Abstract
Large-scale genomics has enabled proteomics by creating sequence infrastructures that can be used with mass spectrometry data to identify proteins. Although protein sequences can be deduced from nucleotide sequences, posttranslational modifications to proteins, in general, cannot. We describe a process for the analysis of posttranslational modifications that is simple, robust, general, and can be applied to complicated protein mixtures. A protein or protein mixture is digested by using three different enzymes: one that cleaves in a site-specific manner and two others that cleave nonspecifically. The mixture of peptides is separated by multidimensional liquid chromatography and analyzed by a tandem mass spectrometer. This approach has been applied to modification analyses of proteins in a simple protein mixture, Cdc2p protein complexes isolated through the use of an affinity tag, and lens tissue from a patient with congenital cataracts. Phosphorylation sites have been detected with known stoichiometry of as low as 10%. Eighteen sites of four different types of modification have been detected on three of the five proteins in a simple mixture, three of which were previously unreported. Three proteins from Cdc2p isolated complexes yielded eight sites containing three different types of modifications. In the lens tissue, 270 proteins were identified, and 11 different crystallins were found to contain a total of 73 sites of modification. Modifications identified in the crystallin proteins included Ser, Thr, and Tyr phosphorylation, Arg and Lys methylation, Lys acetylation, and Met, Tyr, and Trp oxidations. The method presented will be useful in discovering co- and posttranslational modifications of proteins.
The recent explosion in available genomic and protein sequence information is providing a sequence infrastructure for the emerging field of proteomics. A major aspect of many proteomics strategies is the identification of proteins using an analytical “fingerprint” that can be used to search a sequence database. One common “fingerprint” is the tandem mass (MS/MS) spectrum of a peptide. Thus, an MS/MS spectrum can be algorithmically compared with predicted peptide spectra from a sequence database to identify the respective protein (1, 2). The digestion of intact protein mixtures followed by the direct analysis of the resulting peptides by capillary liquid chromatography–MS/MS has facilitated “shotgun” identification of protein mixtures without the need for prior sample fractionation (3). Combined with the recent development of capillary multidimensional liquid chromatography [multidimensional protein identification technology (MudPIT)], this approach is now capable of characterizing proteins directly from entire cell lysates (4, 5). Furthermore, mass spectrometric methods are being developed that not only identify proteins in a mixture but also compare the relative level of protein expression between two different samples (6–9). These proteomic tools are now being used to study a number of biological systems.
Although the identification of proteins in complex mixtures is becoming routine, protein identification alone provides only limited insight into protein function. An important component of protein regulation and function is covalent modifications to protein structures that occur either co- or posttranslationally. Although protein sequences can be deduced from nucleotide sequences, posttranslational modifications to proteins, in general, cannot. Over 200 different modifications have been described (10). Many, such as phosphorylation, have well documented roles in signal transduction and the regulation of cellular processes. In contrast, other modifications are much less well studied but are also likely to play very important roles within the cell. Identifying the type and location of these protein modifications is a first step in understanding their regulatory potential. Despite their importance to cellular function, the methodologies used to study these modifications can be quite involved, are not compatible with protein mixtures, and/or are specific for a given type of posttranslational modification.
Several different strategies have been used to analyze protein modifications, and almost all are targeted to specific types of modifications. The first strategy uses enrichment of the modified peptides. These methods are most highly developed or applied to the area of phosphopeptides. Iron metal affinity chromatography or phosphopeptide-specific antibodies have been used to enrich phosphopeptides for analysis (12). Other methods have used 32P labeling to guide enrichment before analysis by standard phosphopeptide mapping or by mass spectrometry (13, 14). Mass spectrometry methods that use specific fragment ions indicative for phosphorylated peptides have also been used to detect these peptides in mixtures (15). Recently, a software algorithm, using pattern recognition, showed promising results in predicting unanticipated modifications (16).
Recently, three methods for the analysis of protein phosphorylation by mass spectrometry from complex mixtures were reported (17–19). These methods attempt to address the low-stoichiometry and high-complexity problems by selectively enriching phosphorylated peptides before analysis. All three methods use complex multistep chemical derivatization strategies for the enrichment of phosphopeptides. The method of Zhou et al. (19) identified 24 phosphorylated peptides (of which 14 were unambiguous), whereas Oda et al. (17) identified a single phosphorylation site in yeast. Each method is limited to phosphorylated peptides and thus requires a separate analysis to assay other modifications and the many unmodified peptides. Their complexity, application only to protein phosphorylation, and relative inefficiency suggest that these methods will have limited utility, especially when applied to a complex mixture of proteins.
We have addressed the technical challenges associated with measuring protein modifications using a different approach. Our protocol uses the high sensitivity and resolution capability of nanoscale multidimensional liquid chromatography combined with the precise structural specificity of MS/MS spectral data to identify the site and type of modification. By combining high-resolution separations with proteolytic cleavage of different selectivities, overlapping peptides are produced throughout the proteins being studied. The overlapping peptides reduce the ambiguity in mapping modifications while increasing the likelihood of obtaining a peptide resulting in a “quality” MS/MS spectrum. The multiproteolytic cleavage is combined with MudPIT analysis to handle the complexity of protein complexes and lens tissue. Because our procedure is enzymatically controlled, outcome reproducibility is excellent across a range of sample complexities, and losses are minimized. Furthermore, this approach simultaneously measures not only phosphopeptides but also unmodified peptides and peptides containing other modifications amenable to MS/MS. In this report, we describe a robust generalized system for identifying protein modifications in complex protein mixtures.
