Abstract
The RecA protein of Escherichia coli, and all filament-forming homologues identified to date, promote DNA strand exchange by a common, ordered pathway. A filament is first formed on single-stranded DNA, followed by uptake of the duplex substrate. These proteins are thereby targeted to single-strand gaps and tails where recombinational DNA repair is required. The observed course of DNA strand exchange promoted by the RecA protein from the extremely radioresistant bacterium Deinococcus radiodurans is the exact inverse of this established pathway. This reaction lies at the heart of a remarkably efficient system for the repair of DNA damage.
Deinococcus radiodurans (Dr) is part of a small family of seven described bacterial species that are the most radiation-resistant organisms known (1–3). Although this organism efficiently repairs many types of severe DNA damage (3), resistance to ionizing radiation is especially remarkable. Dr is able to survive exposures to gamma radiation in excess of 1.7 Mrads without lethality or induced mutation (4) and can survive hundreds of irradiation-induced DNA double-stranded breaks per haploid genome (4, 5). The mechanisms underlying this organism's very efficient DNA repair seem to be complex but remain poorly understood (6).
Proteins required for recombinational DNA repair in other bacteria have become a focus of attention in Deinococcus. A functional Dr RecA protein is required for expression of the resistance phenotypes of Dr (7). Interestingly, the Dr genome does not include genes for homologues of the Escherichia coli (Ec) RecB and RecC proteins (6). In Ec, these proteins are part of the RecBCD enzyme, which processes duplex DNA ends to produce a single-stranded DNA (ssDNA) tail to which RecA protein is bound (8, 9). RecBCD plays a critical role in recombinational processes in many bacteria. The Dr RecA protein is normally expressed at very low levels in Dr cells, and only transiently at high levels after exposure to heavy DNA damage (10, 11). The ssDNA binding (SSB) gene of Dr is apparently disrupted, containing segments in different reading frames (6). This situation has delayed the accurate cloning of the Dr SSB protein. The Ec SSB is nevertheless highly stimulatory in Dr RecA-mediated DNA strand exchange (10), and the Ec SSB is used in these experiments.
Expression of the Dr RecA protein in Ec seems to be toxic (3, 11, 12). Nevertheless, the Dr RecA protein has been successfully expressed in Ec and purified from that source (10). In many respects, the Dr RecA protein is similar to other bacterial RecA proteins. It forms helical filaments on DNA, hydrolyzes ATP and dATP, and promotes DNA strand exchange (10). Its binding to double-stranded DNA (dsDNA) is more facile than that observed for the Ec RecA protein. The Dr RecA protein hydrolyzes dATP faster than ATP, and dATP facilitates its displacement of Ec SSB protein from ssDNA. However, the hydrolysis of dATP is not coupled well to DNA strand exchange. For example, the Dr RecA protein promotes DNA strand exchange through heterologous insertions in the presence of ATP, but very poorly when dATP is hydrolyzed (10). Further, when both ss- and dsDNA are present in a solution, the Dr RecA protein binds preferentially to the dsDNA (10). This property is in stark contrast to the DNA-binding properties of the Ec RecA protein, and led us to examine the pathway for Dr RecA-mediated DNA strand exchange more closely.
Materials and Methods
Enzymes and Reagents.
The Ec RecA protein was purified as described (13). Its concentration was determined by absorbance at 280 nm with the extinction coefficient ɛ280 = 0.59 A280 mg−1 ml (14). The native Dr RecA protein was purified as described (10) and its concentration determined with the extinction coefficient ɛ280 = 0.372 A280 mg−1 ml (10). Ec SSB was purified as described (15) and stored frozen at −70°C in a buffer containing 20 mM Tris⋅HCl (40% cation, pH 8.4), 0.15 M NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, and 50% glycerol. The concentration of SSB was determined with an extinction coefficient of ɛ280 = 1.5 A280 mg−1ml (16). Restriction endonucleases were purchased from New England Biolabs. ATPγS, Tris buffer, creatine phosphokinase, and phosphocreatine were purchased from Roche Molecular Biochemicals. Phosphoenolpyruvate, pyruvate kinase, lactic dehydrogenase, NADH, ATP, and dATP were purchased from Sigma.
