Abstract
Selective receptor modulators, such as the antiprogestin RU486, are known to exhibit partial agonist activities in a cell-type-dependent manner. Employing an in vitro chromatin transcription system that recapitulates progesterone receptor (PR)-mediated transcription in vivo, we have investigated the molecular basis by which the antiprogestin RU486 regulates transcription in a cell-type-specific manner. We have compared the effects of RU486 on PR-dependent transcription in vitro using T47D and HeLa cell nuclear extracts. RU486 exhibits a differential ability to activate transcription within these two cell types. The differential effect on transcription correlates with different ratios of endogenous coactivators/corepressors in these cells. Unlike agonist-bound PR that interacts only with coactivators such as steroid receptor coactivator-1 (SRC-1), RU486-bound PR binds to both coactivator SRC-1 and corepressor silencing mediator for retinoid and thyroid hormone receptor (SMRT) in vitro. Both SRC-1 and SMRT have the capacity to modulate RU486-dependent activity. Moreover, a change in the relative levels of SRC-1 and SMRT contained in our chromatin transcription system modulates agonist/antagonist effects of RU486 on transcription by PR. Our data indicate that the ability of RU486 to activate transcription is modulated by the ratio of coactivators to corepressors and substantiate the important roles of coregulators in the regulation of steroid receptor mediated transactivation in response to selective receptor modulators.
Biological actions of progesterone are mediated primarily by the progesterone receptor (PR), a member of the nuclear receptor superfamily of transcription factors (1–3). In the absence of progesterone, PR exists in a transcriptionally inactive form associated with heat shock proteins (4). On hormone binding, the receptor undergoes a conformational change, resulting in dissociation from heat shock proteins, translocation to the nucleus, and dimerization and binding to progesterone-responsive elements (PREs) within the promoter regions of target genes (1–3). When bound to the PRE, the receptor can modulate target gene transcription by directly contacting components of the transcriptional machinery (5) or indirectly by means of coactivators, such as steroid receptor coactivator-1 (SRC-1) (6) and p300/CBP (CREB-binding protein) (7).
Progesterone antagonists are synthetic pharmaceutical agents that suppress the transcriptional activity of the natural steroid agonist, progesterone (8, 9). They are used in the treatment of a variety of endocrine disorders as well as certain hormone-dependent tumors (8, 9), and exhibit a spectrum of activity ranging from pure antagonists to mixed antagonists. The pure antagonists, such as onapristone (ZK98299), are generally incapable of activating transcription and fully antagonize PR functions. The molecular basis of the effects of ZK98299 remains unclear (10–14). In contrast, mixed antagonists, known as selective progesterone receptor modulators (SPRMs) (15), may stimulate PR action depending on the cell type, the promoter context, and/or the cellular environment of other signaling pathways (16–19). These mixed antagonists include compounds such as RU486 (mifepristone) (8, 9). As a type I antiprogestin (14), it binds PR with high affinity, and the affinity of RU486-bound PR for DNA is indistinguishable from that of agonist-activated PR (8, 9).
A plausible explanation for mixed agonist/antagonist activity of RU486 and other SPRMs arises from the observation that such mixed antagonists/agonists induce a variety of conformational changes within PR (20, 21). In fact, progesterone and RU486 appear to contact noncoincident, but overlapping, amino acids in the ligand-binding domain (LBD) of PR (22–24). The precise manner in which multiple conformations of PR induced by SPRMs affects its agonist or antagonist activity is open to question. Recent data suggest that the altered conformation in the LBD of PR induced by antagonists impairs the ability of receptors to interact with coactivators (6), perhaps allowing the recruitment of corepressors (25–28), such as the nuclear receptor corepressor (NCoR) (29) and the silencing mediator for retinoid and thyroid hormone receptor (SMRT) (30). NCoR and SMRT are two closely related proteins, which were initially identified as corepressors for unliganded thyroid and retinoid receptors, but their potential roles as corepressors have since been extended to numerous transcription factors, including steroid receptors (31). These findings led us to attempt to directly assess the hypothesis that SPRM-bound PR interacts with both coactivators and corepressors, and that the relative amounts of coactivator and corepressor within a given cell type determine the partial agonist or antagonist activity of a SPRM.
