Abstract
Adenosine deaminases that act on RNA (ADARs) constitute a family of RNA-editing enzymes that convert adenosine to inosine within double-stranded regions of RNA. We previously developed a method to identify inosine-containing RNAs and used it to identify five ADAR substrates in Caenorhabditis elegans. Here we use the same method to identify five additional C. elegans substrates, including three mRNAs that encode proteins known to affect neuronal functions. All 10 of the C. elegans substrates are edited in long stem-loop structures located in noncoding regions, and thus contrast with previously identified substrates of other organisms, in which ADARs target codons. To determine whether editing in noncoding regions was a conserved ADAR function, we applied our method to poly(A)+ RNA of human brain and identified 19 previously unknown ADAR substrates. The substrates were strikingly similar to those observed in C. elegans, since editing was confined to 3′ untranslated regions, introns, and a noncoding RNA. Also similar to what was found in C. elegans, 15 of the 19 substrates were edited in repetitive elements. The identities of the newly identified ADAR substrates suggest that RNA editing may influence many biologically important processes, and that for many metazoa, A-to-I conversion in coding regions may be the exception rather than the rule.
Adenosine deaminases that act on RNA (ADARs) are RNA-editing enzymes that convert adenosine (A) to inosine (I) in double-stranded RNA (dsRNA) (reviewed in refs. 1–4). Mice (5, 6), flies (7), and worms (L. A. Tonkin, L. Saccomanno, D.P.M., T. Brodigan, M. Krause, and B.L.B., unpublished work) that lack ADARs have behavioral defects, emphasizing that ADARs have important roles in the nervous system. However, exactly how the enzymes mediate their functions, and which RNAs they target, is known for only a few cases. For example, in mammals ADARs are known to target codons so that multiple protein isoforms can be synthesized from a single message; in this way ADARs generate isoforms of glutamate receptors (GluRs) (8–10), serotonin receptors (11), and the hepatitis delta antigen (12, 13). In addition, mammalian ADAR2 edits its own pre-mRNA to create a new splice site, which may serve to autoregulate ADAR2 levels (14). Finally, recent data show that RNAs targeted by ADARs at many sites are selectively retained in the nucleus (15), at least in some cases.
In addition to altering the sequence of RNA, ADARs can alter RNA structure. Deamination of an adenosine within an A⋅U base pair produces the less stable I⋅U wobble pair, whereas deamination within A⋅C mismatches produces more stable I⋅C pairs. Therefore, ADARs can either decrease or increase the stability of dsRNA. Sequence or structural changes produced by ADARs within noncoding regions of mRNAs such as untranslated regions (UTRs), or introns, could potentially affect such processes as RNA translation, stability, localization, or splicing. Other types of RNAs such as catalytic RNAs, guide RNAs, small nuclear RNAs, small nucleolar RNAs (snoRNAs), and RNAs involved in dosage compensation and imprinting, indeed any RNA molecule that forms intra- or intermolecular duplexes of sufficient length, are potential functionally important ADAR targets.
The amounts of inosine in mRNA isolated from various rat tissues show that there is far more inosine than can be accounted for by the few known mammalian ADAR substrates (16). Given the large number of undiscovered substrates, and the vast potential for RNA editing to perform regulatory functions, it remains possible that codon changes do not represent the major role for ADARs. Toward the goal of understanding the full scope of ADAR function, we developed a systematic method to identify RNAs that contain inosine (17, 18). In our first application of the method we identified 5 previously unknown ADAR substrates in Caenorhabditis elegans (18); as described here, we have now increased this number to 10. Each of the C. elegans substrates is edited within a stem-loop structure located in a UTR, an intron, or a noncoding RNA. Several of the edited structures are moderately repetitive sequences called IR (inverted repeat) elements that are likely remnants of once-functional transposons (19). These ADAR substrates suggest that editing functions to regulate gene expression in ways other than targeting codons. To further investigate this issue, we asked whether editing in noncoding regions was a conserved ADAR function. Here we show that in human brain, as in C. elegans, noncoding regions of mRNAs appear to be the primary targets of ADARs.
Materials and Methods
Identification of ADAR Substrates in C. elegans and Human Brain.
C. elegans substrates were identified and analyzed as previously described (18). Human brain poly(A)+ RNA used for the initial identification of human substrates was purchased from CLONTECH. Candidate human substrates were identified as described previously (18) except that PCR products amplified from DNA isolated from RNase T1-dependent bands was not cloned, but sequenced directly.
