proteome reveals pathogenic mechanisms shared in human frontotemporal dementia caused by GRN mutations. Acta Neuropathol Commun. 2020;8(1):163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gotzl JK, Colombo AV, Fellerer K, Reifschneider A, Werner G, Tahirovic S, Haass C, Capell A. Early lysosomal maturation deficits in microglia triggers enhanced lysosomal activity in other brain cells of progranulin knockout mice. Mol Neurodegener. 2018;13(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gotzl JK, Mori K, Damme M, Fellerer K, Tahirovic S, Kleinberger G, Janssens J, van der Zee J, Lang CM, Kremmer E, et al. Common pathobiochemical hallmarks of progranulin-associated frontotemporal Lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol. 2014;127(6):845–60. [DOI] [PubMed] [Google Scholar]
- 128.Arrant AE, Onyilo VC, Unger DE, Roberson ED. Progranulin gene therapy improves lysosomal dysfunction and microglial pathology associated with frontotemporal dementia and neuronal ceroid lipofuscinosis. J Neurosci. 2018;38(9):2341–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bacioglu M, Schweighauser M, Gray D, Lovestam S, Katsinelos T, Quaegebeur A, van Swieten J, Jaunmuktane Z, Davies SW, Scheres SHW, et al. Cleaved TMEM106B forms amyloid aggregates in central and peripheral nervous systems. Acta Neuropathol Commun. 2024;12(1):99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Busch JI, Martinez-Lage M, Ashbridge E, Grossman M, Van Deerlin VM, Hu F, Lee VM, Trojanowski JQ, Chen-Plotkin AS. Expression of TMEM106B, the frontotemporal Lobar degeneration-associated protein, in normal and diseased human brain. Acta Neuropathol Commun. 2013;1:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Satoh J, Kino Y, Kawana N, Yamamoto Y, Ishida T, Saito Y, Arima K. TMEM106B expression is reduced in alzheimer’s disease brains. Alzheimers Res Ther. 2014;6(2):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Feng T, Sheng RR, Sole-Domenech S, Ullah M, Zhou X, Mendoza CS, Enriquez LCM, Katz II, Paushter DH, Sullivan PM, et al. A role of the frontotemporal Lobar degeneration risk factor TMEM106B in myelination. Brain. 2020;143(7):2255–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang T, Pang W, Feng T, Guo J, Wu K, Nunez Santos M, Arthanarisami A, Nana AL, Nguyen Q, Kim PJ, et al. TMEM106B regulates microglial proliferation and survival in response to demyelination. Sci Adv. 2023;9(18):eadd2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stagi M, Klein ZA, Gould TJ, Bewersdorf J, Strittmatter SM. Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol Cell Neurosci. 2014;61:226–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brady OA, Zheng Y, Murphy K, Huang M, Hu F. The frontotemporal Lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum Mol Genet. 2013;22(4):685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L, Busch JI, Akle S, Grossman M, Van Deerlin V, et al. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci. 2012;32(33):11213–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Klein ZA, Takahashi H, Ma M, Stagi M, Zhou M, Lam TT, Strittmatter SM. Loss of TMEM106B ameliorates lysosomal and frontotemporal Dementia-Related phenotypes in Progranulin-Deficient mice. Neuron. 2017;95(2):281–96. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Takahashi H, Perez-Canamas A, Lee CW, Ye H, Han X, Strittmatter SM. Lysosomal TMEM106B interacts with galactosylceramidase to regulate Myelin lipid metabolism. Commun Biol. 2024;7(1):1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kang J, Lim L, Song J. TMEM106B, a risk factor for FTLD and aging, has an intrinsically disordered cytoplasmic domain. PLoS ONE. 2018;13(10):e0205856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Timms RT, Zhang Z, Rhee DY, Harper JW, Koren I, Elledge SJ. A glycine-specific N-degron pathway mediates the quality control of protein N-myristoylation. Science 2019, 365(6448). [DOI] [PMC free article] [PubMed]
- 141.Thinon E, Serwa RA, Broncel M, Brannigan JA, Brassat U, Wright MH, Heal WP, Wilkinson AJ, Mann DJ, Tate EW. Global profiling of co- and post-translationally N-myristoylated proteomes in human cells. Nat Commun. 2014;5:4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lacrampe A, Hou D, Perez IG, Gong B, Franco-Hernandez N, Yee A, Chen W, Young-Chapon MA, Lin H, Hu F. Myristoylation of TMEM106B by NMT1/2 regulates TMEM106B trafficking and turnover. J Biol Chem 2025:110322. [DOI] [PMC free article] [PubMed]