In the course of our investigation of SPRM effects, we observed that RU486 acted as an agonist in T47D cells and as an antagonist in HeLa cells. In an attempt to understand the molecular basis of the differential affects of RU486 on transcription in various cell types, we used a cell free biochemical approach. We manipulated the concentrations of coactivator and corepressor present in a cell-free chromatin transcription system, and analyzed RU486 action under varying coactivator/corepressor ratios. The unique feature of our in vitro study is that we are able to analyze the “direct effects” of different coregulators on the mixed antagonist/agonist actions of RU486 in the same controlled biochemical environment and in the absence of cellular squelching and transduction pathway alterations that might occur using cell transfection approaches. Our results provide direct evidence to substantiate the hypothesis that the relative amount of coactivator and corepressor is a primary modulator of RU486-dependent PR activity, ranging from transcriptional activation to repression.
Methods
Purification of Recombinant Proteins.
Purification of His-tagged human PR B isoform (PRB) and full-length Flag-tagged SRC-1 was described (32). His-tagged human SMRT was expressed in Sf9 cells by using baculovirus and purified by using Ni-NTA affinity resin (Qiagen) as specified by the manufacturer.
Chromatin Assembly and in Vitro Transcription.
Nucleosomal arrays were assembled on the plasmid pPRE3-E4 (32) DNA with assembly extracts derived from Drosophila embryos (32). PRB, ligand, and/or coregulators such as SRC-1 and SMRT were added after the chromatin assembly was complete. The reaction mixtures were incubated for an additional 30 min to allow interactions of these proteins with the chromatin templates as well as formation of proper complex among these proteins. In vitro transcription reactions were performed essentially as described (32). Chromatin template (100 ng) was incubated at room temperature with HeLa cell nuclear extract (20 μg) for 30 min to allow the formation of transcription preinitiation complexes. Subsequently, transcription was initiated by the addition of rNTPs (0.5 mM final), and the templates were transcribed for 1 h at 30°C. The resulting transcripts were detected by primer extension analysis. All experiments were performed at least three times to ensure reproducibility. Quantitation of the data were carried out by a PhosphorImager (Molecular Dynamics).
Cell Culture and Transient Transfections.
T47D cells were maintained in RPMI medium 1640 (Life Technologies, Rockville, MA) containing 10% FBS. HeLa cells were maintained in DMEM (Life Technologies) supplemented with 10% FBS. Twenty-four hours before transfection, 3 × 105 cells were seeded per well of a 6-well dish and cultured in medium containing 5% charcoal-stripped serum. Cells were transfected with the indicated DNAs by using FuGENE 6 transfection reagent (Roche Diagnostics) as specified by the manufacturer. After 6 h, appropriate ligands were added to the culture medium, and the cells were incubated continuously at 37°C for 24 h with RU486 (10−7 M) or progesterone (10−8 M). Luciferase assays were performed as described (32). Triplicate samples were measured in each experiment.
Protein–Protein Interactions.