Confirmation and Quantification of Editing, and Identification of Additional Editing Sites.
Regions surrounding RNase T1 cleavage sites were amplified from random hexamer-primed cDNA and from genomic DNA. As indicated in Fig. 2, C. elegans PCR products were either cloned followed by sequencing of individual clones, or sequenced as a population, without cloning. All of the human PCR products were directly sequenced without cloning. For the human samples, we used poly(A)+ RNA and genomic DNA isolated from the same individual (purchased from BioChain Institute, Hayward, CA) to ensure differences between genomic and cDNA sequences were due to RNA editing rather than polymorphisms. When the PCR products were directly sequenced, the extent of editing at each site was determined by using the electropherograms from the sequencing reactions to estimate the relative amounts of A and G (see Fig. 3A). As in our previous work (18), when the PCR products were cloned, editing efficiency was estimated by evaluating the sequence of 10 clones. In the work reported here most PCR products were sequenced directly, and this resulted in more accurate estimates of editing efficiency because the sequences represented the entire population of transcripts.
Figure 2.
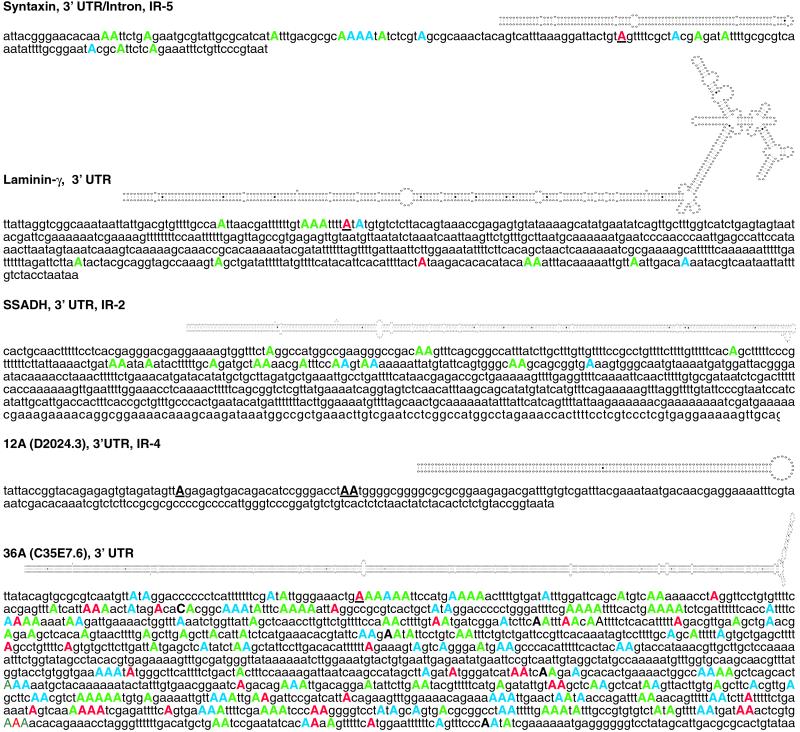
Each of the newly identified C. elegans substrates is designated with a name, the location of editing within the mRNA, and the type of repetitive element, if one exists. Structures predicted by mfold are shown above the unedited sequence. Adenosines determined to be edited by cDNA sequencing are shown in uppercase, color-coded according to the percentage of the cDNA population that was edited at the site (red, >70%; blue, 40–70%; green, <40%); RNase T1 cleavage sites are underlined. Editing in syntaxin occurs within an alternatively spliced region that can be either an intron or a 3′ UTR. SSADH was found by virtue of its IR element, not by T1 cleavage, and only nucleotides 2538–2773 (complementary strand) of cosmid F45H10 were sequenced to determine editing sites. Editing sites were mapped by compiling sequence data from multiple PCR clones (SSADH), uncloned PCR populations (36A), or both types of data (syntaxin; laminin-γ); the editing sites of 12A have not been mapped, but three T1 cleavage sites were detected and are shown as black underlined capitals.
Figure 3.