- 143.Busch JI, Unger TL, Jain N, Tyler Skrinak R, Charan RA, Chen-Plotkin AS. Increased expression of the frontotemporal dementia risk factor TMEM106B causes C9orf72-dependent alterations in lysosomes. Hum Mol Genet. 2016;25(13):2681–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baggen J, Jacquemyn M, Persoons L, Vanstreels E, Pye VE, Wrobel AG, Calvaresi V, Martin SR, Roustan C, Cronin NB, et al. TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry. Cell. 2023;186(16):3427–42. e3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lang CM, Fellerer K, Schwenk BM, Kuhn PH, Kremmer E, Edbauer D, Capell A, Haass C. Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal Lobar degeneration. J Biol Chem. 2012;287(23):19355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schwenk BM, Lang CM, Hogl S, Tahirovic S, Orozco D, Rentzsch K, Lichtenthaler SF, Hoogenraad CC, Capell A, Haass C, et al. The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. EMBO J. 2014;33(5):450–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Colacurcio DJ, Nixon RA. Disorders of lysosomal acidification-The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res Rev. 2016;32:75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kundu ST, Grzeskowiak CL, Fradette JJ, Gibson LA, Rodriguez LB, Creighton CJ, Scott KL, Gibbons DL. TMEM106B drives lung cancer metastasis by inducing TFEB-dependent lysosome synthesis and secretion of cathepsins. Nat Commun. 2018;9(1):2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Forgac M. Vacuolar atpases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8(11):917–29. [DOI] [PubMed] [Google Scholar]
- 150.Jun MH, Han JH, Lee YK, Jang DJ, Kaang BK, Lee JA. TMEM106B, a frontotemporal Lobar dementia (FTLD) modifier, associates with FTD-3-linked CHMP2B, a complex of ESCRT-III. Mol Brain. 2015;8:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, Nielsen JE, Hodges JR, Spillantini MG, Thusgaard T, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37(8):806–8. [DOI] [PubMed] [Google Scholar]
- 152.Edwards GA 3rd, Wood CA, He Y, Nguyen Q, Kim PJ, Gomez-Gutierrez R, Park KW, Xu Y, Zurhellen C, Al-Ramahi I, et al. TMEM106B coding variant is protective and deletion detrimental in a mouse model of tauopathy. Acta Neuropathol. 2024;147(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cabron AS, Borgmeyer U, Richter J, Peisker H, Gutbrod K, Dormann P, Capell A, Damme M. Lack of a protective effect of the Tmem106b protective SNP in the Grn knockout mouse model for frontotemporal Lobar degeneration. Acta Neuropathol Commun. 2023;11(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ito Y, Hartley T, Baird S, Venkateswaran S, Simons C, Wolf NI, Boycott KM, Dyment DA, Kernohan KD. Lysosomal dysfunction in TMEM106B hypomyelinating leukodystrophy. Neurol Genet. 2018;4(6):e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Takahashi H, Strittmatter SM. An unexpected protein aggregate in diseased and ageing brains. Nature. 2022;605(7909):227–8. [DOI] [PubMed] [Google Scholar]
- 156.Schweighauser M, Arseni D, Bacioglu M, Huang M, Lovestam S, Shi Y, Yang Y, Zhang W, Kotecha A, Garringer HJ, et al. Age-dependent formation of TMEM106B amyloid filaments in human brains. Nature. 2022;605(7909):310–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chang A, Xiang X, Wang J, Lee C, Arakhamia T, Simjanoska M, Wang C, Carlomagno Y, Zhang G, Dhingra S, et al. Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases. Cell. 2022;185(8):1346–e13551315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jiang YX, Cao Q, Sawaya MR, Abskharon R, Ge P, DeTure M, Dickson DW, Fu JY, Ogorzalek Loo RR, Loo JA, et al. Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43. Nature. 2022;605(7909):304–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fan Y, Zhao Q, Xia W, Tao Y, Yu W, Chen M, Liu Y, Zhao J, Shen Y, Sun Y, et al. Generic amyloid fibrillation of TMEM106B in patient with parkinson’s disease dementia and normal elders. Cell Res. 2022;32(6):585–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Neumann M, Perneel J, Cheung S, Van den Broeck M, Nygaard H, Hsiung GR, Wynants S, Heeman B, Rademakers R, Mackenzie IRA. Limbic-predominant age-related TDP-43 proteinopathy (LATE-NC) is associated with abundant TMEM106B pathology. Acta Neuropathol. 2023;146(1):163–6. [DOI] [PubMed] [Google Scholar]