Assays to determine interactions between PR and SMRT proteins were carried out with baculovirus-infected Sf9 cells. Cells were coinfected with proper amounts of recombinant viruses for PR and SMRT. The cells were incubated with viruses for 3 days at 27°C under the standard growth conditions. After the incubations, infected cells were collected, washed three times with PBS, and homogenized on ice by using a miniDounce in the binding buffer (50 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS/2 mM 2-mercaptoethanol/2 mM phenylmethylsulfonyl fluoride). The homogenates were centrifuged to removed cell debris. The supernatants were incubated with indicated ligands (1 μM) on ice for 30 min, then mixed with anti-PR antibody. After incubation at 4°C for 1 h with rocking, 10 μl of protein A/G-Sepharose beads were added, and incubation was continued for 1 h at 4°C. Subsequently, the beads were washed five times with the binding buffer. Bound proteins were eluted with 2× SDS loading buffer and analyzed by SDS/PAGE and Western blot with anti-SMRT and anti-PR antibodies. To assess interactions between PR and SRC-1, oocyte lysates containing full-length SRC-1 proteins (32) were incubated with anti-FLAG M2 affinity resin in lysis buffer for 2 h at 4°C. Subsequently, the supernatant was removed and resin was mixed with unfractionated Sf9 cell extracts containing His-tagged PR in lysis buffer (20 mM Hepes, pH 7.9/150 mM KCl/20% glycerol/0.5 mM EDTA, pH 8/0.1% Nonidet P-40/2 mM DTT/0.5 mM phenylmethylsulfonyl fluoride) supplemented with vehicle, progesterone (1 μM), or RU486 (1 μM). After incubation for 2 h, resin was washed with lysis buffer, and bound proteins were analyzed by Western blot with anti-PR and anti-SRC-1 antibodies.
Results
RU486 Acts as an Agonist in T47D Cells and an Antagonist in HeLa Cells.
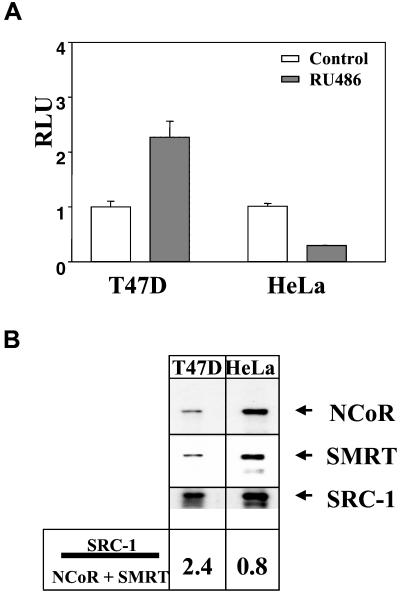
After transfection of a PRE-tk-Luc reporter in the cervical carcinoma HeLa cells or breast cancer T47D cells, we observed that RU486 treatment led to an increase in the reporter gene activity by using T47D cells and a decrease in the reporter gene activity by using HeLa cells (Fig. 1A). Interestingly, we observed that the repressive effect of RU486 in HeLa cells correlated with a high level of expression of the corepressors SMRT and NCoR in these cells compared with T47D cells (Fig. 1B). Moreover, overexpression of SMRT in T47D significantly inhibited RU486-mediated transactivation, whereas SRC-1 overexpression enhanced both RU486- and progesterone-mediated transactivation (data not shown).
Figure 1.
Effect of RU486 in T47D and HeLa cells. (A) RU486 functioned as an agonist in T47D cells, but failed to activate transcription in HeLa cells. T47D cells were transfected with 500 ng of PRE-tk-Luc, whereas HeLa cells were cotransfected with 500 ng of PRE-tk-Luc and 10 ng of PRB. Cells were treated with vehicle (Control) or RU486 for 24 h. Data are presented as the average ± SEM of triplicate experiment. These data are representative of three separate experiments. (B) Expression levels of corepressors were different between T47D and HeLa cells. Nuclear extracts were prepared from T47D and HeLa cells, and identical amounts of proteins concentrations were subjected to SDS/PAGE and Western blotting with various antibodies as indicated. Experiments were performed with three different cell extract preparations with similar results. Quantitation of data were accomplished by using a Molecular Dynamics densitometer.