(A) The electropherograms correspond to the first two editing sites of HsC7-1 mRNA. The two adenosines shown in the genomic DNA sequence appear as a mixture of adenosine and guanosine in the cDNA, which is diagnostic of editing. N indicates that the sequencing software could not distinguish between A and G. (B) Eight of the newly identified human brain substrates are represented as in Fig. 2. (The RNase T1 cleavage site in the CDK8 mRNA was on the bottom strand, which was not sequenced.)
Sequence and Structure Analysis.
Identification of genes and assignment of editing sites to 3′ UTRs, introns, or noncoding RNAs were made by using predicted ORFs in genomic sequences, mRNA and EST (expressed sequence tag) sequences, and data in the published literature. We used sequence data from both the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST/) and Celera (celera.com/). Repetitive elements were identified by using the program RepeatMasker (repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker). Secondary structure predictions were made by using the program mfold (bioinfo.math.rpi.edu/∼mfold/rna/form1.cgi).
Results
Identification of Additional C. elegans Substrates.
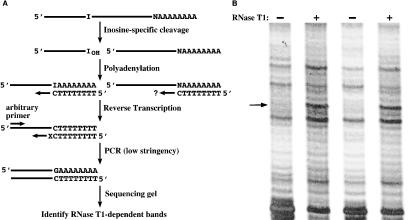
To gain further evidence that the ADAR substrates previously identified in C. elegans represented the norm, we continued screening C. elegans poly(A)+ RNA for inosine-containing RNAs. The method we used has been described (18) and is outlined in Fig. 1A. Briefly, glyoxalated poly(A)+ RNA is treated with RNase T1, which results primarily in cleavage 3′ of inosines, with some background cleavage 3′ of guanosines. In parallel, glyoxalated poly(A)+ RNA is treated identically, but the RNase T1 is omitted. Both samples of RNA are reverse-transcribed, amplified with the PCR, and compared by using a differential display protocol (Fig. 1B). As shown in Fig. 1B, candidate ADAR substrates are identified as RNase T1-dependent bands (arrow). The sequence of the DNA isolated from an RNase T1-dependent band is submitted to a blast search to identify the corresponding genomic DNA sequence. A true ADAR substrate is distinguished from a false positive (G cleavage) by the presence of a genomically encoded adenosine at the RNA cleavage site.
Figure 1.
(A) The method used to detect inosine-containing RNAs. As described (18), inosine-specific cleavage is followed by polyadenylation to create a primer binding site for a subsequent reverse transcription step. N, nucleotide 5′ of poly(A) tail; the question mark indicates that efficiency of synthesis from uncleaved RNAs will vary depending on the nucleotide 5′ of the poly(A) tail. X, nucleotide(s) on the 3′ end of the downstream PCR primer was G, A, C, TG, TA, TT, or TC. (B) An example of an RNase T1-dependent band. Duplicate samples of human poly(A)+ RNA were subjected to the protocol of A. The arrow points to the RNase T1-dependent band (in the + lanes) whose sequence revealed that it derived from HsC7-I mRNA cleaved in its 3′ UTR.
We identified 5 additional ADAR substrates in C. elegans, and these are listed along with our previously identified C. elegans substrates in Table 1. Like the previously isolated substrates, all editing sites in the new substrates were located in noncoding regions. Our recent work indicates that in C. elegans, as in other organisms, ADARs are highly expressed in the nervous system (L. A. Tonkin, L. Saccomanno, D.P.M., T. Brodigan, M. Krause, and B.L.B., unpublished work). Consistent with this, three of the mRNAs identified as new substrates, unc-64 syntaxin (20), laminin-γ (21), and succinate semialdehyde dehydrogenase (SSADH) (22) are important for proper function of the nervous system.
Table 1.
C. elegans ADAR substrates
| RNA* (Wormbase ID) | Editing | Repeat | Function |
|---|---|---|---|
| M05B5.3 | 5′ UTR | IR-5 | Unknown |
| 52G (F55A4.9) | Noncoding | None | Unknown |
| pop-1 (W10C8.2) | 3′ UTR | None | Transcription factor (39) |
| 9A (ZC239.6) | 3′ UTR | IR-3 | Unknown |
| 16G (Y6D11A.1) | 3′ UTR | None | Unknown |
| Syntaxin (F56A8.7a) | 3′ UTR/ intron† | IR-5 | Secretion of neurotransmitters (20) |
| Laminin-γ (C54D1.5) | 3′ UTR | None | Component of basement membranes (21) |
| SSADH (F45H10.1) | 3′ UTR | None | γ-Aminobutyrate turnover (22) |
| 12A (D2024.3) | 3′ UTR | IR-2 | Unknown |
| 36A (C35E7.6) | 3′ UTR | IR-4 | Unknown |
Identification of ADAR Substrates in Human Brain.