- 161.Hoq MR, Bharath SR, Hallinan GI, Fernandez A, Vago FS, Ozcan KA, Li D, Garringer HJ, Vidal R, Ghetti B, et al. Cross-beta helical filaments of Tau and TMEM106B in Gray and white matter of multiple system tauopathy with presenile dementia. Acta Neuropathol. 2023;145(5):707–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ishikawa R, Yamazaki Y, Nakamori M, Takahashi T, Maruyama H. Antibody-recognizing residues 188–211 of TMEM106B exhibit immunohistochemical reactivity with the TMEM106B C-terminal fragment. Front Neurosci. 2023;17:1250547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Perneel J, Neumann M, Heeman B, Cheung S, Van den Broeck M, Wynants S, Baker M, Vicente CT, Faura J, Rademakers R, et al. Accumulation of TMEM106B C-terminal fragments in neurodegenerative disease and aging. Acta Neuropathol. 2023;145(3):285–302. [DOI] [PubMed] [Google Scholar]
- 164.Marks JD, Ayuso VE, Carlomagno Y, Yue M, Todd TW, Hao Y, Li Z, McEachin ZT, Shantaraman A, Duong DM, et al. TMEM106B core deposition associates with TDP-43 pathology and is increased in risk SNP carriers for frontotemporal dementia. Sci Transl Med. 2024;16(730):eadf9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.C TV, Perneel J, Wynants S, Heeman B, Van den Broeck M, Baker M, Cheung S, Faura J, Mackenzie IRA, Rademakers R. C-terminal TMEM106B fragments in human brain correlate with disease-associated TMEM106B haplotypes. Brain. 2023;146(10):4055–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Brady OA, Zhou X, Hu F. Regulated intramembrane proteolysis of the frontotemporal Lobar degeneration risk factor, TMEM106B, by signal peptide peptidase-like 2a (SPPL2a). J Biol Chem. 2014;289(28):19670–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Held S, Erck C, Kemppainen S, Bleibaum F, Giridhar NJ, Feederle R, Krenner C, Juopperi SP, Calliari A, Mentrup T, et al. Physiological shedding and C-terminal proteolytic processing of TMEM106B. Cell Rep. 2024;44(1):115107. [DOI] [PubMed] [Google Scholar]
- 168.Ghetti B, Schweighauser M, Jacobsen MH, Gray D, Bacioglu M, Murzin AG, Glazier BS, Katsinelos T, Vidal R, Newell KL, et al. TMEM106B amyloid filaments in the Biondi bodies of ependymal cells. Acta Neuropathol. 2024;148(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Oksche A, Liesner R, Tigges J, Tigges M. Intraepithelial inclusions resembling human Biondi bodies in the choroid plexus of an aged chimpanzee. Cell Tissue Res. 1984;235(2):467–9. [DOI] [PubMed] [Google Scholar]
- 170.Riordan R, Saxton A, Han M, McMillan PJ, Kow RL, Liachko NF, Kraemer BC. TMEM106B C-terminal fragments aggregate and drive neurodegenerative proteinopathy in Transgenic Caenorhabditis elegans. Alzheimers Dement 2024. [DOI] [PMC free article] [PubMed]
- 171.Filiano AJ, Martens LH, Young AH, Warmus BA, Zhou P, Diaz-Ramirez G, Jiao J, Zhang Z, Huang EJ, Gao FB, et al. Dissociation of frontotemporal dementia-related deficits and neuroinflammation in progranulin haploinsufficient mice. J Neurosci. 2013;33(12):5352–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Arrant AE, Filiano AJ, Warmus BA, Hall AM, Roberson ED. Progranulin haploinsufficiency causes biphasic social dominance abnormalities in the tube test. Genes Brain Behav. 2016;15(6):588–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cook AK, Greathouse KM, Manuel PN, Cooper NH, Eberhardt JM, Freeman CD, Weber AJ, Herskowitz JH, Arrant AE. Dendritic spine head diameter is reduced in the prefrontal cortex of progranulin haploinsufficient mice. Mol Brain. 2024;17(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ghoshal N, Dearborn JT, Wozniak DF, Cairns NJ. Core features of frontotemporal dementia recapitulated in progranulin knockout mice. Neurobiol Dis. 2012;45(1):395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yin F, Dumont M, Banerjee R, Ma Y, Li H, Lin MT, Beal MF, Nathan C, Thomas B, Ding A. Behavioral deficits and progressive neuropathology in progranulin-deficient mice: a mouse model of frontotemporal dementia. FASEB J. 2010;24(12):4639–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tanaka Y, Chambers JK, Matsuwaki T, Yamanouchi K, Nishihara M. Possible involvement of lysosomal dysfunction in pathological changes of the brain in aged progranulin-deficient mice. Acta Neuropathol Commun. 2014;2:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ward ME, Taubes A, Chen R, Miller BL, Sephton CF, Gelfand JM, Minami S, Boscardin J, Martens LH, Seeley WW, et al. Early retinal neurodegeneration and impaired Ran-mediated nuclear import of TDP-43 in progranulin-deficient FTLD. J Exp Med. 2014;211(10):1937–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang J, Velmeshev D, Hashimoto K, Huang YH, Hofmann JW, Shi X, Chen J, Leidal AM, Dishart JG, Cahill MK, et al. Neurotoxic microglia promote TDP-43 proteinopathy in progranulin deficiency. Nature. 