Experimental
Preparation of Test Sample.
The effectiveness of our protocol was tested by using a simple mixture containing five proteins. A 1-pmol aliquot of phosphorylated glycogen phosphorylase (phosphorylase A; Sigma) was mixed with a protein mixture (Bio-Rad) containing approximately 10 pmol each of unphosphorylated glycogen phosphorylase (phosphorylase B), myosin heavy chain, β-galactosidase, serum albumin, and ovalbumin. The mixture was diluted in 8 M urea/100 mM Tris⋅HCl, pH 8.5, to a final volume of 30 μl. The protein disulfide bonds were reduced with 0.8 μl of 100 mM DTT and incubated at 50°C for 25 min. The sample was cooled to room temperature and the resulting free thiols alkylated with 1.7 μl of 100 mM iodoacetamide.
Tandem Affinity Purification of cdc2.
A Schizosaccharomyces pombe strain in which the endogenous cdc2 locus expressed a tandem affinity purification (TAP) tagged version of the protein (cdc2-TAP) was used to isolate cdc2 and associated proteins (20, 21). Proteins were affinity purified from 8 liters of S. pombe cells grown to an OD of ≈0.9 at 595 nm as described previously (21). The resulting protein mixture, approximately 15 μg, was resuspended directly in 40 μl of 8.0 M urea/100 mM Tris, pH 8.5, and reduced and alkylated as described above except that 100 mM Tris(2-carboxyethyl)-phosphine (Pierce) at room temperature was used for reduction instead of DTT.
Preparation of Human Cataract Lens.
Lens tissue was obtained from a 4-year-old congenital cataract patient and homogenized with microtube pestle in 0.1 ml of 20 mM phosphate buffer (1 mM EGTA, pH 7.0) at 4°C. This suspension was centrifuged at 10,000 × g for 30 min at 4°C to remove insoluble material. The pH of the soluble fraction was adjusted to 8.5 with 1 M ammonium bicarbonate. The sample, containing <1 mg of total protein, was sequentially solubilized in 8 M urea, reduced by adding DTT to 2 mM, and carboxyamidomethylated in 20 mM iodoacetamide.
Triple Digest Protocol.
Reduced and alkylated protein samples were split into three equal fractions and digested with three different proteases by using a modified protocol described previously (11).
Fraction one.
The first fraction was diluted 3-fold with 100 mM Tris⋅HCl, pH 8.5, to bring the total urea concentration to ≈2 M. An aliquot of 100 mM CaCl2 was added to the protein sample resulting in a 1 mM final solution. Modified trypsin (Roche Diagnostics) was added at an estimated enzyme-to-substrate ratio of 1:50 (wt/wt) and incubated by mixing for 12–24 h at 37°C. After incubation, the reaction was quenched with 90% formic acid to 4% final and stored at −20°C until analysis.
Fraction two.
The second fraction was diluted 3-fold with buffer containing 4.8 M urea/100 mM Tris⋅HCl at pH 8.5. Subtilisin (Boehringer Mannheim) was added at an enzyme-to-substrate ratio of 1:50 (wt/wt) and incubated for 2–3 h at 37°C. The subtilisin digest was then quenched with formic acid and frozen at −20°C until analysis.
Fraction three.
The third fraction was diluted 3× with 100 mM Tris⋅HCl, pH 8.5, and elastase (Boehringer Mannheim) was added in a 1:50 enzyme-to-substrate ratio (wt/wt). After incubation for 12 h at 37°C, the reaction was quenched with formic acid and frozen at −20°C. The individual fractions from the test mixture were pooled before analysis by MudPIT, whereas the three individual proteolytic fractions from the tandem affinity purification of cdc2 and the human lens tissue were analyzed individually by MudPIT.
MudPIT.
A triphasic microcapillary column was constructed from 100-μm i.d. fused silica capillary tubing pulled to a 5-μm i.d. tip by using a Sutter Instruments P-2000 CO2 laser puller (Novato, CA). The protein test mixture and TAP-cdc2 digests were loaded directly onto separate capillary columns, each slurry packed with 7 cm of 5 μm Polaris C18-A material (Metachem, Ventura, CA), 3 cm of 5 μm Partisphere strong cation exchanger (Whatman), followed by another 3 cm of Polaris C18-A. The lens tissue digests were loaded onto separate fused silica capillary desalting columns containing 3 cm of Polaris C18-A packed into a 250-μm i.d. capillary with a 2-μm filtered union (UpChurch Scientific, Oak Harbor, WA). The desalting columns were washed with buffer containing 95% water, 5% MeCN, and 0.1% acetic acid. After desalting, a 100-μm i.d. column packed with 7 cm of 5-μm Polaris C18-A material, 6 cm of 5 μm Partisphere strong cation exchanger, followed by 3 cm of 5-μm hydrophilic interaction chromatography material (PolyLC) was attached to the filtered union, and the lens tissue peptides were eluted onto the triphasic column by using 20% water, 80% MeCN, and 0.1% formic acid. After loading the peptide digests, analysis was performed by using either a 6- or 18-step multidimensional separation with a modified protocol described previously (5).