To determine if editing of noncoding regions was a conserved ADAR function, we applied our method to human brain RNA. We first attempted to detect inosine in two mRNAs already known to be ADAR substrates in human brain. Human brain poly(A)+ RNA was treated as outlined in Fig. 1A, and the PCR products were analyzed by differential display (e.g., Fig. 1B). Using primers designed to anneal upstream of known editing sites, we successfully detected the Q/R editing site in the mRNA encoding subunit B of glutamate-gated ion channels (GluR-B; ref. 8) and editing site C in the serotonin 2C receptor mRNA (11) (data not shown). Our previous applications of our method indicated that new ADAR substrates could be identified with a minimum of 8 complementary positions between the 13-nucleotide arbitrary primer and the new ADAR substrate (18). Thus, to approximate a real experiment, the primers used in the differential display step were not identical to the target mRNAs but contained 8 of 13 identities (GluR-B) or 9 of 13 identities (serotonin receptor). In addition, like the arbitrary primers used for the real experiments, each 13-mer had a G+C content of ≈50%. Our ability to detect editing sites within the GluR-B and serotonin receptor sequences showed that our method was capable of detecting mRNAs edited in coding regions, and instilled confidence that our starting RNA pool was not overly biased for highly abundant or highly structured RNAs. [GluR-B mRNA is a low-abundance mRNA (16), and although ADARs target double-stranded regions in both GluR-B and serotonin receptor pre-mRNAs, the structured regions are removed by splicing and not present in the sequences we amplified.]
We next used our collection of arbitrary primers (18) to identify previously unknown human brain substrates. The search for human substrates was far more efficient than our search in C. elegans. When we used C. elegans RNA, 700 PCRs were required to find 100 candidate substrates, only 10 of which were true positives (ref. 18 and this study). In contrast, only 130 PCRs were required to identify 44 candidate substrates in human brain RNA, 19 of which were true positives. Of the remaining 25 human candidates, 12 did not give a unique sequence, 8 were not found in any database, and, significantly, only 5 were false positives cleaved after guanosine.
Sequence Analysis of Human RNAs.
To determine the identities of the previously unknown human ADAR substrates, we searched GenBank [nonredundant and HTGS (high-throughput genomic sequence) databases] and the Celera human genome sequence (23). Seven of the 19 substrates encoded gene products of known function, and 2 encoded proteins with sequence similarities to known proteins (see Table 2). As observed in C. elegans, editing within these 9 transcripts occurred exclusively in noncoding regions. Five were edited in introns, 3 in 3′ UTRs, and 1 of the new substrates was a noncoding RNA (PAR-SN). The remaining 10 substrates were found in regions of the genome not yet annotated (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org), and we could identify no reasonably long ORFs at or near the editing sites. By inference, these RNAs are probably also edited in noncoding regions, but we cannot yet determine whether editing occurs in introns, UTRs, or noncoding RNAs.
Table 2.
Human brain ADAR substrates with blastp hits
| Gene product | Accession nos., RNA/gene | Editing | Repeat* | Function |
|---|---|---|---|---|
| Similar to CDK8 | AY028424/Z84480 | Intron | AluSx(+)†, AluJo(−)† | Unknown |
| NrCAM | AJ001057/AF172277 | Intron | None | Cell adhesion, axon guidance (40) |
| PDE8A | AF056490/AC012064 | Intron | None | cAMP hydrolysis (41) |
| Similar to ubiquitin hydrolase | AK001647/AC019221 | Intron | LINE1 | Unknown |
| HsC7-I | D26599‡/AL357035 | 3′ UTR | AluSc(+), AluJb(−) | Proteasome subunit (42) |
| NADH:ubiquinone oxidoreductase subunit B14.5B | BC007323/AC023532 | 3′ UTR | AluJb(+), AluSc(−) | Mitochondrial electron transport (43) |
| Tankyrase | AF082557/AC055869 | 3′ UTR | MER53 | ADP-ribosylation of TRF (44) |
| PP2C-β | AF294792/hCG15783§ | Intron | AluY(+)¶ | Protein phosphatase (45) |
| PAR-SN | U55937/AC009696 | Noncoding | AluJo(−), AluSx(+), AluSx(+) | snoRNA host (HBII-13) (30, 31) |
(+) indicates that Alu is in sense orientation, (−) indicates antisense orientation.