2020;588(7838):459–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nguyen AD, Nguyen TA, Zhang J, Devireddy S, Zhou P, Karydas AM, Xu X, Miller BL, Rigo F, Ferguson SM, et al. Murine knockin model for progranulin-deficient frontotemporal dementia with nonsense-mediated mRNA decay. Proc Natl Acad Sci U S A. 2018;115(12):E2849–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Frew J, Nygaard HB. Neuropathological and behavioral characterization of aged Grn R493X progranulin-deficient frontotemporal dementia knockin mice. Acta Neuropathol Commun. 2021;9(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Smith DM, Aggarwal G, Niehoff ML, Jones SA, Banerjee S, Farr SA, Nguyen AD. Biochemical, biomarker, and behavioral characterization of the Grn(R493X) mouse model of frontotemporal dementia. Mol Neurobiol. 2024;61(11):9708–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Du H, Wong MY, Zhang T, Santos MN, Hsu C, Zhang J, Yu H, Luo W, Hu F. A multifaceted role of progranulin in regulating amyloid-beta dynamics and responses. Life Sci Alliance 2021, 4(7). [DOI] [PMC free article] [PubMed]
- 183.Hosokawa M, Tanaka Y, Arai T, Kondo H, Akiyama H, Hasegawa M. Progranulin haploinsufficiency reduces amyloid beta deposition in alzheimer’s disease model mice. Exp Anim. 2018;67(1):63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Minami SS, Min SW, Krabbe G, Wang C, Zhou Y, Asgarov R, Li Y, Martens LH, Elia LP, Ward ME, et al. Progranulin protects against amyloid beta deposition and toxicity in alzheimer’s disease mouse models. Nat Med. 2014;20(10):1157–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hosokawa M, Arai T, Masuda-Suzukake M, Kondo H, Matsuwaki T, Nishihara M, Hasegawa M, Akiyama H. Progranulin reduction is associated with increased Tau phosphorylation in P301L Tau Transgenic mice. J Neuropathol Exp Neurol. 2015;74(2):158–65. [DOI] [PubMed] [Google Scholar]
- 186.Nies SH, Takahashi H, Herber CS, Huttner A, Chase A, Strittmatter SM. Spreading of alzheimer Tau seeds is enhanced by aging and template matching with limited impact of amyloid-beta. J Biol Chem. 2021;297(4):101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhou X, Nicholson AM, Ren Y, Brooks M, Jiang P, Zuberi A, Phuoc HN, Perkerson RB, Matchett B, Parsons TM, et al. Loss of TMEM106B leads to myelination deficits: implications for frontotemporal dementia treatment strategies. Brain. 2020;143(6):1905–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Stroobants S, D’Hooge R, Damme M. Aged Tmem106b knockout mice display gait deficits in coincidence with purkinje cell loss and only limited signs of non-motor dysfunction. Brain Pathol. 2021;31(2):223–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rademakers R, Nicholson AM, Ren Y, Koga S, Nguyen HP, Brooks M, Qiao W, Quicksall ZS, Matchett B, Perkerson RB, et al. Loss of Tmem106b leads to cerebellum purkinje cell death and motor deficits. Brain Pathol. 2021;31(3):e12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Feng T, Luan L, Katz II, Ullah M, Van Deerlin VM, Trojanowski JQ, Lee EB, Hu F. TMEM106B deficiency impairs cerebellar myelination and synaptic integrity with purkinje cell loss. Acta Neuropathol Commun. 2022;10(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhou X, Sun L, Brady OA, Murphy KA, Hu F. Elevated TMEM106B levels exaggerate Lipofuscin accumulation and lysosomal dysfunction in aged mice with progranulin deficiency. Acta Neuropathol Commun. 2017;5(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Arrant AE, Nicholson AM, Zhou X, Rademakers R, Roberson ED. Partial Tmem106b reduction does not correct abnormalities due to progranulin haploinsufficiency. Mol Neurodegener. 2018;13(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dominguez SL, Laufer BI, Ghosh AS, Li Q, Ruggeri G, Emani MR, Phu L, Friedman BA, Sandoval W, Rose CM, et al. TMEM106B reduction does not rescue GRN deficiency in iPSC-derived human microglia and mouse models. iScience. 2023;26(11):108362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Perez-Canamas A, Takahashi H, Lindborg JA, Strittmatter SM. Fronto-temporal dementia risk gene TMEM106B has opposing effects in different lysosomal storage disorders. Brain Commun. 2021;3(1):fcaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhou X, Brooks M, Jiang P, Koga S, Zuberi AR, Baker MC, Parsons TM, Castanedes-Casey M, Phillips V, Librero AL, et al. Loss of Tmem106b exacerbates FTLD pathologies and causes motor deficits in progranulin-deficient mice. EMBO Rep. 2020;21(10):e50197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Feng T, Minevich G, Liu P, Qin HX, Wozniak G, Pham J, Pham K, Korgaonkar A, Kurnellas M, Defranoux NA et al. AAV-GRN partially corrects motor deficits and ALS/FTLD-related pathology in Tmem106b(-/-)Grn(-/-) mice. iScience 2023, 26(7):107247. [DOI] [PMC free article] [PubMed]
