Abstract
Mammalian tRNA synthetases form a macromolecular complex with three nonenzyme factors: p43, p38, and p18. Here we introduced a mutation within the mouse p38 gene to understand its functional significance for the formation of the multi-tRNA synthetase complex. The complex was completely disintegrated by the deficiency of p38. In addition, the protein levels and catalytic activities of the component enzymes and cofactors were severely decreased. A partial truncation of the p38 polypeptide separated the associated components into different subdomains. The mutant mice showed lethality within 2 days of birth. Thus, this work provides the first evidence, to our knowledge, that p38 is essential for the structural integrity of the multi-tRNA synthetase complex and mouse viability.
Keywords: aminoacyl-tRNA synthetase‖macromolecular protein complex‖gene trap‖protein–protein interaction
Aminoacyl-tRNA synthetases (ARSs) are essential enzymes catalyzing the ligation of their cognate amino acids and tRNAs. Eight different enzymes of higher eukaryotes form a macromolecular complex with three nonsynthetase factors: p43, p38, and p18 (1–3). Although this complex was first reported more than two decades ago, the functional reason for their molecular assembly and the structural organization of the components still remain unknown.
Recently, much information has been obtained about the associations and interactions of the component proteins. The assembly of the complex is mediated by heat-shock protein 90 (4) and involves protein–protein interactions via the unique noncatalytic peptides attached to each of the component enzymes (5–7) and their catalytic core domains (8). It was expected that the three associating factors would also contribute to the complex formation. Among the three auxiliary factors, the interaction and function of p43 have been best elucidated. The p43 protein is located in the middle of the complex (9) and is associated with arginyl-tRNA synthetase via its N-terminal region (10). Its C-terminal domain contains an OB-fold, which is responsible for the interaction with tRNA (11, 12) and facilitates the catalytic activity of the bound enzyme (10). Interestingly, p43 is also secreted to work as a proinflammatory cytokine (13, 14). The functions of the two other factors are less understood. p38 interacts with many components of the complex (2, 15). To understand the in vivo functional significance of p38 in the formation of the multi-ARS complex, we mutated the p38 structural gene in the mouse and investigated its effects on the cellular stability of the multi-ARS complex and the component proteins. Deletion analyses of p38 were also conducted to map the organization of the component proteins within the complex.
Materials and Methods
Generation and Characterization of the p38 Mutant.
The mutation in the mouse genomic DNA was generated by the gene trap method (16). The gene trap vector was used to generate a random mutant library (OmniBank Library, Lexicon Genetics, The Woodlands, TX) of embryonic stem cells derived from the 129/SvEvBrd mouse. In the library, we identified the OST7994 clone, in which the p38 gene was disrupted. By using this clone, the C57/BL6 albino heterozygous mice were generated by following the standard protocol of Lexicon Genetics. The heterozygous mice were then crossed to obtain homozygous mutant mice. The mouse embryonic fibroblast (MEF) cells were obtained from the 13.5-day mouse embryos.
The mutation was confirmed by Southern blot and PCR analyses by using the genomic DNAs isolated from MEF cells. p38 expression was determined by Northern and Western blot analyses. The location of the gene trap insertion was determined by genomic DNA sequencing. For Southern blotting, genomic DNA was isolated from the mouse tail and digested with SacI. The cleaved DNA fragments were separated by denaturing gel electrophoresis and hybridized with the radioactively labeled 1.2-kb PCR product of the p38 gene generated with the primers, f3 (5′-GAGCATCTGAGTTTGACCCCTGGAATCTAC-3′) and r2 (5′-TTGCAGGATGTTGGTTACGTCCAAGTCTGCATCTG-3′). The hybridized bands were detected with a PhosphorImage analyzer. The isolated genomic DNA was also used for PCR analysis with the specific pairs of primers, f6 (5′-GTCACCGAATGTAACTGTGCTGGCTTCGAAG-3′) or pKOf2 (5′-GGTAGGTCCAGAGTCTTCAGAGATCAAGTC-3′) and r6 (5′-CCATAACCCAGGTTGACGGGTTTAGTGCCC-3′). For Northern analysis, total cellular RNA was prepared from MEF cells by using the RNeasy Midi kit (Qiagen, Chatsworth, CA), according to the manufacturer's instructions. The isolated RNAs (30 μg) were separated by electrophoresis in 1.2% agarose/2.2 M formaldehyde gels, transferred to Hybond N membranes (Amersham Pharmacia), and then fixed to the membrane by UV irradiation. The RNAs on the membrane were hybridized with the specific probes generated by PCR of the p38 cDNA by using the ExpressHyb hybridization solution (CLONTECH). The proteins were extracted from MEF cells, separated by SDS/PAGE, and subjected to Western blotting with an anti-p38 antibody, as described (8, 10).