Only AluSx was sequenced, but RNase T1 cleavage site was in AluJo.
mRNA sequence does not extend to edited region; a composite cDNA was constructed from expressed sequence tags to connect edited region to upstream ORF.
Accession no. for gene is from the Celera database.
There are three antisense Alus within 1 kb of this Alu but RNA was not sequenced to determine if other Alus are edited.
As in C. elegans, most (15 of 19) of the human substrates were edited within repetitive elements that were embedded within the mRNA. One of these is a MER53 element (a Mariner-like transposon; ref. 24) and the other 14 are retroelements. Among the retroelements are 3 LINE elements, 1 MaLR element, and 10 Alu repeats.
Structure Analysis.
ADARs are dsRNA-binding proteins, and consequently, deaminate RNAs in regions that fold to create long stretches of dsRNA. To identify potential secondary structures in the substrates, we submitted sequences surrounding the edited regions to the RNA folding program mfold (25, 26). We found good candidate structures for each of the new worm substrates (Fig. 2) and for 10 of the 19 human substrates, including 4 of the substrates shown in Fig. 3. As observed for the C. elegans substrates, the structures of some of the human substrates involved pairing between the repetitive elements found in the mRNAs. For example, 9 human substrates contained edited Alu sequences that could potentially form long stem-loop structures because they were located within 1 kb of a second Alu repeat oriented in the opposite direction (such pairs of Alu sequences are referred to as “inverted Alus”; Fig. 3B).
The initial sequencing of DNA isolated from our differential display gels provided only short stretches of sequence upstream of the RNase T1 cleavage sites. To confirm that sequences required to form the predicted secondary structures were actually part of the identified transcripts, we attempted to amplify the entire structured region of each substrate by using primer pairs that flanked the putative stem-loop. If no reverse transcription–PCR product was obtained, we attempted to amplify the regions of interest in two overlapping pieces. Using these two strategies, followed by direct sequencing of the PCR products, we confirmed the presence of the complementary sequences for 3 of the 5 newly described worm substrates (syntaxin, laminin, and 36A; Fig. 2) and for the 4 structures shown in Fig. 3B.
Analysis of Editing Patterns.
ADARs typically deaminate multiple adenosines in their substrates. Consistent with this, in some cases we isolated multiple RNase T1-dependent bands for a single substrate (e.g., Fig. 2, 12A). However, T1 cleavage at closely spaced inosines would yield small fragments that are difficult to amplify, except for the fragment corresponding to the region upstream of the most 5′ cleavage site. In fact, for most substrates we isolated a single T1-dependent band that corresponded to cleavage at the most 5′ editing site, or the most efficiently edited site nearest to the 5′ end (see Figs. 2 and 3). To analyze the editing pattern throughout the base-paired region, we sequenced cDNA clones, or directly sequenced reverse transcription–PCR products. (A-to-I conversion at the RNA level appears as an A-to-G change in the corresponding cDNA.) We determined the pattern and efficiency of editing in 4 of the 5 new worm substrates (Fig. 2) and for 8 of the 9 human substrates listed in Table 2 (Fig. 3B). In some structures only 1 or 2 editing sites were found, whereas others showed numerous sites (see Figs. 2 and 3). This result suggested that our protocol was not biased for substrates that contained a particular number of editing sites.