- 197.Reich M, Simon MJ, Polke B, Paris I, Werner G, Schrader C, Spieth L, Davis SS, Robinson S, de Melo GL, et al. Peripheral expression of brain-penetrant progranulin rescues pathologies in mouse models of frontotemporal Lobar degeneration. Sci Transl Med. 2024;16(750):eadj7308. [DOI] [PubMed] [Google Scholar]
- 198.Feng T, Du H, Yang C, Wang Y, Hu F. Loss of TMEM106B exacerbates Tau pathology and neurodegeneration in PS19 mice. Acta Neuropathol. 2024;147(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Perneel J, Lastra Osua M, Alidadiani S, Peeters N, De Witte L, Heeman B, Manzella S, De Rycke R, Brooks M, Perkerson RB, et al. Increased TMEM106B levels lead to lysosomal dysfunction which affects synaptic signaling and neuronal health. Mol Neurodegener. 2025;20(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Almeida S, Zhang Z, Coppola G, Mao W, Futai K, Karydas A, Geschwind MD, Tartaglia MC, Gao F, Gianni D, et al. Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep. 2012;2(4):789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Raitano S, Ordovas L, De Muynck L, Guo W, Espuny-Camacho I, Geraerts M, Khurana S, Vanuytsel K, Toth BI, Voets T, et al. Restoration of progranulin expression rescues cortical neuron generation in an induced pluripotent stem cell model of frontotemporal dementia. Stem Cell Rep. 2015;4(1):16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bossolasco P, Cimini S, Maderna E, Bardelli D, Canafoglia L, Cavallaro T, Ricci M, Silani V, Marucci G, Rossi G. GRN-/- iPSC-derived cortical neurons recapitulate the pathological findings of both frontotemporal Lobar degeneration and neuronal ceroidolipofuscinosis. Neurobiol Dis. 2022;175:105891. [DOI] [PubMed] [Google Scholar]
- 203.Lee C, Frew J, Weilinger NL, Wendt S, Cai W, Sorrentino S, Wu X, MacVicar BA, Willerth SM, Nygaard HB. hiPSC-derived GRN-deficient astrocytes delay spiking activity of developing neurons. Neurobiol Dis. 2023;181:106124. [DOI] [PubMed] [Google Scholar]
- 204.de Majo M, Koontz M, Marsan E, Salinas N, Ramsey A, Kuo YM, Seo K, Li H, Drager N, Leng K, et al. Granulin loss of function in human mature brain organoids implicates astrocytes in TDP-43 pathology. Stem Cell Rep. 2023;18(3):706–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Prudencio M, Humphrey J, Pickles S, Brown AL, Hill SE, Kachergus JM, Shi J, Heckman MG, Spiegel MR, Cook C, et al. Truncated stathmin-2 is a marker of TDP-43 pathology in frontotemporal dementia. J Clin Invest. 2020;130(11):6080–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhou X, Paushter DH, Pagan MD, Kim D, Nunez Santos M, Lieberman RL, Overkleeft HS, Sun Y, Smolka MB, Hu F. Progranulin deficiency leads to reduced glucocerebrosidase activity. PLoS ONE. 2019;14(7):e0212382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jian J, Tian QY, Hettinghouse A, Zhao S, Liu H, Wei J, Grunig G, Zhang W, Setchell KDR, Sun Y, et al. Progranulin recruits HSP70 to beta-Glucocerebrosidase and is therapeutic against gaucher disease. EBioMedicine. 2016;13:212–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Arrant AE, Roth JR, Boyle NR, Kashyap SN, Hoffmann MQ, Murchison CF, Ramos EM, Nana AL, Spina S, Grinberg LT, et al. Impaired beta-glucocerebrosidase activity and processing in frontotemporal dementia due to progranulin mutations. Acta Neuropathol Commun. 2019;7(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Vieira SRL, Schapira AHV. Glucocerebrosidase mutations and Parkinson disease. J Neural Transm (Vienna). 2022;129(9):1105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Logan T, Simon MJ, Rana A, Cherf GM, Srivastava A, Davis SS, Low RLY, Chiu CL, Fang M, Huang F, et al. Rescue of a lysosomal storage disorder caused by Grn loss of function with a brain penetrant progranulin biologic. Cell. 2021;184(18):4651–68. e4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Valdez C, Ysselstein D, Young TJ, Zheng J, Krainc D. Progranulin mutations result in impaired processing of prosaposin and reduced glucocerebrosidase activity. Hum Mol Genet 2019. [DOI] [PMC free article] [PubMed]