Quantitative Reverse Transcription–PCR (RT-PCR).
The transcript levels of the components for the multi-ARS complex were determined by quantitative RT-PCR. The total RNAs were isolated from the MEF cells of the wild-type and mutant mice and then converted to cDNAs by using M-MLV reverse transcriptase (GIBCO/BRL) and an anchored oligo-dT primer set. The resulting cDNAs were used as templates for PCR amplification with specific primer pairs (sequences provided on request) specific to the cDNA sequences of MRS, QRS, RRS, p43, p38, and p18. The PCR reactions of the MRS, QRS, RRS, p43, p38, and p18 cDNAs generate 345-, 600-, 649-, 714-, 890-, and 486-bp fragments, respectively. The reactions were run at 94°C (1 min), 50°C (40 sec), and 72°C (1 min) for 30 cycles, which was within the linear reaction window. The transcript of glyceraldehyde 3 phosphate dehydrogenase (GAPDH) was also quantified by RT-PCR from the same RNA samples and was used as an internal control.
Aminoacylation Activities.
The cultivated MEF cells of the wild-type and mutant mice were harvested, suspended in hypotonic solution (10 mM Hepes, pH 7.6/10 mM KCl/1.5 mM MgCl2/0.5 mM EGTA/10 mM NaF/1 mM PMSF/100 units/ml of aprotinin), and lysed by passage through a 23-gauge syringe. After centrifugation, the supernatant containing the extracted proteins was used to compare the aminoacylation activities of different ARSs. The reaction conditions were optimized for each enzyme, and the activities of the enzymes were determined by using a [3H]-labeled amino acid substrate. [35S] methionine was used for the assay of MRS. For the PRS and IRS assays, 50 mM Hepes buffer (pH 7.4) containing 25 mM MgCl2, 20 mM KCl, 200 μg/ml of BSA, 20 mM β-mercaptoethanol, and 5 mM ATP was used. For the LRS assay, 55 mM Hepes buffer (pH 7.4), containing 30 mM KCl, 200 μg of BSA, 10 mM ATP, 10 mM MgCl2, and 1.2 mM reduced glutathione, was used. For the MRS assay, 30 mM Hepes buffer (pH 7.4) containing 100 mM KOAc, 10 mM Mg(OAc)2, and 100 mM ATP, was used. For the RRS assay, 125 mM Tris buffer (pH 7.4), containing 200 μg/ml of BSA, 5 mM ATP, 4 mM EDTA, and 50 mM MgCl2, was used. For the WRS assay, 50 mM Hepes buffer (pH 7.4), containing 25 mM MgCl2, 20 mM KCl, 200 μg/ml of BSA, 20 mM β-mercaptoethanol, and 5 mM ATP, was used. For the TRS assay, 100 mM Tris buffer (pH 7.4), containing 2 mM ATP, 20 mM β-mercaptoethanol, and 3 mM MgCl2, was used. The charged tRNAs were precipitated by trichloroacetic acid and quantified by liquid scintillation.
Gel Filtration Chromatography.
The proteins extracted from the MEF cells were subjected to sizing chromatography by using Sephacryl S-300 (separation range 10–1,500 kDa) in an AKTA-FPLC system (Amersham Pharmacia), as described previously (8). The separated proteins were then resolved by SDS/PAGE and subjected to Western blotting with the antibodies specific to each of the components.
Coimmunoprecipitation.
Each of the Myc-tagged full length and partial fragments of p38 was expressed in human embryonic kidney 293 cells by transient transfection. The proteins extracted from the lysed cells were mixed with the anti-Myc antibody (9E10, Santa Cruz Biotechnology), bound to Protein A agarose (Sigma), and then precipitated. The components of the multi-ARS complex coprecipitated with each of the p38 fragments were determined by Western blotting with their specific antibodies, as described (8).
Results and Discussion
Generation of the p38 Mutation in Mice.