The structures found in the various substrates exhibited a wide range of stabilities. Further, as previously observed (4), the fraction of adenosines edited within each structure correlated with the stability of the structure, with the longest, most stable, structures showing the highest number of editing sites. For example, the long structures formed by the inverted Alus of the human substrates were edited at multiple sites, and in each case, editing was detected in both strands of the predicted hairpin. Because ADARs are specific for dsRNA, this provided further evidence that the inverted Alus pair in these RNAs (Fig. 3B). Unlike the inverted Alus, the short structure formed by the MER53 element in the tankyrase 3′ UTR was edited with moderate efficiency at only a single site (Fig. 3B). We also sequenced cDNAs derived from human NrCAM, PDE8A, and ubiquitin carboxyl-terminal hydrolase RNAs, for which no convincing secondary structure was detected. Each of these substrates contained only 2 to 4 editing sites in a 400- to 500-nucleotide region surrounding the originally detected RNase T1 cleavage site (Fig. 3B). Possibly the structures formed in these RNAs were difficult to predict because they are of low stability or because the complementary regions lie far from the detected editing sites; alternatively, these RNAs could pair in trans to an antisense RNA. Finally, we sequenced PAR-SN RNA and found multiple editing sites, verifying that this molecule is edited. A structure is not shown for PAR-SN RNA because it contains an antisense Alu and two sense Alus, all of which are edited, making structure prediction ambiguous (the antisense Alu could pair with either sense Alu).
Discussion
Using our previously described method, we identified 5 previously unknown ADAR substrates of C. elegans, as well as 19 of human brain. All of the newly discovered substrates are edited in noncoding regions, similar to what we found in a previous application of our method to C. elegans (18). Whereas previous studies clearly show that some mammalian substrates are edited in coding sequences, our data suggest that these substrates are more rare than those edited in noncoding sequences, and thus, less likely to be identified by our protocol. Codon changes may be the exception rather than the rule in C. elegans and human brain, and by extrapolation, noncoding regions may be the major ADAR targets in most other metazoa and tissues.
Another interpretation of our results is that our protocol is somehow biased for the selection of substrates edited in noncoding regions. Although we cannot definitively rule this out, many of our results are inconsistent with this possibility. For example, in our control experiments we readily detected editing in coding regions of glutamate and serotonin receptor mRNAs (data not shown), demonstrating that, in theory, our protocol can identify inosines in coding sequences. Further, the substrates we identified are quite diverse, arguing against some of the more obvious potential biases. They exhibit structures of a wide range of stabilities and editing frequencies, and according to Northern analyses, represent low- to high-abundance mRNAs (data not shown). Finally, in support of our observations, an ongoing human brain cDNA sequencing project recently found additional cDNAs with clustered A-to-G changes, all in 3′ UTRs (O. Ohara, personal communication and ref. 27). The cDNAs of this project typically included both the complete coding region and the 3′ UTR, and thus should not be biased toward finding A-to-G changes in noncoding sequences.
At present we can only speculate about the function of editing in the new substrates. There are two questions pertinent to our speculations. First, do the RNA secondary structures in noncoding regions have functions? Second, does editing influence these functions?
Possible Functions for Editing in Introns.
Since incompletely spliced RNAs normally constitute only a small fraction of the poly(A)+ pool, it is noteworthy that we detected five edited introns in human brain RNA. Either our method is very sensitive, or these introns are spliced unusually slowly. ADARs sometimes target regions of pairing between exons and introns, and possibly such structures decrease the rate of splicing. Consistent with this idea, an exon/intron structure in para pre-mRNA that is targeted by the Drosophila ADAR must be resolved by an RNA helicase before proper splicing can occur (28). This raises the possibility that one function of ADARs is to regulate splicing by targeting pre-mRNAs with exon/intron structures. By changing A·U base pairs to I·U mismatches, ADARs could reduce the stability of such structures and promote their unfolding. Whereas previous studies show that ADARs can change an AA dinucleotide to AI, creating a new 3′ splice site (14), the intronic editing sites we detected do not fit with such a scenario.
Possible Functions for Editing of 3′ UTRs.
UTR sequences sometimes regulate the translation, stability, or localization of an mRNA (29). The stem-loop structures we identified could mediate these functions by recruiting proteins, or controlling access to regulatory sequences. Editing could regulate the function of a stem-loop by changing its sequence to destroy (or create) a protein-binding site or to alter its structure so as to reveal (or mask) a nearby protein-binding site.
Possible Functions for Editing of PAR-SN RNA.