Analysis of MS/MS Spectra.
MS/MS spectra were analyzed sequentially by using the following protocol. First, a software algorithm called 2TO3 (22) was used to determine the appropriate charge state (either +2 or +3) of multiply charged peptide mass spectra, delete spectra of poor quality, and identify spectra containing a prominent 98-Da (−H3PO4) neutral loss from the precursor. The MS/MS spectra after 2TO3 were searched by using a parallel virtual machine version of sequest (1) running on a 31-node Beowulf computer cluster against a protein database of the appropriate organism. The resulting sequest output files were filtered by using the program dtaselect (23). A subset database was made containing just the proteins identified. This subset database was then used to expedite all subsequent differential modification searches (24). The MS/MS data were then researched five times against the subset database to consider modifications of: (i) +80 on STY (phosphorylation); (ii) +42 on K (acetylation); (iii) static +42 modifications on N-terminal residues (acetylation); (iv) +14 on KR (methylation); and (v) +16 on MWY (oxidation). The spectra containing the prominent 98-Da neutral loss were also searched against a subset database by using a modified version of sequest that considers the unique MS/MS fragmentation patterns of phosphorylated Ser and Thr containing peptides (sequest-phos). The sequest-phos software algorithm will be described in detail elsewhere.
Results and Discussion
Identification of Low Stoichiometry Modifications in a Simple Protein Mixture.
Phosphorylase A was chosen as a protein to test our methodology because it contains a single known phosphorylation site near the N terminus of the protein, does not give a “true-tryptic” peptide of appropriate length to produce unambiguous MS/MS sequence data, and also has a commercially available unphosphorylated form (phosphorylase B). Phosphorylase A was mixed in a 1:10 molar ratio with phosphorylase B, myosin heavy chain, β-galactosidase, serum albumin, and ovalbumin to mimic the measurement of a substoichiometric modification in a small protein complex.
Fig. 1 shows the sequence coverage for glycogen phosphorylase and ovalbumin from the peptide MS/MS spectra obtained. Because of the relative simplicity of this sample, the peptides generated from the multiple enzyme digestion were pooled before analysis, more than tripling the complexity of the mixture. Sequence coverage was excellent for both glycogen phosphorylase and ovalbumin with 83.7 and 95.1% of the respective proteins' sequence covered by peptide MS/MS spectra. The high amino acid sequence coverage of proteins within this mixture demonstrates MudPIT's resolving capacity for individual peptide components and the concomitant automated acquisition of MS/MS spectra. Furthermore, the high sequence coverage confirms our ability to produce peptides from the multiple digest that produce “quality” MS/MS spectra across the entire protein.
Figure 1.
The identification of glycogen phosphorylase and ovalbumin protein modifications within a simple protein mixture. The protein modifications identified with overlapping peptide coverage are displayed with phosphorylation in blue, acetylation in yellow, and oxidation in red.
The multiple enzyme digest produces overlapping peptides that cover the entire protein sequence and increases the chances of identifying a modification on a specific amino acid residue (Table 1). With this method, we identify with three overlapping peptides the known S15 phosphorylation site of glycogen phosphorylase at a 10% stoichiometry in a mixture (Fig. 1). sequest also matched multiple overlapping peptide MS/MS spectra to the two known phosphorylation sites of ovalbumin (S69 and S345; Fig. 1). Both phosphorylase and ovalbumin are modified at their N terminus (25), and we found numerous sites where methionine was oxidized to the sulfoxide. Because these methionine oxidations can occur during the sample preparation process, we have difficulty discriminating between artifacts and those with bona fide physiological significance. The above sequest search results were confirmed by manual evaluation of the spectra using previously reported criteria (4). Multiple overlapping peptides allow more confidence in sequest output from only fair spectra, reduce ambiguity in an assignment of a modified amino acid between nearby residues, and minimize the possibility of false positives. For these reasons, we required overlapping peptides to validate any previously unreported protein modification.
Table 1.
Protein sequence coverage of selected proteins using the triple digestion protocol
| Protein | Trypsin*
|
Subtilisin*
|
Elastase*
|
Combined
|
||||
|---|---|---|---|---|---|---|---|---|
| % Coverage | Peptides | % Coverage | Peptides | % Coverage | Peptides | % Coverage | Peptides | |
| Cdc2 | 74.5 | 75 | 44.8 | 41 | 61.7 | 62 | 88.2 | 171 |
| Cdc13 | 56.0 | 67 | 58.3 | 65 | 53.1 | 56 | 91.7 | 169 |
| Cig1 | 28.9 | 38 | 43.9 | 26 | 24.6 | 18 | 57.8 | 77 |
| Crystallin, α A chain | 65.9 | 45 | 69.9 | 43 | 64.7 | 104 | 90.2 | 192 |
| Crystallin, α B chain | 69.7 | 66 | 58.9 | 46 | 62.9 | 65 | 94.9 | 177 |
The proteases trypsin, subtilisin, and elastase were chosen because they consistently produced peptides with different specificity resulting in high total sequence coverage by tandem mass spectrometry.