These observations led us to focus on the hypothesis that RU486-mediated transcription depends on the relative level of expression of coactivators and corepressors present in the two different cell types. To test this hypothesis, we used an in vitro assay that we recently developed (32) to analyze the “direct effects” of the different coregulators on the mixed antagonist/agonist actions of RU486 in a context of the same controlled biochemical environment and in the absence of cellular squelching and transduction pathway alterations.
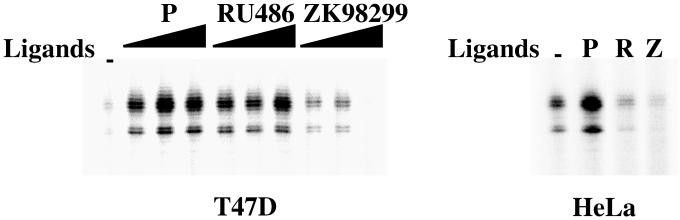
As observed by using tissue culture cells, RU486 induced in vitro transcriptional activation of the reporter gene by using T47D nuclear extracts whereas RU486 failed to enhance PR-dependent transcription by using HeLa nuclear extracts (Fig. 2). Thus, in a context of the identical promoter, RU486 acts as an antagonist in the presence of HeLa cell extract and an agonist in the presence of T47D cell extract. PR transcriptional activity was greatly reduced in the presence of the pure antagonist ZK98299 at low concentrations, and no significant transcription was observed at higher concentrations in the presence of nuclear extracts prepared from either cell type (Fig. 2).
Figure 2.
RU486 was an agonist with T47D nuclear extracts, and an antagonist with HeLa nuclear extracts in vitro. Chromatin assembly and in vitro transcription reactions were performed with T47D nuclear extracts (NE) as described in Materials and Methods in the presence of increasing concentrations (10−9, 10−8, 10−7 M) of P, RU486, or ZK98299 as indicated. Where noted, when HeLa NE was used, purified PR was included. The final concentration of progesterone (P), RU486 (R), or ZK98299 (Z) was 10−7 M. Experiments were performed at least three times and had similar results.
Coregulators Modulate the Ability of RU486 to Activate Transcription.
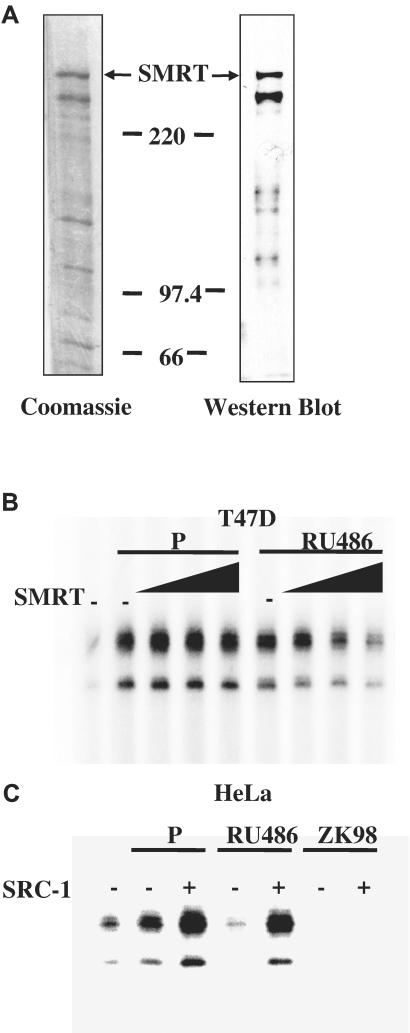
To directly test whether the relative levels of coregulators altered the effect of RU486 on PR-mediated transcription, we attempted to manipulate the mixed agonist/antagonist activity of RU486 by adding exogenous corepressor to the T47D nuclear extract used in the in vitro transcription assay. Full-length His-tagged SMRT was isolated and purified from a baculovirus expression system (Fig. 3A), and added to the cell-free chromatin transcription system along with T47D nuclear extract. As shown in Fig. 3B, exogenous SMRT inhibited RU486-induced transcription in a dose-dependent manner. In contrast, SMRT did not affect progesterone-dependent transcription. This result indicates that the ability of RU486 to modulate PR-mediated transcription is influenced by the levels of corepressors in a ligand-specific context.