DNA.
Circular duplex and ssDNA from M13mp8 were prepared with described methods (13). Circular ss, supercoiled ds, and nicked circular φX174 DNAs were purchased from New England Biolabs. The concentrations of ss- and dsDNA were determined by absorbance, with A260 nm = 1 to be equivalent to 36 and 50 μg ml−1, respectively. Molar concentrations of DNA are given in terms of total nucleotides. Linear duplex φX174 DNA was prepared by digestion of supercoiled DNA with PstI endonuclease. M13mp8 dsDNA was linearized by digestion with EcoRI. The 1.9-kbp linear fragment of φX174 DNA (1,957 bp) was prepared by cleavage of circular duplex φX174 DNA with NarI restriction endonuclease, followed by purification of the smaller fragment on an agarose gel.
DNA Strand Exchange Reactions.
The RecA-dependent DNA strand exchange reaction was carried out as described (17, 18) between circular ssDNA and the linear dsDNA (derived from either φX174 or M13mp8). Unless otherwise stated, all reactions were carried out at 37°C in solutions containing 25 mM Tris-acetate (80% cation or pH 7.5 for Ec RecA protein; 50% cation or pH 8.1 for Dr RecA protein reactions), 1 mM DTT, 5% glycerol, 3 mM potassium glutamate, 10 mM magnesium acetate, and an ATP-regenerating system (10 units/ml of pyruvate kinase/3.3 mM phosphoenolpyruvate or 10 units/ml creatine kinase/12 mM phosphocreatine). DNA, SSB, ATP (or dATP), and Ec RecA or Dr RecA protein concentrations are indicated for each experiment. The standard protocol (modified as described in figure legends) began with a preincubation of ssDNA with Dr RecA protein at 37°C for 5 min. This preincubation was followed by addition of ATP and SSB. After an additional 5-min incubation, linear duplex DNA was added to start the DNA strand exchange reactions. Aliquots (20 μl unless otherwise indicated) of the strand exchange reactions were removed at each time point, and the reactions were stopped by addition of 5 μl of gel loading buffer (0.125% bromophenol blue/25 mM EDTA/25% glycerol/5% SDS). These aliquots were stored on ice until after the last time point was taken. Samples were electrophoresed in a 0.8% agarose gel with TAE buffer (19), stained with GelStar nucleic acid gel stain (FMC), or ethidium bromide photographed with UV light using a NucleoVision (NucleoTech, San Mateo, CA) gel documentation camera. The DNA bands were quantified with imagequant software (version 4.2; Molecular Probes). To correct for variability in sample loading onto the agarose gel, the band corresponding to full-length circular hybrid duplex product was quantified as the fraction of the total fluorescing DNA in a given gel lane, excluding only the band corresponding to the ssDNA.
Results
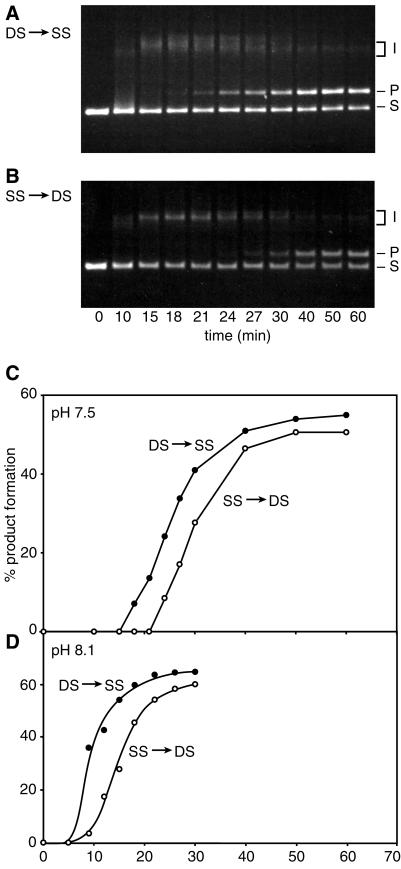
The first clue that the Dr RecA protein promoted DNA strand exchange in an unusual manner came in order of addition experiments as shown in Fig. 1. Full-length nicked circular DNA products of DNA strand exchange appeared nearly 10 min earlier when the Dr RecA protein was preincubated with dsDNA, as opposed to the standard protocol featuring preincubation with ssDNA. Reactions at pH 7.5 are shown in Fig. 1 A and B, and quantified in C. A similar phenomenon is observed at pH 8.1 (Fig. 1D). Both sets of conditions are within the optimal pH range for Dr RecA protein reactions in the presence of ATP (10).