PAR-SN RNA is encoded within the Prader–Willi syndrome/Angelman syndrome (PWS/AS) region on human chromosome 15 (30, 31). A number of genes within this 2- to 3-Mb region are imprinted and are transcribed exclusively from either the maternal or paternal chromosome (for reviews see refs. 32 and 33). PAR-SN RNA is paternally expressed, and possibly editing of PAR-SN RNA influences this imprinting. In addition, genes for a novel class of brain-specific C/D-box snoRNAs have been found in the PWS/AS region (34, 35). One of them (HBII-13) is embedded near the 3′ end of the ≈3.4-kb PAR-SN transcript (30, 31, 36), downstream of the three edited Alus we identified. Interestingly, it has been suggested that HBII-52 may negatively regulate editing of the serotonin 2C receptor mRNA by ADAR, by base-pairing to the edited region (34). If editing influences processing of snoRNAs in the PWS/AS region, and if these snoRNAs regulate editing of ADAR substrates, it suggests that ADARs regulate themselves by facilitating the processing of inhibitory snoRNAs.
Do Stem-Loops in Noncoding Regions Activate Other dsRNA Pathways?
dsRNA-binding proteins (dsRBPs) are not sequence-specific, and thus, the long double-stranded regions we observe may interact with other dsRBPs, in addition to ADARs. For example, in mammalian cells, the structured regions could activate the dsRNA-dependent protein kinase (PKR) to trigger phosphorylation of the α subunit of eiF2 and inhibition of protein synthesis (reviewed in ref. 37). Similarly, the base-paired regions could bind dsRBPs involved in the RNA interference (RNAi; ref. 38) pathway and trigger gene silencing. In these scenarios, deamination of the dsRNA by ADARs could provide further regulation by modulating the RNA structure and sequence.
Supplementary Material
Acknowledgments
We thank Osamu Ohara for communicating unpublished results and offering helpful advice, and Jana Brubaker for assistance in preparing figures. This work was supported by funds to B.L.B. from the National Institute of General Medical Sciences (GM44073). Oligonucleotide synthesis and DNA sequencing were performed by the Howard Hughes Medical Institute oligonucleotide synthesis facility at the University of Utah supported by the National Cancer Institute (Grant 42014) and the Howard Hughes Medical Institute. B.L.B. is a Howard Hughes Medical Institute Associate Investigator.
Abbreviations
- ADARs
adenosine deaminases that act on RNA
- dsRNA
double-stranded RNA
- GluR
glutamate receptor
- UTR
untranslated region
- snoRNAs
small nucleolar RNAs
- IR
inverted repeat
- SSADH
succinate semialdehyde dehydrogenase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hough R F, Bass B L. In: RNA Editing, Frontiers in Molecular Biology. Bass B L, editor. Vol. 34. Oxford: Oxford Univ. Press; 2001. pp. 77–108. [Google Scholar]
- 2.Emeson R B, Singh M. In: RNA Editing, Frontiers in Molecular Biology. Bass B L, editor. Vol. 34. Oxford: Oxford Univ. Press; 2001. pp. 109–138. [Google Scholar]
- 3.Rueter S M, Emeson R B. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 343–361. [Google Scholar]
- 4.Bass B L. Trends Biochem Sci. 1997;22:157–162. doi: 10.1016/s0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- 5.Higuchi M, Maas S, Single F N, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg P H. Nature (London) 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q, Khillan J, Gadue P, Nishikura K. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 7.Palladino M J, Keegan L P, O'Connell M A, Reenan R A. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 8.Higuchi M, Single F N, Kohler M, Sommer B, Sprengel R, Seeburg P H. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 9.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger J R, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg P H. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 10.Herb A, Higuchi M, Sprengel R, Seeburg P H. Proc Natl Acad Sci USA. 1996;93:1875–1880. doi: 10.1073/pnas.93.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Nature (London) 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 12.Casey J L, Gerin J L. J Virol. 1995;69:7593–7600. doi: 10.1128/jvi.69.12.7593-7600.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polson A G, Bass B L, Casey J L. Nature (London) 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 14.Rueter S M, Dawson T R, Emeson R B. Nature (London) 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Carmichael G G. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 16.Paul M S, Bass B L. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morse D P, Bass B L. Biochemistry. 1997;36:8429–8434. doi: 10.1021/bi9709607. [DOI] [PubMed] [Google Scholar]