Figure 3.
Coregulators modulated the ability of RU486 to regulate PR transactivation. (A) Purification of human SMRT from baculovirus infected Sf9 cells. The full-length His-6-tagged SMRT was overexpressed in Sf9 cells by using a baculovirus expression system and purified by Ni-NTA affinity chromatography. The recombinant protein was subjected to staining with Coomassie brilliant blue R-250 or Western blot analysis with an antibody specific for SMRT. (B) SMRT preferentially inhibited RU486-stimulated PR transcriptional activity. Chromatin assembly and in vitro transcription reactions were performed with T47D nuclear extracts (NE), P, or RU486 in the presence of increasing concentrations (0, 1, 2.5, or 5 nM) of purified SMRT proteins. (C) SRC-1 enhanced both P- and RU486-dependent PR transcriptional activity. Transcription reactions were carried out by using PR, HeLa NE, P, RU486, or ZK98299 in the presence of purified SRC-1 (1 nM). The final concentration of P, RU486, or ZK98299 was 10−7 M. Results in B and C are representative of three independent experiments. SRC-1 was produced and purified as described.
As a complementary experiment, we tested whether the addition of coactivator would promote the agonist activity of RU486 in the presence of HeLa nuclear extract. As expected, in the presence of progesterone, purified SRC-1 potentiated PR-dependent transcription from chromatin templates (Fig. 3C). Importantly, addition of SRC-1 was able also to increase the transcriptional activity of RU486-bound PR significantly, whereas ZK98299 failed to activate transcription, whether or not exogenous SRC-1 was present (Fig. 3C).
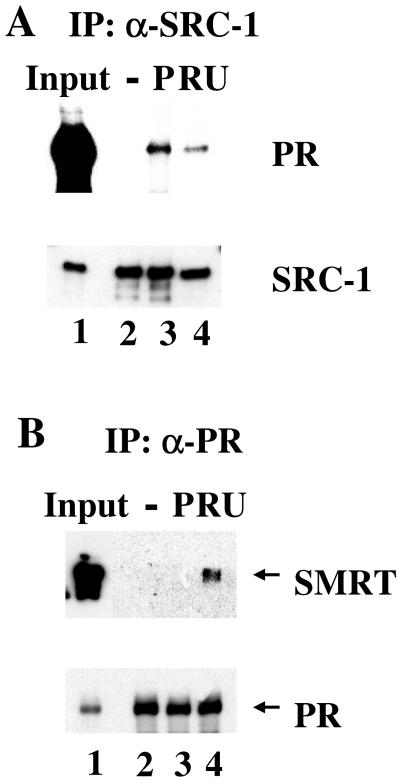
To confirm that RU486-bound PR is capable of recruiting both coactivators and corepressors, we examined the interactions of PR with SRC-1 and SMRT in the presence of either progesterone or RU486 (Fig. 4). SRC-1 was coprecipitated with PR in the presence of either progesterone or RU486, but not with ligand-free PR. As expected, RU486-bound PR recruited SRC-1 although less efficiently than progesterone-bound PR (Fig. 4A). As shown in Fig. 4B, RU486 induced the association of SMRT with PR. In contrast, progesterone was unable to induce this association. These data collectively indicate that the RU486-induced conformation within PR is accessible to both coactivators and corepressors, whereas PR occupied by the full agonist progesterone is accessible only to coactivators.
Figure 4.