Figure 1.
Preincubation of Dr RecA protein with dsDNA leads to faster production of DNA strand exchange products. Reactions contained 2 μM Dr RecA protein, 6 μM circular ss M13mp8 DNA, 12 μM linear duplex M13mp8 DNA (PstI-cut), 0.6 μM Ec SSB, 2 mM ATP, and an ATP-regenerating system. Reactions in A–C were carried out at pH 7.5, whereas those in D were done at pH 8.1. In the DS → SS reactions, the Dr RecA protein was preincubated with the linear dsDNA with the ATP for 40 min. The ssDNA was then added to start the reaction, and the SSB was added 5 min later. In the SS → DS reactions, the Dr RecA protein was preincubated with the ssDNA for 5 min, followed by the addition of SSB and ATP, with the reaction initiated after another 5 min by addition of the linear dsDNA. The labels S, I, and P denote the linear duplex DNA, the joint molecule reaction intermediates, and the nicked circular products of DNA strand exchange, respectively. The reaction itself is illustrated in Fig. 4.
The design of the experiment in Fig. 1 reflects the characterization of Dr RecA protein reported elsewhere (10). Although the Dr RecA protein binds preferentially to dsDNA when both ssDNA and dsDNA are present in the solution (10), direct binding to dsDNA is not fast. There is a 15–20-min lag observed in the binding of the Dr RecA protein to dsDNA under most conditions (10). Direct binding to ssDNA is actually much faster than binding to dsDNA, and for this reason the preincubation times in Fig. 1 are longer in the experiments where dsDNA was included in the preincubation mix. We note that when both ssDNA and dsDNA are present (with the dsDNA in excess relative to the Dr RecA protein), the ssDNA seems to potentiate binding to dsDNA. The Dr RecA ends up on the dsDNA, but the lag is largely abolished (ref. 10, and J.-I.K. and M.M.C., unpublished data).
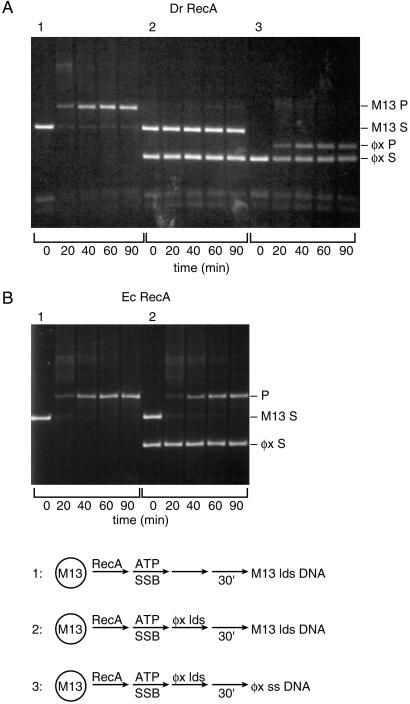
To explore further the Dr RecA protein-mediated strand exchange pathway, a series of challenge experiments were carried out in which heterologous dsDNA was added to the reaction before the homologous dsDNA. Thus, if the Dr RecA is transferred to the challenging dsDNA, a subsequent strand exchange will be blocked. In Fig. 2A, a normal DNA strand exchange reaction is shown, with DNA substrates derived from bacteriophage M13mp8. If dsDNA from bacteriophage φX174 is added before the M13mp8 dsDNA, the reaction is almost completely inhibited (Fig. 2A, reaction 2). We interpret this inhibition to indicate that much of the Dr RecA protein has been sequestered on the φX174 dsDNA. If, after addition of the φX174 dsDNA, ssDNA from φX174 is added instead of the dsDNA from M13mp8, a strong reaction is restored—but now it is a reaction between the φX174 DNA substrates (Fig. 2A, reaction 3).