- 18.Morse D P, Bass B L. Proc Natl Acad Sci USA. 1999;96:6048–6053. doi: 10.1073/pnas.96.11.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devine S E, Chissoe S L, Eby Y, Wilson R K, Boeke J D. Genome Res. 1997;7:551–563. doi: 10.1101/gr.7.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saifee O, Wei L, Nonet M L. Mol Biol Cell. 1998;9:1235–1252. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Wadsworth W G. Science. 2000;288:150–154. doi: 10.1126/science.288.5463.150. [DOI] [PubMed] [Google Scholar]
- 22.Chambliss K L, Caudle D L, Hinson D D, Moomaw C R, Slaughter C A, Jakobs C, Gibson K M. J Biol Chem. 1995;270:461–467. doi: 10.1074/jbc.270.1.461. [DOI] [PubMed] [Google Scholar]
- 23.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 24.Kapitonov V V, Jurka J. DNA Seq. 1998;8:277–288. doi: 10.3109/10425179809034073. [DOI] [PubMed] [Google Scholar]
- 25.Zuker M, Mathews D H, Turner D H. In: RNA Biochemistry and Biotechnology, NATO ASI Series. Barciszewski J, Clark B F C, editors. Dordrecht, The Netherlands: Kluwer; 1999. pp. 11–43. [Google Scholar]
- 26.Mathews D H, Sabina J, Zuker M, Turner D H. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 27.Kikuno R, Nagase T, Waki M, Ohara O. Nucleic Acids Res. 2002;30:166–168. doi: 10.1093/nar/30.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reenan R A, Hanrahan C J, Barry G. Neuron. 2000;25:139–149. doi: 10.1016/s0896-6273(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 29.Decker C J, Parker R. Curr Opin Cell Biol. 1995;7:386–392. doi: 10.1016/0955-0674(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 30.Buiting K, Dittrich B, Endele S, Horsthemke B. Genomics. 1997;40:132–137. doi: 10.1006/geno.1996.4571. [DOI] [PubMed] [Google Scholar]
- 31.Ning Y, Roschke A, Christian S L, Lesser J, Sutcliffe J S, Ledbetter D H. Genome Res. 1996;6:742–746. doi: 10.1101/gr.6.8.742. [DOI] [PubMed] [Google Scholar]
- 32.Mann M R, Bartolomei M S. Hum Mol Genet. 1999;8:1867–1873. doi: 10.1093/hmg/8.10.1867. [DOI] [PubMed] [Google Scholar]
- 33.Nicholls R D, Saitoh S, Horsthemke B. Trends Genet. 1998;14:194–200. doi: 10.1016/s0168-9525(98)01432-2. [DOI] [PubMed] [Google Scholar]
- 34.Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan C I, Horsthemke B, Bachellerie J P, Brosius J, Huttenhofer A. Proc Natl Acad Sci USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de los Santos T, Schweizer J, Rees C A, Francke U. Am J Hum Genet. 2000;67:1067–1082. doi: 10.1086/303106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirth J, Back E, Huttenhofer A, Nothwang H G, Lich C, Gross S, Menzel C, Schinzel A, Kioschis P, Tommerup N, et al. Hum Mol Genet. 2001;10:201–210. doi: 10.1093/hmg/10.3.201. [DOI] [PubMed] [Google Scholar]
- 37.Samuel C E. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernstein E, Denli A M, Hannon G J. RNA. 2001;7:1509–1521. [PMC free article] [PubMed] [Google Scholar]
- 39.Lin R, Thompson S, Priess J R. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 40.Wang B, Williams H, Du J S, Terrett J, Kenwrick S. Mol Cell Neurosci. 1998;10:287–295. doi: 10.1006/mcne.1997.0658. [DOI] [PubMed] [Google Scholar]
- 41.Fisher D A, Smith J F, Pillar J S, St Denis S H, Cheng J B. Biochem Biophys Res Commun. 1998;246:570–577. doi: 10.1006/bbrc.1998.8684. [DOI] [PubMed] [Google Scholar]
- 42.Nothwang H G, Tamura T, Tanaka K, Ichihara A. Biochim Biophys Acta. 1994;1219:361–368. doi: 10.1016/0167-4781(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 43.Loeffen J L, Triepels R H, van den Heuvel L P, Schuelke M, Buskens C A, Smeets R J, Trijbels J M, Smeitink J A. Biochem Biophys Res Commun. 1998;253:415–422. doi: 10.1006/bbrc.1998.9786. [DOI] [PubMed] [Google Scholar]
- 44.Smith S, Giriat I, Schmitt A, de Lange T. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 45.Cheng A, Kaldis P, Solomon M J. J Biol Chem. 2000;275:34744–34749. doi: 10.1074/jbc.M006210200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.