As evidence of the power of this approach, we found several additional sites of modification. In ovalbumin, we found two sites of phosphorylation, S237 and S241 (Fig. 1), and in myosin heavy chain, an arginine methylation at site R652 (data not shown). Because these proteins were purified for use as protein standards, the physiological significance of these modifications is not clear, but our data suggest that these sites should be further evaluated for possible biological roles. Furthermore, identification of these sites affirms the value of producing peptides across the entire sequence of the protein, using high-resolution separations combined with MS/MS spectrometry to collect data for all peptides, and then mining those data algorithmically for possible sites of modification.
Characterization of Protein Modifications from Cdc2-TAP Purified Complexes.
To extend the application of these strategies to a more biologically relevant sample, we chose to analyze the mixture of proteins that associates with the cell cycle regulating cyclin-dependent kinase in S. pombe—Cdc2p. It has well characterized sites of phosphorylation (26) and multiple identified binding partners, at least some of which are phosphorylated (Gould lab, unpublished results). Furthermore, Cdc2p has already proven amenable to purification via the tandem affinity purification (TAP) tag (21). Cdc2p-TAP complexes from 8 liters of S. pombe cells were precipitated, one-quarter set aside for evaluation by SDS/PAGE, and the remainder subjected to the triple-digest protocol. Because the sample was more complex (Fig. 2A) than our “test” mixture, each individual digest was analyzed in a separate six-cycle MudPIT.
Figure 2.
The identification of protein modifications in S. pombe cdc2-TAP purified complexes. (A) Many proteins were copurified with cdc2-TAP as observed by SDS/PAGE by using silver stain. (B) The total sequence coverage and modifications measured by MudPIT for cdc13, cig1, and cdc2 are shown. Modifications are displayed with phosphorylation in blue, acetylation in yellow, and oxidation in red.
Of the over 200 proteins present in the mixture, 20 showed greater than 40% sequence coverage, again attesting to the utility of the triple digest protocol for generating higher sequence coverage than with any of the single enzymes alone (Table 1). The high-percentage sequence coverage for this number of proteins is impressive, because in this mixture only five proteins appear to be in a similar stoichiometric range (Fig. 2A). The expected Cdc2p phosphorylations at Y15 and T167 were both detected with multiple overlapping peptides (Fig. 2B). As with the test mixture, several oxidized methionines (methionine sulfoxide) were identified. Again, it is difficult to determine whether the oxidized methionines are artifactual or have some physiological relevance.
Interestingly, we were able to identify novel sites of phosphorylation in two of the cyclin partners of Cdc2p, Cdc13p and Cig1p. Although we obtained only 57.8% sequence coverage for Cig1p, we were able to identify with multiple overlapping peptides a phosphorylation at S108. These data are consistent with unpublished observations showing that Cig1p is indeed a phosphoprotein. At least one phosphorylated serine was also detected in Cdc13p. Because of their proximity to each other in the primary sequence, there remains some ambiguity as to which of three serines (S176, S180, or S183) is phosphorylated (Fig. 2B). From our data, it is possible that each site is phosphorylated on different protein molecules and/or more than one site is phosphorylated at a time. Although we did not find evidence for multiple phosphorylations on individual peptides in our data set, the fragmentation patterns of these peptides may have complicated their identification. The position of these sites is consistent with previous phosphopeptide mapping of in vivo 32P-labeled Cdc13p. We had determined that the phosphorylation site(s) within Cdc13p were located between amino acids 98 and 198 (data not shown). Because physiologically important roles for cyclin phosphorylation have been previously determined (27), it is likely that phosphorylation of sites within Cig1p and Cdc13p will also prove important for regulating their function.
A most intriguing result from this analysis was the finding of multiple methylation sites within Cdc2p (Fig. 2B). There is no precedent for this type of modification in the regulation of cyclin-dependent kinase activity, but methylation is being increasingly implicated as a regulator of protein function (28, 29). It will be interesting to determine the role of these modifications in regulating Cdc2p activity. An important aspect to this study is in coupling the ability to identify proteins from TAP-purified protein complexes with determining sites modifications to their major protein constituents. As more efforts begin to isolate and characterize large numbers of protein complexes, it is now feasible to identify sites of modification in addition to their protein composition.
Mapping Protein Modification Sites in Human Lens Tissue Without Protein Enrichment.
The modification analysis was next extended to a whole tissue. Eye lens tissue was chosen for several reasons. One is that it is not an extremely complex tissue; by mass, most of the protein in lens tissue is from a relatively small number of individual proteins. In fact, a family of structural proteins, crystallins, constitutes about 90% of the total protein mass within the lens. Not only are these proteins important for normal lens development, but they have also been implicated in aging and disease progression—especially relating to cataract formation. Furthermore, these proteins do not appear to turn over during aging, and thus any changes in their function will be through some posttranslational mechanism. The described modifications to this family of proteins include: truncation of N and C termini (30–34), deamidation (35–37), racemization (38), phosphorylation (33, 35, 39), oxidation (33, 37, 38, 40), acetylation (41, 42), carbamylation (43), disulfide formation (35, 44), and glycation (42). Finally, because these proteins are long lived, they are prone to adventitious modification; therefore, it is necessary to have the ability to survey many possible modifications during a single experiment.