- 212.Root J, Mendsaikhan A, Taylor G, Merino P, Nandy S, Wang M, Araujo LT, Ryu D, Holler C, Thompson BM, et al. Granulins rescue inflammation, lysosome dysfunction, lipofuscin, and neuropathology in a mouse model of progranulin deficiency. Cell Rep. 2024;43(12):114985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Khrouf W, Saracino D, Rucheton B, Houot M, Clot F, Rinaldi D, Vitor J, Huynh M, Heng E, Schlemmer D, et al. Plasma lysosphingolipids in GRN-related diseases: monitoring lysosomal dysfunction to track disease progression. Neurobiol Dis. 2023;181:106108. [DOI] [PubMed] [Google Scholar]
- 214.Zhao X, Lin Y, Liou B, Fu W, Jian J, Fannie V, Zhang W, Setchell KDR, Grabowski GA, Sun Y, et al. PGRN deficiency exacerbates, whereas a brain penetrant PGRN derivative protects, GBA1 mutation-associated pathologies and diseases. Proc Natl Acad Sci U S A. 2023;120(1):e2210442120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Boland S, Swarup S, Ambaw YA, Malia PC, Richards RC, Fischer AW, Singh S, Aggarwal G, Spina S, Nana AL, et al. Deficiency of the frontotemporal dementia gene GRN results in gangliosidosis. Nat Commun. 2022;13(1):5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sevigny J, Uspenskaya O, Heckman LD, Wong LC, Hatch DA, Tewari A, Vandenberghe R, Irwin DJ, Saracino D, Le Ber I, et al. Progranulin AAV gene therapy for frontotemporal dementia: translational studies and phase 1/2 trial interim results. Nat Med. 2024;30(5):1406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Colella P, Sayana R, Suarez-Nieto MV, Sarno J, Nyame K, Xiong J, Pimentel Vera LN, Arozqueta Basurto J, Corbo M, Limaye A, et al. CNS-wide repopulation by hematopoietic-derived microglia-like cells corrects progranulin deficiency in mice. Nat Commun. 2024;15(1):5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Medoh UN, Abu-Remaileh M. The Bis(monoacylglycero)-phosphate hypothesis: from lysosomal function to therapeutic avenues. Annu Rev Biochem. 2024;93(1):447–69. [DOI] [PubMed] [Google Scholar]
- 219.Chen Y, Jian J, Hettinghouse A, Zhao X, Setchell KDR, Sun Y, Liu CJ. Progranulin associates with hexosaminidase A and ameliorates GM2 ganglioside accumulation and lysosomal storage in Tay-Sachs disease. J Mol Med (Berl). 2018;96(12):1359–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wilkening G, Linke T, Uhlhorn-Dierks G, Sandhoff K. Degradation of membrane-bound ganglioside GM1. Stimulation by bis(monoacylglycero)phosphate and the activator proteins SAP-B and GM2-AP. J Biol Chem. 2000;275(46):35814–9. [DOI] [PubMed] [Google Scholar]
- 221.Onal G, Kutlu O, Gozuacik D, Dokmeci Emre S. Lipid droplets in health and disease. Lipids Health Dis. 2017;16(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, Pluvinage JV, Mathur V, Hahn O, Morgens DW, et al. Lipid-droplet-accumulating microglia represent a dysfunctional and Proinflammatory state in the aging brain. Nat Neurosci. 2020;23(2):194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Haney MS, Palovics R, Munson CN, Long C, Johansson PK, Yip O, Dong W, Rawat E, West E, Schlachetzki JCM, et al. APOE4/4 is linked to damaging lipid droplets in alzheimer’s disease microglia. Nature. 2024;628(8006):154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhang T, Feng T, Wu K, Guo J, Nana AL, Yang G, Seeley WW, Hu F. Progranulin deficiency results in sex-dependent alterations in microglia in response to demyelination. Acta Neuropathol. 2023;146(1):97–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sung W, Noh MY, Nahm M, Kim YS, Ki CS, Kim YE, Kim HJ, Kim SH. Progranulin haploinsufficiency mediates cytoplasmic TDP-43 aggregation with lysosomal abnormalities in human microglia. J Neuroinflammation. 2024;21(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Evers BM, Rodriguez-Navas C, Tesla RJ, Prange-Kiel J, Wasser CR, Yoo KS, McDonald J, Cenik B, Ravenscroft TA, Plattner F, et al. Lipidomic and transcriptomic basis of lysosomal dysfunction in progranulin deficiency. Cell Rep. 2017;20(11):2565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Levine TP. TMEM106B in humans and Vac7 and Tag1 in yeast are predicted to be lipid transfer proteins. Proteins. 2022;90(1):164–75. [DOI] [PubMed] [Google Scholar]
- 228.Marcus J, Popko B. Galactolipids are molecular determinants of Myelin development and axo-glial organization. Biochim Biophys Acta. 2002;1573(3):406–13. [DOI] [PubMed] [Google Scholar]