Figure 2.
Heterologous dsDNA inhibits the DNA strand exchange reactions promoted by Dr RecA protein. Reactions contained 2 μM Dr RecA protein, 6 μM circular ss M13mp8 DNA, 12 μM linear duplex M13mp8 DNA (or, where indicated, the same amount of φX174 linear dsDNA), 0.6 μM Ec SSB, 2 mM ATP, and an ATP-regenerating system. Reactions were done at pH 8.1. Reactions were (1) normal DNA strand exchange reaction with M13mp8 DNA substrates. Dr RecA protein was preincubated with the ssDNA for 5 min, ATP and SSB were added, and the preincubation continued for 30 min, and the reaction was initiated by addition of the dsDNA. (2) Same protocol as reaction 1, but the φX174 linear dsDNA was added 5 min after the ATP and SSB (30 min before the M13mp8 dsDNA). (3) Same protocol as reaction 2, except that 6 μM φX174 circular ssDNA replaced the M13mp8 dsDNA as the final addition. A and B show reactions with the Dr and Ec RecA proteins, respectively. The labels S and P denote the substrates (linear duplex DNA) and products (nicked circular DNA), respectively, derived from M13mp8 (M13) or φX174 (φX) as indicated.
The properties of the Dr and Ec RecA proteins contrast strikingly in this experiment. In a similar trial with the Ec RecA protein, no detectable inhibition of DNA strand exchange is observed because of addition of φX174 dsDNA before the M13mp8 dsDNA (Fig. 2B). The Ec RecA protein remains on the ssDNA and reacts with the M13mp8 dsDNA despite the heterologous dsDNA challenge.
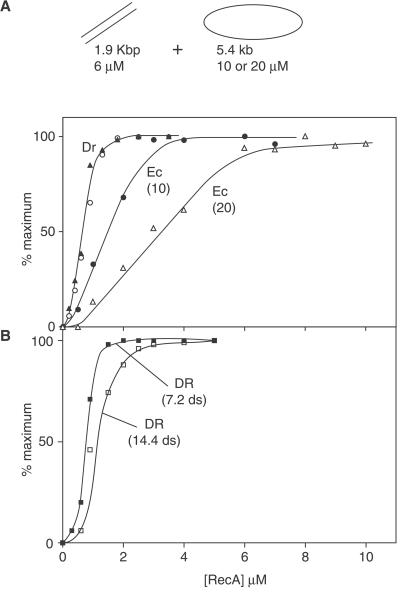
We next set up a DNA strand exchange reaction with a short (1.9 kbp) fragment derived from φX174 DNA. The much longer ssDNA circles and the dsDNA fragments were present at nearly a 1:1 molecular ratio. We reasoned that a maximal DNA strand exchange should require an amount of RecA protein stoichiometric with the DNA on which it initiated DNA strand exchange. The reaction with the Ec RecA protein is maximized when sufficient Ec RecA is present to saturate the binding sites on the ssDNA, as shown in Fig. 3. When the ssDNA concentration is doubled without changing the dsDNA concentration, the amount of Ec RecA required also doubles. The result seen with the Dr RecA protein is much different. The maximum reaction is seen when sufficient Dr RecA protein is present to saturate only the shorter dsDNA. This situation is not altered when the ssDNA concentration is doubled. In contrast, when the duplex DNA concentration is varied, the amount of Dr RecA protein required to obtain the maximum reaction also varies in kind (Fig. 3B). Thus, unlike the Ec RecA protein, the requirements for Dr RecA protein in DNA strand exchange seem to be stoichiometric with the dsDNA substrate.