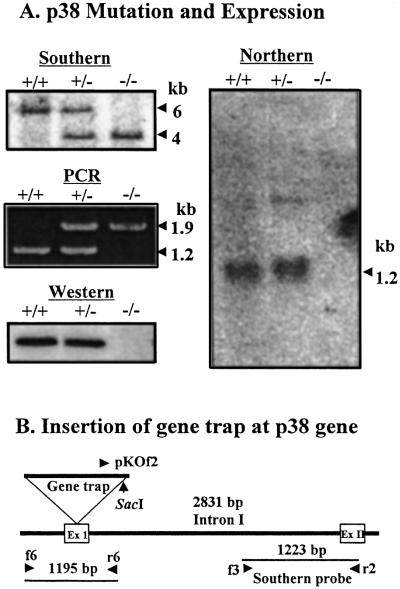
The mutation of the p38 gene was generated by the insertion of the gene trap vector, as described previously (16). The mutation within the structural gene of p38 in mice was determined by Southern blot and PCR analysis of the genomic DNA isolated from mouse embryonic fibroblast cells. Although the wild type generated an approximately 6-kb SacI genomic DNA fragment that hybridized with the p38 specific probe, the p38 homozygous mutant cells yielded about a 4-kb fragment (Fig. 1A Top Left). Because the wild-type and mutant alleles generate PCR fragments of ≈1.2 and 1.9 kb, respectively, the PCR results of the wild-type hetero- and homozygous mutant mice also confirmed the mutation at the p38 gene (Fig. 1A Middle Left). The expression of p38 was then determined by Northern and Western blot analysis. The p38 transcript and protein were not observed in the homozygous mutant cells, as determined by Northern (Fig. 1A Right) and Western blotting (Fig. 1A Bottom Left), respectively. The sequence determination of the mutated p38 allele revealed that the gene trap vector was inserted at the first exon of the p38 gene (Fig. 1B).
Figure 1.
Characterization of p38 mutant mice. (A) The mutation within the p38 gene was determined by Southern blot and PCR analyses. The 1,223-bp PCR fragment was generated from the intron I region of the mouse p38 gene with the primers f3 and r2 and was used as a probe for the hybridization of SacI fragments of mouse genomic DNA. A mixture of the primers, f6, r6, and pKOf2, was used for PCR analysis. The wild-type and mutated alleles generate PCR products of ≈1.2- and 1.9-kb, respectively. The mutational effect on the p38 expression was determined by Northern blotting with the p38 cDNA probe and Western blotting with a polyclonal rabbit antibody raised against the purified human p38. The same amount of actin was detected in each lane of Northern blot (data not shown). The symbols, +/+, +/−, and −/−, represent the wild-type, heterozygous, and homozygous mutant mice. (B) The location of the gene trap insertion was determined by sequencing of the genomic DNA of the mutated allele.
Effect of the p38 Mutation on the Formation of the Multi-tRNA Synthetase Complex.
After confirming the mutation and its effect on the expression of p38 in the homozygous mutant mice, we first investigated how the p38 mutation affects the cellular concentrations of the components for the multi-ARS complex. Fibroblast cells were obtained from 13.5-day embryos of wild-type and homozygous mutant mice. The proteins were extracted from these cells, and the levels of the components for the multi-ARS complex were compared between wild type and mutant by Western blotting with their specific antibodies. The protein levels of the component enzymes, IRS, MRS, and RRS, and the other cofactors, p43 and p18, were significantly decreased in the mutant cells, although the extent of the reduction varied, depending on the protein (Fig. 2A).
Figure 2.
The effect of the p38 mutation on the protein level, catalytic activity, and complex formation of ARSs. (A) Western blot of the components for the multi-ARS complex. The proteins extracted from MEF cells of the wild-type and homozygous mutant mice were subjected to Western blot analysis. The polyclonal rabbit antibodies specific to each of the components were prepared as previously described (8) and were used for Western blotting. Polyclonal rabbit antibodies for human KRS and hamster DRS were kindly provided by K. Shiba (Cancer Institute, Tokyo) and M. Mirande (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France). (B) Comparison of the transcript levels of the components between the wild-type and mutant mice. The isolated total RNAs were converted to cDNAs, and the transcripts of three enzymes (MRS, QRS, and RRS) and three cofactors (p43, p38, and p18) were amplified by PCR with their specific primers as described in the Materials and Methods. GAPDH was used as an internal control. (C) The aminoacylation activities of seven different ARSs (five complex-forming enzymes—PRS, IRS, LRS, MRS, and RRS—and two free-form enzymes—WRS, and TRS) were compared between the wild-type and homozygous mutant MEF cells. The reactions were repeated three times, and the detailed reaction conditions for each enzyme are described in Materials and Methods. (D) The proteins extracted from MEF cells of the wild-type and mutant mice were subjected to gel filtration chromatography. The eluted proteins within each fraction were separated by SDS/PAGE, and each component was detected by Western blotting with the specific antibody.