- 229.Hinderer C, Miller R, Dyer C, Johansson J, Bell P, Buza E, Wilson JM. Adeno-associated virus serotype 1-based gene therapy for FTD caused by GRN mutations. Ann Clin Transl Neurol. 2020;7(10):1843–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Amado DA, Rieders JM, Diatta F, Hernandez-Con P, Singer A, Mak JT, Zhang J, Lancaster E, Davidson BL, Chen-Plotkin AS. AAV-Mediated progranulin delivery to a mouse model of progranulin deficiency causes T Cell-Mediated toxicity. Mol Ther. 2019;27(2):465–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kariolis MS, Wells RC, Getz JA, Kwan W, Mahon CS, Tong R, Kim DJ, Srivastava A, Bedard C, Henne KR et al. Brain delivery of therapeutic proteins using an Fc fragment blood-brain barrier transport vehicle in mice and monkeys. Sci Transl Med 2020, 12(545). [DOI] [PubMed]
- 232.Kurnellas M, Mitra A, Schwabe T, Paul R, Arrant AE, Roberson ED, Ward M, Yeh F, Long H, Rosenthal A. Latozinemab, a novel progranulin-elevating therapy for frontotemporal dementia. J Transl Med. 2023;21(1):387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Miyakawa S, Sakuma H, Warude D, Asanuma S, Arimura N, Yoshihara T, Tavares D, Hata A, Ida K, Hori Y, et al. Anti-sortilin1 antibody Up-Regulates progranulin via Sortilin1 Down-Regulation. Front Neurosci. 2020;14:586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ek M, Nilvebrant J, Nygren PA, Stahl S, Lindberg H, Lofblom J. An anti-sortilin affibody-peptide fusion inhibits sortilin-mediated progranulin degradation. Front Immunol. 2024;15:1437886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Lee WC, Almeida S, Prudencio M, Caulfield TR, Zhang YJ, Tay WM, Bauer PO, Chew J, Sasaguri H, Jansen-West KR, et al. Targeted manipulation of the sortilin-progranulin axis rescues progranulin haploinsufficiency. Hum Mol Genet. 2014;23(6):1467–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ward M, Carter LP, Huang JY, Maslyar D, Budda B, Paul R, Rosenthal A. Phase 1 study of Latozinemab in progranulin-associated frontotemporal dementia. Alzheimers Dement (N Y). 2024;10(1):e12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Aggarwal G, Banerjee S, Jones SA, Benchaar Y, Belanger J, Sevigny M, Smith DM, Niehoff ML, Pavlack M, de Vera IMS, et al. Antisense oligonucleotides targeting the miR-29b binding site in the GRN mRNA increase progranulin translation. J Biol Chem. 2023;299(12):105475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sha SJ, Miller ZA, Min SW, Zhou Y, Brown J, Mitic LL, Karydas A, Koestler M, Tsai R, Corbetta-Rastelli C, et al. An 8-week, open-label, dose-finding study of nimodipine for the treatment of progranulin insufficiency from GRN gene mutations. Alzheimers Dement (N Y). 2017;3(4):507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Capell A, Liebscher S, Fellerer K, Brouwers N, Willem M, Lammich S, Gijselinck I, Bittner T, Carlson AM, Sasse F, et al. Rescue of progranulin deficiency associated with frontotemporal Lobar degeneration by alkalizing reagents and Inhibition of vacuolar ATPase. J Neurosci. 2011;31(5):1885–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Alberici A, Archetti S, Pilotto A, Premi E, Cosseddu M, Bianchetti A, Semeraro F, Salvetti M, Muiesan ML, Padovani A, et al. Results from a pilot study on Amiodarone administration in Monogenic frontotemporal dementia with granulin mutation. Neurol Sci. 2014;35(8):1215–9. [DOI] [PubMed] [Google Scholar]
- 241.Cenik B, Sephton CF, Dewey CM, Xian X, Wei S, Yu K, Niu W, Coppola G, Coughlin SE, Lee SE, et al. Suberoylanilide hydroxamic acid (vorinostat) up-regulates progranulin transcription: rational therapeutic approach to frontotemporal dementia. J Biol Chem. 2011;286(18):16101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.She A, Kurtser I, Reis SA, Hennig K, Lai J, Lang A, Zhao WN, Mazitschek R, Dickerson BC, Herz J, et al. Selectivity and kinetic requirements of HDAC inhibitors as progranulin enhancers for treating frontotemporal dementia. Cell Chem Biol. 2017;24(7):892–906. e895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Almeida S, Gao F, Coppola G, Gao FB. Suberoylanilide hydroxamic acid increases progranulin production in iPSC-derived cortical neurons of frontotemporal dementia patients. Neurobiol Aging. 2016;42:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ljubenkov PA, Edwards L, Iaccarino L, La Joie R, Rojas JC, Koestler M, Harris B, Boeve BF, Borroni B, van Swieten JC, et al. Effect of the histone deacetylase inhibitor FRM-0334 on progranulin levels in patients with progranulin gene haploinsufficiency: A randomized clinical trial. JAMA Netw Open. 2021;4(9):e2125584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Holler CJ, Taylor G, McEachin ZT, Deng Q, Watkins WJ, Hudson K, Easley CA, Hu WT, Hales CM, Rossoll W, et al. Trehalose upregulates progranulin expression in human and mouse models of GRN haploinsufficiency: a novel therapeutic lead to treat frontotemporal dementia. Mol Neurodegener. 