Because of the complexity of the lens sample, each digest was analyzed separately by using an 18-step MudPIT analysis. After combining the MS/MS data generated from all three digests, 270 proteins were identified, 52 of which showed more than 40% sequence coverage. We will focus this report on crystallins only because of their relative abundance and numerous reported modifications. For this family of proteins, a total of 73 modifications were found in 11 different crystallin proteins. Considering phosphorylation, methylation, oxidation, and acetylation, our method found 13 of the 18 reported sites of posttranslational modifications reported from both bovine and human samples (Table 2). The modifications not found in this analysis could be the result of species–specific differences, from the analysis having been performed on a different developmental time point (young patient), from the disease state (congenital cataract), or from a limit in our methods. Beyond those reported modifications, we found an additional 60 unambiguous modifications (total of 73).
Table 2.
Protein modifications of crystallins from a 4-year congenital cataract
| Protein/accession no. | %* | Phosphorylation | Oxidation | Acetylation† | Methylation |
|---|---|---|---|---|---|
| Crystallin, α A chain gi|1706112|sp|P02489| | 90.2 | T13, S45‡, S122‡, T140 | Y18, Y34, M138‡ | K70‡, K78, K88, K145 | R21, K88 |
| Crystallin, α B chain gi|117385|sp|P02511| | 94.9 | S19‡, S21‡, S43‡§, S45‡, S53, S59‡, S76 | Y48, W60‡, M68‡ | K92‡§ | R22, R50 |
| Crystallin, β A1 gi|4885155|ref|NP_005199.1| | 65.6 | T127, S160 | M126†§ | K122, K125, K131 | R137 |
| Crystallin, β A4 gi|4503059|ref|NP_001877.1| | 78.1 | S35, T43 | W149 | ||
| Crystallin, β B1 gi|4503061|ref|NP_001878.1| | 88.9 | S10, T12 | W216, M226 | K6, K160 | R230, R231, K235 |
| Crystallin, β B2 gi|1169091|spl|P43320| | 85.4 | T118 | W59, M122, W151 | K76, K121 | K42, K68, K121 |
| Crystallin, β B3 gi|4758074|ref|NP_004067.1| | 54.0 | Y29 | M129 | K128 | K128 |
| Crystallin, γ B gi|4885157|ref|NP_005201.1| | 60.6 | Y63, Y66 | W69, M70 | ||
| Crystallin, γ C gi|10518338|ref|NP_066269.1| | 61.5 | Y63, Y66 | Y56, W69, M70, W131 | ||
| Crystallin, γ D gi|2506321|sp|P07320| | 58.0 | Y46 | |||
| Crystallin, γ S gi|1362852|pir∥S55263 | 80.6 | M41, M101, M106 | K6 |
Database (human, Homosapiens) was downloaded from www.ncbi.nlm.gov on 11/29/00 and has 57,847 entries.
Percentage of amino acid sequence coverage obtained for proteins using the DTASelect filter cutoff values chosen for this study.
Acetylation [M+42]+ was indistinguishable from carbamylation [M+43]+ in this study.
Modification sites reported in literature (33, 35, 42, 44, 45, and 58).
Modification site identified by a single peptide only.
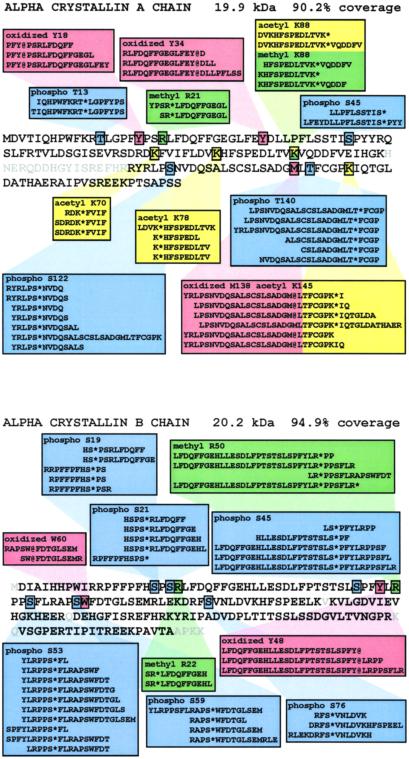
α-Crystallin, composed of two polypeptide subunits αA- and αB-crystallin, is the most abundant soluble protein in mammalian eye lens cytoplasm. Fig. 3 summarizes all modifications observed with multiple overlapping peptides for both αA- and αB-crystallin. αA-crystallin, as expected, showed phosphorylation at S45 and S122. In addition, two more phosphorylation sites, T13 and T140, were observed (Fig. 3). Past studies from both bovine and human lenses have shown that Ser-122 is a major site of in vivo phosphorylation (35, 45, 46), and elevated phosphorylation at this residue is believed to be a developmentally regulated event (39). S45 is reported to be unique to human αA-crystallin (35). For αB-crystallin, all three sites, S19, S45, and S59, reported earlier in both human and bovine lens (33, 35, 47), and two sites, S21 and S43, reported only in bovine lens (46, 48, 49), were confirmed by multiple overlapping peptides. In addition, two new sites, S53 and S76, were identified. α-Crystallin has been shown to possess a chaperone-like activity and to bind to ATP (50, 51). Kamei et al. showed that monophosphorylation of αB-crystallin markedly reduced this activity (33). There are also suggestions that phosphorylated α-crystallin may be involved in the interaction of crystallins with membranes and matrix structures/intermediate filament proteins (52, 53).