In Vitro interaction of PR with SRC-1 or SMRT. (A) Interactions between PR and SRC-1. The full-length SRC-1 synthesized in oocytes was first incubated with anti-FLAG M2 resin, then incubated with His-tag PR in the presence of vehicle, P, or RU486. The bound proteins were subjected to Western blotting of PR and SRC-1. (B) Interactions between PR and SMRT. Sf9 cells were coinfected with PR and SMRT baculovirus. Whole cell extracts were prepared and incubated with anti-PR antibody conjugated to protein A argarose beads in the presence of vehicle, P, or RU486. The proteins bound to the beads were subjected to the Western blot analysis with antibodies against SMRT or PR.
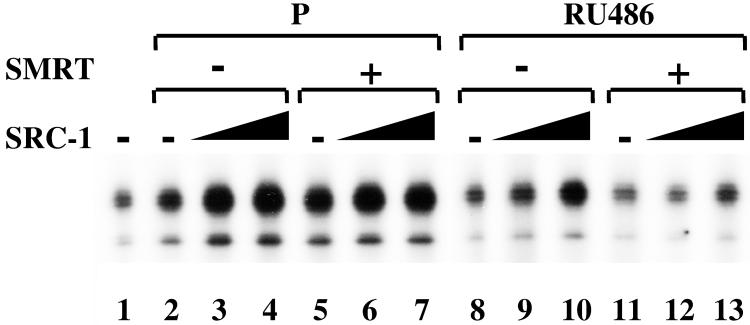
Different Coactivator/Corepressor Concentration Ratios Affect RU486 Action.
To further explore the importance of the relative coactivator/corepressor ratio in the regulation of RU486-bound PR activity, we tested the effects of RU486 in the presence of varying ratios of exogenous coactivators and corepressors in the cell-free chromatin transcription system (Fig. 5). In the presence of progesterone, SRC-1 enhanced PR transcriptional activity to a comparable degree in the absence (lanes 2–4) or presence (lanes 5–7) of recombinant SMRT protein. In contrast, the dose-dependent enhancement by SRC-1 of RU486-bound PR (lanes 8–10) was attenuated by the addition of exogenous SMRT (lanes 11–13). These results indicate that the relative amounts of SRC-1 and SMRT modulate the ability of the SPRM (RU486) to activate PR-dependent transcription on chromatin.
Figure 5.
Modulation of RU486 activity by varying ratio of coactivator to corepressor. Transcription reactions were performed by using PR, HeLa NE, P, or RU486 in the absence or presence of exogenous SMRT (2.5 nM) or SRC-1 (0.5–1 nM) as indicated. Data described here were a representative of three independent experiments.
Discussion
The mixed antagonist RU486, which is used clinically as an antiprogestin, has a number of pharmacological properties that can be attributable to agonist effects (8). Depending on the cell and the promoter context, RU486 can function as either an agonist or an antagonist. Accumulating evidence reveals that mixed antagonists induce a unique PR conformation that is different from those conformations induced by either pure agonists or pure antagonists (22), and that ligand-induced conformational changes in a receptor may regulate the interactions of steroid receptors with coregulators (25, 28). To explain the capacity of mixed antagonists to manifest different activities in different cellular contexts, we hypothesized that a differential ratio of coactivator to corepressor can directly modulate the ability of RU486 to activate transcription in different cellular contexts.
In this study, we compared the effects of RU486 on PR function in two different cell lines by using a cell-free chromatin transcription assay. We observed that RU486 exerted significant agonist activity in T47D cells, whereas it failed to activate transcription in HeLa cells. Similar results have been reported for RU486 regulation of GR activity, and tamoxifen modulation of ER transactivation in a cell-dependent manner (13, 28). Interestingly, we noted a general correlation between the cellular concentrations of corepressors/coactivators and RU486 activity. Substantially higher levels of endogenous corepressors (SMRT and NCoR) were detected in HeLa cells relative to T47D cells, whereas the levels of endogenous coactivators (SRC-1 and p300) in both cell types were comparable. Consistent with predictions, RU486-bound PR was able to interact with both SRC-1 and SMRT in vitro.