2016;11(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Tesla R, Guhl C, Werthmann GC, Dixon D, Cenik B, Addepalli Y, Liang J, Fass DM, Rosenthal Z, Haggarty SJ, et al. Benzoxazole-derivatives enhance progranulin expression and reverse the aberrant lysosomal proteome caused by GRN haploinsufficiency. Nat Commun. 2024;15(1):6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Rosenthal ZC, Fass DM, Payne NC, She A, Patnaik D, Hennig KM, Tesla R, Werthmann GC, Guhl C, Reis SA, et al. Epigenetic modulation through BET bromodomain inhibitors as a novel therapeutic strategy for progranulin-deficient frontotemporal dementia. Sci Rep. 2024;14(1):9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon readthrough. Annu Rev Genomics Hum Genet. 2014;15:371–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Kuang L, Hashimoto K, Huang EJ, Gentry MS, Zhu H. Frontotemporal dementia non-sense mutation of progranulin rescued by aminoglycosides. Hum Mol Genet. 2020;29(4):624–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Frew J, Baradaran-Heravi A, Balgi AD, Wu X, Yan TD, Arns S, Shidmoossavee FS, Tan J, Jaquith JB, Jansen-West KR, et al. Premature termination codon readthrough upregulates progranulin expression and improves lysosomal function in preclinical models of GRN deficiency. Mol Neurodegener. 2020;15(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Smith DM, Niehoff ML, Ling K, Jafar-Nejad P, Rigo F, Farr SA, Wilkinson MF, Nguyen AD. Targeting nonsense-mediated RNA decay does not increase progranulin levels in the Grn R493X mouse model of frontotemporal dementia. PLoS ONE. 2023;18(3):e0282822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Guan Z, Chen Z, Fu S, Dai L, Shen Y. Progranulin administration attenuates beta-Amyloid deposition in the Hippocampus of 5xFAD mice through modulating BACE1 expression and microglial phagocytosis. Front Cell Neurosci. 2020;14:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Van Kampen JM, Kay DG. Progranulin gene delivery reduces plaque burden and synaptic atrophy in a mouse model of alzheimer’s disease. PLoS ONE. 2017;12(8):e0182896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Van Kampen JM, Baranowski D, Kay DG. Progranulin gene delivery protects dopaminergic neurons in a mouse model of parkinson’s disease. PLoS ONE. 2014;9(5):e97032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Arechavaleta-Velasco F, Perez-Juarez CE, Gerton GL, Diaz-Cueto L. Progranulin and its biological effects in cancer. Med Oncol. 2017;34(12):194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ventura E, Ducci G, Benot Dominguez R, Ruggiero V, Belfiore A, Sacco E, Vanoni M, Iozzo RV, Giordano A, Morrione A. Progranulin oncogenic network in solid tumors. Cancers (Basel) 2023, 15(6). [DOI] [PMC free article] [PubMed]
- 257.Kao AW, Eisenhut RJ, Martens LH, Nakamura A, Huang A, Bagley JA, Zhou P, de Luis A, Neukomm LJ, Cabello J, et al. A neurodegenerative disease mutation that accelerates the clearance of apoptotic cells. Proc Natl Acad Sci U S A. 2011;108(11):4441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Gotzl JK, Brendel M, Werner G, Parhizkar S, Sebastian Monasor L, Kleinberger G, Colombo AV, Deussing M, Wagner M, Winkelmann J et al. Opposite microglial activation stages upon loss of PGRN or TREM2 result in reduced cerebral glucose metabolism. EMBO Mol Med 2019, 11(6). [DOI] [PMC free article] [PubMed]
- 259.Schmitz K, Wilken-Schmitz A, Vasic V, Brunkhorst R, Schmidt M, Tegeder I. Progranulin deficiency confers resistance to autoimmune encephalomyelitis in mice. Cell Mol Immunol. 2020;17(10):1077–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhao X, Liberti R, Jian J, Fu W, Hettinghouse A, Sun Y, Liu CJ. Progranulin associates with Rab2 and is involved in autophagosome-lysosome fusion in gaucher disease. J Mol Med (Berl). 2021;99(11):1639–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Santos MN, Paushter DH, Zhang T, Wu X, Feng T, Lou J, Du H, Becker SM, Fragoza R, Yu H, et al. Progranulin-derived granulin E and lysosome membrane protein CD68 interact to reciprocally regulate their protein homeostasis. J Biol Chem. 2022;298(9):102348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.