Figure 3.
Optimal DNA strand exchange requires Dr RecA protein in amounts stoichiometric to the dsDNA. (A) Reactions were carried out under standard reaction conditions and contained either 10 or 20 μM circular ss φX174 DNA, 6 μM linear duplex φX174 DNA [a 1.9-kbp fragment; molecular stoichiometry is 0.83:1 with 10 μM ssDNA, so that the ssDNA is in slight excess and the maximum reaction in all cases is 100% conversion of linear dsDNA (ldsDNA) to gapped DNA (gDNA)], 1.0 μM or 2.0 μM Ec SSB (for 10 or 20 μM ssDNA, respectively), 2 mM ATP, an ATP-regenerating system, and the indicated amount of Dr or Ec RecA protein. The circular and triangular symbols denote Dr RecA reactions with 10 or 20 μM ssDNA, respectively, whereas the Ec RecA reactions are labeled with the ssDNA concentration (in μM) in parentheses. Reactions with Ec or Dr RecA protein were carried out at pH values 7.5 and 8.1, respectively. For all reactions, the RecA proteins were preincubated with ssDNA and ATP for 5 min, followed by addition of SSB and another 5-min preincubation. Reactions were initiated with dsDNA. Reactions were generally completed with these short duplex DNAs in 15 min or less. Reaction extents were determined by averaging 2 or 3 points taken between 20 and 60 min after the reaction was initiated. In no case did averaged individual measurements disagree by more than 10% of the measured value. Each reaction extent was then plotted as a fraction of the maximum extent observed for any of the reactions (seen at one of the higher RecA concentrations). Maximum reactions (conversion of ldsDNA to gDNA, expressed as gDNA/gDNA + ldsDNA + detectable intermediates) were 60% and 80% for the Dr RecA reactions at 10 and 20 μM ssDNA, respectively. The same reactions for Ec RecA protein produced a maximum of 75% and 60% conversion to product, respectively. (B) Reactions were identical to those in A, except that all reactions contained Dr RecA protein, the concentration of ssDNA was 10 μM, and the concentration of the dsDNA fragment was either 7.2 (■) or 14.4 (□) μM. The ssDNA and dsDNA are present at 1:1 and 1:2 molecular ratios, respectively. The maximum reactions for these experiments were 46% and 29% for the 7.2 and 14.4 μM reactions, respectively, which is 46% and 58% of the theoretical maximums.
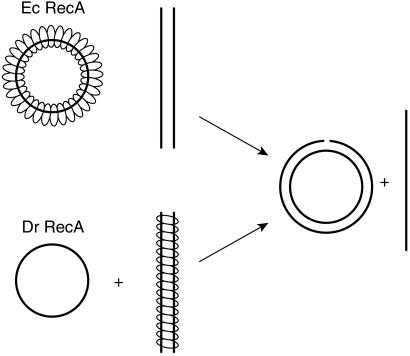
The conclusion that the Dr RecA protein promotes DNA strand exchange via a pathway that is the inverse of that used by other recombinases in this class (Fig. 4) is thus supported by three separate observations. (i) When Dr RecA protein is preincubated with dsDNA, the lag observed before products seen in the subsequent DNA strand exchange is shorter than the lag observed after preincubation with ssDNA. (ii) Heterologous dsDNA, introduced before the homologous dsDNA, strongly inhibits the subsequent DNA strand exchange. The same heterologous DNA has no effect on reactions of Ec RecA protein. We interpret this finding to mean that the Dr RecA protein is being sequestered on the heterologous dsDNA. (iii) In a DNA strand exchange reaction between a short linear dsDNA and a long circular ssDNA, the amount of Dr RecA protein required for optimal reaction is stoichiometric with the duplex DNA and is unaffected by changes in ssDNA concentration. In contrast, the Ec RecA reactions exhibit a stoichiometric relationship to the ssDNA.
Figure 4.