Figure 3.
The identification of protein modification sites in α-crystallin from a 4-year-old congenital cataract. For α-crystallin, modifications for phosphorylation are displayed in blue, acetylation in yellow, methylation in green, and oxidation in red. Only modification sites evidenced by multiple overlapping peptidesare shown.
In αA-crystallin, we found oxidation at three residues, namely Y18 and Y34 and M138. In αB-crystallin, Y48, W60, and M68 were oxidized. Hanson et al. previously reported the oxidation of methionine-138 and -68 in αA- and αB-crystallin, respectively (37). Finley et al. have reported oxidation of W9 and W60 in αA- and αB-crystallin, respectively, in bovine lens (40). Although our current methodology does not allow us to determine unequivocally whether these sites of oxidation occur during sample preparation or in vivo, all of these sites of oxidation have been previously reported in the literature. This appears to be especially true for oxidation of tryptophan and tyrosine residues that were not observed in either the test mixture or the cdc2 mixture experiments. Oxidation of methionine residues has been observed when lens crystallins are exposed to hydroxyl radicals, for example in a reaction with Fe+3 and H2O2 (54), and this exposure strongly inhibits chaperone activity (55). Also, it has been suggested that oxidation of tryptophan and tyrosine is initiated by UV radiation (56, 57) and can contribute to changes in lens crystallins.
We were able to detect several methylations, acetylations, and/or carbamylations to α-crystallin. In αA-crystallin, K70, K78, K88, and K145 were acetylated/carbamylated, and R21 and K88 were methylated. For αB-crystallin, K92 was acetylated/carbamylated, and R22 and R50 were methylated. Because urea was used in our sample preparation, we cannot rule out the possibility that some of these previously unreported acetylations are artifactual carbamylations (indistinguishable from acetylation using this approach) from the sample preparation. However, because only fresh urea solutions were used, the absence of widespread lysine modification in our samples, and previous reports of carbamylation occurring in vivo in the lens (43), it is unlikely that these modifications are artifacts of the sample preparation.
Both acetylation and methylation have been shown to affect protein activity and are emerging as important signaling molecules; they have been proposed to play roles in signal transduction similar to those of phosphorylation (58–60). Lin et al. reported acetylation of ≈5% of Lys-70 in αA-crystallin and suggested that it decreases the chaperone activity of crystallins (41). Lapko reported that K92 in αB-crystallin is ≈1% acetylated and ≈2% carbamylated (43). Although our current technique does not allow us to determine the relative stoichiometry of modified versus unmodified peptides, the reported stoichiometries attest to the potential for this strategy to detect very low-level modifications from within a complex mixture. The relative ease with which modifications can be discovered in a complex mixture of proteins will allow a comparison of a large number of lenses from different developmental stages, including age-related cataracts.
Conclusion
Because of the pivotal role that posttranslational modifications play in regulating protein activity, identifying the type of modification and its location can be essential to understanding the function of a given protein and ultimately the cell as a whole. In general, techniques for studying protein modifications are specific for a given type of modification and usually require extensive purification of the protein of interest. In vivo studies often necessitate the use of radioactive labeling, which itself could perturb the system being studied. Newer techniques (see Introduction) provide the potential for looking at much more complicated mixtures but are limited both by the extensive chemistry and the ability to detect only one type of modification. We report a strategy that uses multidimensional liquid chromatography and MS/MS spectrometry combined with multiple separate proteolytic digests to yield higher sequence coverage (Table 1). Significant overlap is achieved for peptides containing modifications providing internal validation for the assignment of the modification to a specific site. These data can then be “mined” algorithmically to identify modification sites within the mixture of proteins. We demonstrate the effectiveness of the strategy on mixtures that vary in complexity from only a few proteins to hundreds. Thus, not only is it possible to assay complicated protein mixtures, but also a variety of posttranslational modifications can be found from a single experiment. Further optimizations to digestion, peptide separation, data collection, and analysis should extend the capabilities and sensitivity of this strategy.
There are two striking results of this study. First, modifications can be discovered by the direct analysis of complex protein mixtures. Clearly, this will enable the analysis of proteins that may be difficult to purify to homogeneity, the analysis of modifications of proteins in complexes, and more complicated systems like the lens. Second, a remarkable number of different modifications are found on proteins in these mixtures. This finding clearly suggests that our view of the complexity of protein structure and modification may be limited by the current set of techniques to study them. Thus, the analysis of protein modifications will warrant an increasingly higher priority in the postgenome era. Last, this study suggests that a more comprehensive analysis of proteins will reveal a more complete picture of protein modification and provide a better context to their roles in regulating physiological activity.
Acknowledgments
We gratefully acknowledge financial support from National Institutes of Health Grants R33CA81665, RO1EY13288, RR11823-05, F32DK59731 (M.J.M.), and GM47728 (K.L.G.). This project was also supported in part by the Howard Hughes Medical Institute (K.L.G.) and the Merck Genome Research Institute (J.R.Y.).