To see whether the reduction of the component concentrations in the mutant cells resulted from the decreased level of their mRNAs, we compared the transcript levels of several components between wild-type and mutant cells by quantitative RT-PCR with their specific primers. In the mutant cells, no PCR product was detected for the p38 transcript, whereas the same amount of the GAPDH transcript was obtained as the wild-type cells (Fig. 2B). In the cases of MRS, RRS, p43, and p18, in which the protein levels were significantly decreased by the deficiency of p38, their transcript levels were similar both in the mutant and wild-type cells. This result suggests that the reduction of the protein levels in the mutant cells was due to the decreased stability of the component proteins. Then, the aminoacylation activities of the five complex-forming ARSs (PRS, IRS, LRS, MRS, and RRS) and the two free-form enzymes (WRS and TRS) were also compared between the wild-type and mutant cells. All of the five complex-forming enzymes in the mutant cells showed reduced catalytic activities as compared with their corresponding enzymes in the wild type, whereas the activities of the two other ARSs were only slightly affected in the mutant cells (Fig. 2C; see supporting information on the PNAS web site, www.pnas.org). The protein levels of WRS and TRS were similar between the wild-type and mutant cells (data not shown). The reduction of the PRS and LRS activities is interesting, because their cellular concentrations were not significantly decreased in the mutant cells (Fig. 2A). In these cases, the complex formation may help to maintain their efficient catalytic activities.
We then analyzed how the p38 mutation affected the complex formation of the multi-ARS complex by sizing chromatography. The components bound to the intact macromolecular ARS complex would elute in the void volume of the gel filtration column, whereas the dissociated components would be detected in the following fractions according to their molecular sizes. The components of the complex from the wild-type cells were coeluted in the void volume, as expected (Fig. 2D Upper). However, none of the components in the p38 mutant cells were detected in the void volume. Instead, they were either absent or detected in the later fractions (Fig. 2D Lower). This result indicates that the complex was completely disintegrated into individual proteins in the p38 mutant cells.
Deletion Mapping with p38.
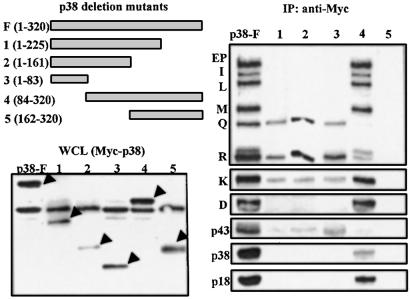
The above results suggest that p38 serves as a template on which other components are assembled. We thus thought that the structural organization of the components with respect to p38 could be determined by using deletion fragments of p38. The Myc-tagged full or partial fragments of p38 were expressed in human embryonic kidney 293 cells (Fig. 3 Left) and were immunoprecipitated with the anti-Myc antibody. The components of the multi-ARS complex that coprecipitated with each of the fragments were determined by Western blotting with their specific antibodies. The QRS, RRS, and p43 proteins coprecipitated with the N-terminal 83-aa peptide of p38 (Fig. 3). The longer N-terminal fragments (the N-terminal 1–161 and 1–225 amino acids) did not pull down any additional components, indicating that the N-terminal 83-aa peptide is sufficient for the association of QRS, RRS, and p43. The rest of the components were precipitated with the C-terminal 225-aa peptide (84–320) but not with the shorter peptide (162–320) (Fig. 3). Thus, the central peptide, from 84 to 225, does not seem necessary for the anchoring of p43, RRS, and QRS, but is needed to hold the rest of the components. Although the majority of KRS was precipitated with the C-terminal part of p38, its small portion was also detected with the components precipitated with the N-terminal domain of p38. It is known that KRS makes a direct interaction with p38 (15) and is also associated with RRS (7). Thus, KRS appears to be linked to both domains of p38. The endogenous p38 was precipitated with either the exogenous full-length or the C-terminal peptide of p38, suggesting that it may form a homodimer via the interaction of its C-terminal domain. This result is consistent with the previous report (2).