Normal vs. inverse DNA strand exchange. In the normal reaction pathway promoted by most RecA family proteins, RecA protein forms a filament on ssDNA first and then takes up a homologous duplex for DNA strand exchange. The Dr RecA protein forms a filament first on the dsDNA and then takes up a homologous single strand to initiate the same reaction. This latter path has been defined as inverse DNA strand exchange to avoid confusion of terms used to describe other directional properties of the reaction (20), although this may be considered the forward and normal path for the Dr RecA protein.
Discussion
We conclude that the Ec and Dr RecA proteins promote DNA strand exchange by quite different pathways. Unlike the Ec RecA protein, the Dr RecA protein initiates DNA strand exchange with a filament bound to the dsDNA. Although this is clearly the major strand exchange pathway (and thus the forward path) for the Dr RecA protein under all of the conditions we have examined to date, this situation is thus far unprecedented among the reactions promoted by bacterial RecA proteins and their archaeal and eukaryotic homologues. The Dr RecA-mediated pathway is the inverse of the major pathway seen with other proteins of the RecA family.
The Ec RecA protein will promote a limited inverse DNA strand exchange with short oligonucleotides if its binding to short segments of dsDNA is stabilized by the presence of a long contiguous ssDNA (20). Inverse DNA strand exchange is essentially the reverse of the standard reaction, although the optimal conditions for the standard and the much weaker inverse reaction for Ec RecA are somewhat different (20). However, the DNA strand exchange reactions promoted by the Ec RecA protein proceed overwhelmingly by the standard pathway. The strict ordered binding of an ssDNA substrate before a dsDNA has biological importance, helping to ensure that the Ec RecA protein does not bind at random to chromosomal DNA. Instead, nucleation of RecA filament formation is directed to gaps present at stalled replication forks (21, 22) or to DNA ends processed by the RecBCD enzyme (8, 9). The Dr RecA protein has apparently evolved to carry out strand exchange predominantly in the inverse manner. The significance of inverse DNA strand exchange may be related to the absence of the recBC genes in the Dr genome. When subjected to high levels of radiation, the Deinococcus genome is reduced to fragments. RecA protein may play a role in finding overlapping fragments and splicing them together. RecBC is not needed to prepare ss tails, because the Dr RecA protein can bind directly to the dsDNA and promote the repair of ds breaks. When combined with nuclease and polymerase activities, DNA fragments could be rapidly spliced together in this way to reconstruct the bacterial chromosomes. Within this scenario, there may be a mechanism to direct the Dr RecA protein to the ends of duplex DNA. It is not yet clear whether the Dr RecA protein itself exhibits a specificity for DNA ends or whether another protein is required for this function.
Even though the Dr RecA protein initiates DNA strand exchange while bound to the dsDNA, an SSB protein (in this case the Ec SSB) has a very large stimulatory effect on the reaction (10). For the Ec RecA protein, and related recombinases like the eukaryotic Rad51 protein, an ssDNA binding protein (bacterial SSB or eukaryotic RPA) is required to remove secondary structure in the ssDNA and allow complete recombinase filament formation (23–25). The bacterial SSB, and possibly the eukaryotic RPA, also has a postsynaptic role in the reaction, binding to the displaced single strand (24, 25). In the case of the DNA strand exchange promoted by Dr RecA protein, the postsynaptic function of SSB must be critical to the observed stimulation of strand exchange, as there is no evident role of SSB in establishing an active Dr RecA filament on dsDNA.
In Ec, the primary function of RecA protein seems to be its action in the recombinational DNA repair of stalled replication forks (26–28). The Dr RecA protein seems poorly suited to such a role. Instead, the protein seems to have evolved to ameliorate the effects of chromosomal breaks, a byproduct of life in extreme environments. In particular, the Dr RecA protein seems optimized to promote a key step in the repair of ds breaks. This unusual specialization at the apparent expense of other roles may reflect the existence of a particularly facile path for double-stranded break repair in Dr.
Acknowledgments
This work was supported by National Institutes of Health Grant GM32335.