Abbreviations
- MS/MS
tandem mass
- MudPIT
multidimensional protein identification technology
- TAP
tandem affinity purification
References
- 1.Eng J K, McCormack A L, Yates J R., III J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 2.Yates J R, III, Eng J K, McCormack A L. Anal Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- 3.McCormack A L, Schieltz D M, Goode B, Yang S, Barnes G, Drubin D, Yates J R., III Anal Chem. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 4.Link A J, Eng J K, Schieltz D M, Carmack E, Mize G J, Morris D R, Garvik B M, Yates J R., III Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 5.Washburn M P, Wolters D, Yates J R., III Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 6.Oda Y, Huang K, Cross F R, Cowburn D, Chait B T. Proc Natl Acad Sci USA. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paša-Tolic L, Jensen P K, Anderson G A, Lipton M S, Peden K K, Martinovic S, Tolic N, Bruce J E, Smith R D. J Am Chem Soc. 1999;121:7950. [Google Scholar]
- 8.Gygi S P, Rist B, Gerber S A, Turecek F, Gelb M H, Aebersold R. Nat Biotechnol. 2000;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 9.Ji J, Chakraborty A, Geng M, Zhang X, Amini A, Bina M, Renier F. J Chromatrogr B. 2000;745:197–210. doi: 10.1016/s0378-4347(00)00192-4. [DOI] [PubMed] [Google Scholar]
- 10.Krishna R G, Wold F. Adv Enzymol Relat Areas Mol Biol. 1993;67:265–298. doi: 10.1002/9780470123133.ch3. [DOI] [PubMed] [Google Scholar]
- 11.Gatlin C L, Eng J K, Cross S T, Detter J C, Yates J R., III Anal Chem. 2000;72:757–763. doi: 10.1021/ac991025n. [DOI] [PubMed] [Google Scholar]
- 12.Michel H, Hunt D F, Shabanowitz J, Bennett J. J Biol Chem. 1988;263:1123–1130. [PubMed] [Google Scholar]
- 13.Watts J D, Affolter M, Krebs D L, Wange R L, Samelson L E, Aebersold R. J Biol Chem. 1994;269:29520–29529. [PubMed] [Google Scholar]
- 14.de Carvalho M G, McCormack A L, Olson E, Ghomashchi F, Gelb M H, Yates J R, III, Leslie C C. J Biol Chem. 1996;271:6987–6997. doi: 10.1074/jbc.271.12.6987. [DOI] [PubMed] [Google Scholar]
- 15.Annan R S, Huddleston M J, Verma R, Deshaies R J, Carr S A. Anal Chem. 2001;73:393–404. doi: 10.1021/ac001130t. [DOI] [PubMed] [Google Scholar]
- 16.Liebler D C, Hansen B T, Davey S W, Tiscareno L, Mason D E. Anal Chem. 2002;74:203–210. doi: 10.1021/ac0155512. [DOI] [PubMed] [Google Scholar]
- 17.Oda Y, Nagasu T, Chait B T. Nat Biotechnol. 2001;19:379–382. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
- 18.Goshe M B, Conrads T P, Panisko E A, Angell N H, Veenstra T D, Smith R D. Anal Chem. 2001;73:2578–2586. doi: 10.1021/ac010081x. [DOI] [PubMed] [Google Scholar]
- 19.Zhou H, Watts J D, Aebersold R. Nat Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 20.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 21.Tasto J J, Carnahan R H, McDonald W H, Gould K L. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- 22. Sadygov, R. G., Eng, J. K., Durr, E., Saraf, A., McDonald, W. H., MacCoss, M. J. & Yates, J. R., III (2002) J. Proteome Res., in press. [DOI] [PubMed]
- 23.Tabb D L, McDonald W H, Yates J R., III J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yates J R, III, Eng J K, McCormack A L, Schieltz D M. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 25.Tsunasawa S, Narita K. J Biochem (Tokyo) 1982;92:607–613. doi: 10.1093/oxfordjournals.jbchem.a133971. [DOI] [PubMed] [Google Scholar]
- 26.Berry L D, Gould K L. Prog Cell Cycle Res. 1996;2:99–105. doi: 10.1007/978-1-4615-5873-6_10. [DOI] [PubMed] [Google Scholar]
- 27.Ciechanover A, Schwartz A L. Revis Biol Cellular. 1989;20:217–234. [PubMed] [Google Scholar]
- 28.McBride A E, Silver P A. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Huang Z Q, Xia L, Feng Q, Erdjument-Bromage H, Strahl B D, Briggs S D, Allis C D, Wong J, Tempst P, et al. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 30.Ajaz M S, Ma Z, Smith D L, Smith J B. J Biol Chem. 1997;272:11250–11245. doi: 10.1074/jbc.272.17.11250. [DOI] [PubMed] [Google Scholar]
- 31.Jimenez-Asensio J, Colvis C M, Kowalak J A, Duglas-Tabor Y, Datiles M B, Moroni M, Mura U, Rao C M, Balasubramanian D, Janjani A, et al. J Biol Chem. 1999;274:32287–32294. doi: 10.1074/jbc.274.45.32287. [DOI] [PubMed] [Google Scholar]