Figure 3.
Coimmunoprecipitation of the components of the multi-ARS complex with the peptide fragments of p38. The Myc-tagged full and partial fragments of p38 (Upper Left) were expressed in 293 cells. The expression of the deletion fragments of p38 was confirmed by Western analysis of the whole cell lysates (WCL) with an anti-Myc antibody (arrows, Lower Left). These fragments were immunoprecipitated with the anti-Myc antibody, and the coprecipitated components were determined by Western blotting with their specific antibodies, as above (Right).
Arrangement of the Components with Respect to p38.
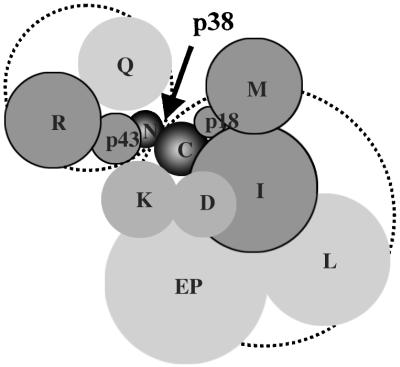
On the basis of the data obtained from this study and previous work, the components of the complex can be grouped within two subdomains by their association with p38 (Fig. 4). One subdomain would consist of p43, RRS, and QRS anchored to the N-terminal region of p38. In this domain, the interactions of p43-RRS (10) and p43-p38 (2) were previously shown, and the weak interactions of QRS and RRS with p38 were also reported (15). Among these components, RRS and p43 depend on p38 for their protein stability (Fig. 2A). The other components, EPRS, IRS, LRS, MRS, KRS, DRS, and p18, appear to be directly or indirectly associated with the C-terminal 236-aa peptide of p38. KRS is also linked to the N-terminal domain of p38 in part (Fig. 3). Although KRS and DRS are strongly and directly associated with p38 (15), they do not seem to depend on p38 for the maintenance of their cellular concentration (Fig. 2 A and B). The interaction between p18 and MRS was previously shown (2). EPRS, IRS, and LRS were proposed to form one subcomplex (17). MRS, IRS, and p18 require p38 to maintain their protein levels (Fig. 2A). All of these in vivo and in vitro data clearly suggest that p38 is a critical factor for the assembly of the multi-ARS complex, and that the components are organized by using p38 as a template. It is interesting that the assembly and stability of a multicomponent protein complex are controlled by a single critical component.
Figure 4.
Schematic arrangement of the components of the multi-ARS complex with respect to p38. The components are grouped into two subdomains by their association with p38 (dotted circles). One subcomplex, consisting of RRS, QRS, and p43, is anchored to the N-terminal 83-aa peptide of p38, whereas the others are associated with the C-terminal 237-aa peptide of p38. Although KRS is assembled to the complex mainly via its association with the C-terminal domain of p38, it is also partly linked to the subdomain anchored to the N-terminal part of p38 (see Fig. 3). Five components (p43, p18, RRS, MRS, and IRS) depend on p38 for their stability (dark gray). Among these five components, RRS and MRS make tight pairs with p43 and p18, respectively (refs. 2 and 10 and unpublished data). The two-dimensional display of the components does not necessarily reflect their relative positions in the multi-ARS complex. The dimer formation of some components was not taken into account, for simplicity.
Functional Implications.
Intercrosses of the p38 heterozygous mutant mice generated offspring of the wild-type, heterozygous, and homozygous mutant mice at a ratio similar to the Mendelian segregation. This result suggests no embryonic lethality of the mutants, and the mutant mice were born at normal shape and size. However, the homozygous mutant mice died within 2 days of birth, implying the physiological significance of p38 in the postnatal stage. Because the p38 mutation affected multiple proteins, as shown in this work, the direct cause for the lethality of the mutant mice at the postnatal stage and the reason why the mutation did not significantly damage the mutant cells and embryos are not clear at this moment. The answers to these questions require further investigation of the mutant mice. Here we focused only on the functional significance of p38 in terms of the formation of the multi-tRNA synthetase complex. Our results have clearly proved the critical role of p38 in the assembly and stability of this complex and provide an insight into the structural organization of the component enzymes and other associated factors.