Abbreviations
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
References
- 1.Battista J R, Earl A M, Park M J. Trends Microbiol. 1999;7:362–365. doi: 10.1016/s0966-842x(99)01566-8. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira A C, Nobre M F, Rainey F A, Silva M T, Wait R, Burghardt J, Chung A P, da Costa M S. Int J Syst Bacteriol. 1997;47:939–947. doi: 10.1099/00207713-47-4-939. [DOI] [PubMed] [Google Scholar]
- 3.Minton K W. Mol Microbiol. 1994;13:9–15. doi: 10.1111/j.1365-2958.1994.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 4.Daly M J, Ouyang L, Fuchs P, Minton K W. J Bacteriol. 1994;176:3508–3517. doi: 10.1128/jb.176.12.3508-3517.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin J Y, Qi R, Aston C, Jing J P, Anantharaman T S, Mishra B, White O, Daly M J, Minton K W, Venter J C, Schwartz D C. Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 6.Makarova K S, Aravind L, Wolf Y I, Tatusov R L, Minton K W, Koonin E V, Daly M J. Microbiol Mol Biol Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minton K W. Mutat Res. 1996;363:1–7. doi: 10.1016/0921-8777(95)00014-3. [DOI] [PubMed] [Google Scholar]
- 8.Anderson D G, Kowalczykowski S C. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 9.Churchill J J, Anderson D G, Kowalczykowski S C. Genes Dev. 1999;13:901–911. doi: 10.1101/gad.13.7.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J-I, Sharma A K, Abbott S N, Wood E A, Dwyer D W, Jambura A, Minton K W, Inman R B, Daly M J, Cox M M. J Bacteriol. 2002;184:1649–1660. doi: 10.1128/JB.184.6.1649-1660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll J D, Daly M J, Minton K W. J Bacteriol. 1996;178:130–135. doi: 10.1128/jb.178.1.130-135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutman P D, Carroll J D, Masters C I, Minton K W. Gene. 1994;141:31–37. doi: 10.1016/0378-1119(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 13.Shan Q, Cox M M, Inman R B. J Biol Chem. 1996;271:5712–5724. doi: 10.1074/jbc.271.10.5712. [DOI] [PubMed] [Google Scholar]
- 14.Craig N L, Roberts J W. J Biol Chem. 1981;256:8039–8044. [PubMed] [Google Scholar]
- 15.Lohman T M, Green J M, Beyer R S. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 16.Lohman T M, Overman L B. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 17.Cox M M, Lehman I R. Proc Natl Acad Sci USA. 1981;78:3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bedale W A, Cox M. J Biol Chem. 1996;271:5725–5732. doi: 10.1074/jbc.271.10.5725. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 20.Zaitsev E N, Kowalczykowski S C. Genes Dev. 2000;14:740–749. [PMC free article] [PubMed] [Google Scholar]
- 21.Cordeiro-Stone M, Makhov A M, Zaritskaya L S, Griffith J D. J Mol Biol. 1999;289:1207–1218. doi: 10.1006/jmbi.1999.2847. [DOI] [PubMed] [Google Scholar]
- 22.Robu M E, Inman R B, Cox M M. Proc Natl Acad Sci USA. 2001;98:8211–8218. doi: 10.1073/pnas.131022698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kowalczykowski S C, Krupp R A. J Mol Biol. 1987;193:97–113. doi: 10.1016/0022-2836(87)90630-9. [DOI] [PubMed] [Google Scholar]
- 24.Lavery P E, Kowalczykowski S C. J Biol Chem. 1992;267:9315–9320. [PubMed] [Google Scholar]
- 25.Sugiyama T, Zaitseva E M, Kowalczykowski S C. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 26.Cox M M, Goodman M F, Kreuzer K N, Sherratt D J, Sandler S J, Marians K J. Nature (London) 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 27.Cox M M. Proc Natl Acad Sci USA. 2001;98:8173–8180. doi: 10.1073/pnas.131004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowalczykowski S C. Trends Biochem Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]