- 32.Takemoto L J. Curr Eye Res. 1995;14:837–841. doi: 10.3109/02713689508995806. [DOI] [PubMed] [Google Scholar]
- 33.Kamei A, Hamaguchi T, Matsuura N, Iwase H, Masuda K. Biol Pharm Bull. 2000;23:226–230. doi: 10.1248/bpb.23.226. [DOI] [PubMed] [Google Scholar]
- 34.Kamei A, Iwase H, Masuda K. Biochem Biophys Res Commun. 1997;231:373–378. doi: 10.1006/bbrc.1997.6105. [DOI] [PubMed] [Google Scholar]
- 35.Miesbauer L R, Zhou X, Yang Z, Sun Y, Smith D L, Smith J B. J Biol Chem. 1994;269:12494–502. [PubMed] [Google Scholar]
- 36.Hanson S R, Smith D L, Smith J B. Exp Eye Res. 1998;67:301–312. doi: 10.1006/exer.1998.0530. [DOI] [PubMed] [Google Scholar]
- 37.Hanson S R, Hasan A, Smith D L, Smith J B. Exp Eye Res. 2000;71:195–207. doi: 10.1006/exer.2000.0868. [DOI] [PubMed] [Google Scholar]
- 38.Fujii N, Ishibashi Y, Satoh K, Fujino M, Harada K. Biochim Biophys Acta. 1994;1204:157–163. doi: 10.1016/0167-4838(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 39.Takemoto L J. Exp Eye Res. 1996;62:499–504. doi: 10.1006/exer.1996.0060. [DOI] [PubMed] [Google Scholar]
- 40.Finley E L, Dillon J, Crouch R K, Schey K L. Protein Sci. 1998;7:2391–2397. doi: 10.1002/pro.5560071116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin P P, Barry R C, Smith D L, Smith J B. Protein Sci. 1998;7:1451–1457. doi: 10.1002/pro.5560070622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groenen P J, Merck K B, de Jong W W, Bloemendal H. Eur J Biochem. 1994;225:1–19. doi: 10.1111/j.1432-1033.1994.00001.x. [DOI] [PubMed] [Google Scholar]
- 43.Lapko V N, Smith D L, Smith J B. Protein Sci. 2001;10:1130–1136. doi: 10.1110/ps.40901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z, Chamorro M, Smith D L, Smith J B. Curr Eye Res. 1994;13:415–421. doi: 10.3109/02713689408999869. [DOI] [PubMed] [Google Scholar]
- 45.Voorter C E, Mulders J W, Bloemendal H, de Jong W W. Eur J Biochem. 1986;160:203–210. doi: 10.1111/j.1432-1033.1986.tb09958.x. [DOI] [PubMed] [Google Scholar]
- 46.Chiesa R, Gawinowicz-Kolks M A, Kleiman N J, Spector A. Biochem Biophys Res Commun. 1987;144:1340–1347. doi: 10.1016/0006-291x(87)91457-4. [DOI] [PubMed] [Google Scholar]
- 47.Lund A L, Smith J B, Smith D L. Exp Eye Res. 1996;63:661–672. doi: 10.1006/exer.1996.0160. [DOI] [PubMed] [Google Scholar]
- 48.Chiesa R, Gawinowicz-Kolks M A, Kleiman N J, Spector A. Exp Eye Res. 1988;46:199–208. doi: 10.1016/s0014-4835(88)80077-0. [DOI] [PubMed] [Google Scholar]
- 49.Smith J B, Sun Y, Smith D L, Green B. Protein Sci. 1992;1:601–608. doi: 10.1002/pro.5560010506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muchowski P J, Hays L G, Yates J R, III, Clark J I. J Biol Chem. 1999;274:30190–30195. doi: 10.1074/jbc.274.42.30190. [DOI] [PubMed] [Google Scholar]
- 51.Muchowski P J, Clark J I. Proc Natl Acad Sci USA. 1998;95:1004–1009. doi: 10.1073/pnas.95.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spector A, Chiesa R, Sredy J, Garner W. Proc Natl Acad Sci USA. 1985;82:4712–4716. doi: 10.1073/pnas.82.14.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muchowski P J, Valdez M M, Clark J I. Invest Ophthalmol Vis Sci. 1999;40:951–958. [PubMed] [Google Scholar]
- 54.Smith J B, Jiang X, Abraham E C. Free Radic Res. 1997;26:103–111. doi: 10.3109/10715769709097789. [DOI] [PubMed] [Google Scholar]
- 55.Cherian M, Smith J B, Jiang X Y, Abraham E C. J Biol Chem. 1997;272:29099–29103. doi: 10.1074/jbc.272.46.29099. [DOI] [PubMed] [Google Scholar]
- 56.Andley U P, Clark B A. Photochem Photobiol. 1989;50:97–105. doi: 10.1111/j.1751-1097.1989.tb04134.x. [DOI] [PubMed] [Google Scholar]
- 57.Li D Y, Borkman R F, Wang R H, Dillon J. Exp Eye Res. 1990;51:663–669. doi: 10.1016/0014-4835(90)90050-5. [DOI] [PubMed] [Google Scholar]
- 58.Kouzarides T. EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Magnaghi-Jaulin L, Ait-Si-Ali S, Harel-Bellan A. Prog Cell Cycle Res. 2000;4:41–47. doi: 10.1007/978-1-4615-4253-7_4. [DOI] [PubMed] [Google Scholar]
- 60.Sterner D E, Berger S L. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]