Supplementary Material
Acknowledgments
We thank Dr. Han Woong Lee for technical assistance with the mouse analyses. This work was supported by a grant from the National Creative Research Initiatives from the Ministry of Science and Technology, Korea.
Abbreviations
- ARSs
aminoacyl-tRNA synthetases
- XRS
the aminoacyl-tRNA synthetase of the substrate amino acid X
- MEF
mouse embryonic fibroblast
- RT-PCR
reverse transcription–PCR
- GAPDH
glyceraldehyde 3 phosphate dehydrogenase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Quevillon S, Agou F, Robinson J-C, Mirande M. J Biol Chem. 1997;272:32573–32579. doi: 10.1074/jbc.272.51.32573. [DOI] [PubMed] [Google Scholar]
- 2.Quevillon S, Robinson J-C, Berthonneau E, Siatecka M, Mirande M. J Mol Biol. 1999;285:183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- 3.Quevillon S, Mirande M. FEBS Lett. 1996;395:63–67. doi: 10.1016/0014-5793(96)01005-8. [DOI] [PubMed] [Google Scholar]
- 4.Kang J, Kim T, Ko Y G, Rho S B, Park S G, Kim M J, Kwon H J, Kim S. J Biol Chem. 2000;275:31682–31688. doi: 10.1074/jbc.M909965199. [DOI] [PubMed] [Google Scholar]
- 5.Rho S B, Lee K H, Kim J W, Shiba K, Jo Y J, Kim S. Proc Natl Acad Sci USA. 1996;93:10128–10133. doi: 10.1073/pnas.93.19.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rho S B, Lee J S, Jeong E J, Kim K S, Kim Y G, Kim S. J Biol Chem. 1998;273:11267–11273. doi: 10.1074/jbc.273.18.11267. [DOI] [PubMed] [Google Scholar]
- 7.Rho S B, Kim M J, Lee J S, Seol W, Motegi H, Kim S, Shiba K. Proc Natl Acad Sci USA. 1999;96:4488–4493. doi: 10.1073/pnas.96.8.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim T, Park S G, Kim J E, Seol W, Ko Y G, Kim S. J Biol Chem. 2000;275:21768–21772. doi: 10.1074/jbc.M002404200. [DOI] [PubMed] [Google Scholar]
- 9.Norcum M T, Warrington J A. J Biol Chem. 2000;275:17921–17924. doi: 10.1074/jbc.C000266200. [DOI] [PubMed] [Google Scholar]
- 10.Park S G, Jung K H, Lee J S, Jo Y J, Motegi H, Kim S, Shiba K. J Biol Chem. 1999;274:16673–16676. doi: 10.1074/jbc.274.24.16673. [DOI] [PubMed] [Google Scholar]
- 11.Kim Y, Shin J, Li R, Cheong C, Kim K, Kim S. J Biol Chem. 2000;275:27062–27068. doi: 10.1074/jbc.C000216200. [DOI] [PubMed] [Google Scholar]
- 12.Renault L, Kerjan P, Pasqualato S, Menetrey J, Robinson J C, Kawaguchi S, Vassylyev D G, Yokoyama S, Mirande M, Cherfils J. EMBO J. 2001;20:570–578. doi: 10.1093/emboj/20.3.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko Y-G, Park H, Kim T, Lee J-W, Park S G, Seol W, Kim J E, Lee W-H, Kim S-H, Park J E, Kim S. J Biol Chem. 2001;276:23028–32303. doi: 10.1074/jbc.M101544200. [DOI] [PubMed] [Google Scholar]
- 14.Park H, Park S G, Lee J-W, Kim T, Kim G, Ko Y-G, Kim S. J Leukocyte Biol. 2002;71:223–230. [PubMed] [Google Scholar]
- 15.Robinson J C, Kerjan P, Mirande M. J Mol Biol. 2000;304:989–994. doi: 10.1006/jmbi.2000.4242. [DOI] [PubMed] [Google Scholar]
- 16.Zambrowicz B P, Friedrich G A, Buxton E C, Lilleberg S L, Person C, Sands A T. Nature (London) 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 17.Norcum M T, Warrington J A. Protein Sci. 1998;7:79–87. doi: 10.1002/pro.5560070108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.