To directly examine the basis of this cell-specific regulation, we used our in vitro chromatin transcription assays to show that addition of exogenous SMRT to T47D cells resulted in the repression of RU486 agonist activity, but not progesterone activity. In addition, an increase in exogenous SRC-1 added to the HeLa cell extracts led to a switch of RU486 from an antagonist to a partial agonist, but did not affect the activity of the pure antagonist onapristone (ZK98299). Our data favor the model that cellular coactivators and corepressors have the potential to modulate the ability of a mixed antagonist to regulate PR transactivation, and that the combined effects of coactivators and corepressors modulate the inhibitory or stimulatory efficacy of a mixed antagonist. The molecular basis for such a differential effect is likely caused by the ability of RU486 to promote interaction with both coactivators and corepressors, as demonstrated (Fig. 4). Consequently, our results predict that the ability of mixed antagonists to activate transcription will vary among tissues depending on the cellular concentrations and availability of endogenous coregulators (33). It is likely also that promoter context and chromatin remodeling could play roles in the modulation of RU486 activity (13). It is noteworthy that endogenous chromatin remodeling complexes are present in the chromatin assembly extracts used in our experiments.
Collectively, our data are in concert with proposed mechanisms in cells treated with Tamoxifen (28, 34). Our present study highlights the general importance of understanding the roles of both coactivators and corepressors in transcriptional regulation induced by selective receptor modulators, and should provide a more substantial basis for design of future pharmaceutical discovery and validation of SRMs.
Acknowledgments
We thank N. Weigel and D. Edwards for the PR antibody. This work was supported by National Institutes of Health Grant HD-08188 and the Welch Foundation (to B.W.O.), and National Institutes of Health Grant DK-52542 (to J.D.C.).
Abbreviations
- PR
progesterone receptor
- PRE
progesterone-responsive element
- SRC-1
steroid receptor coactivator-1
- SPRM
selective progesterone receptor modulators
- NCoR
nuclear receptor corepressor
- SMRT
silencing mediator for retinoid and thyroid hormone receptor
References
- 1.Tsai M J, O'Malley B W. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 2.Beato M, Herrlich P, Schutz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 3.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith D F, Toft D O. Mol Endocrinol. 1993;7:4–11. doi: 10.1210/mend.7.1.8446107. [DOI] [PubMed] [Google Scholar]
- 5.Ing N H, Beekman J M, Tsai S, Y, Tsai M-J, O'Malley B W. J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 6.Onate S A, Tsai S Y, Tsai M-J, O'Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 7.Smith C L, Onate S A, Tsai M-J, O'Malley B W. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadepond F, Ulmann A, Baulieu E-E. Annu Rev Med. 1997;48:129–156. doi: 10.1146/annurev.med.48.1.129. [DOI] [PubMed] [Google Scholar]
- 9.Horwitz K B. Endocr Rev. 1992;13:146–163. doi: 10.1210/edrv-13-2-146. [DOI] [PubMed] [Google Scholar]
- 10.Edwards D P, Altmann M, DeMarzo A, Zhang Y, Weigel N L, Beck C A. J Steroid Biochem Mol Biol. 1995;53:449–458. doi: 10.1016/0960-0760(95)00091-d. [DOI] [PubMed] [Google Scholar]
- 11.Allan G F, Lombardi E, Haynes-Johnson D, Palmer S, Kiddoe M, Kraft P, Campen C, Rybczynski P, Combs D W, Phillips A. Mol Endocrinol. 1996;10:1206–1213. doi: 10.1210/mend.10.10.9121488. [DOI] [PubMed] [Google Scholar]
- 12.Gass E K, Leonhardt S A, Nordeen S K, Edwards D P. Endocrinology. 1998;139:1905–1919. doi: 10.1210/endo.139.4.5944. [DOI] [PubMed] [Google Scholar]
- 13.Fryer C J, Nordeen S K, Archer T K. J Biol Chem. 1998;273:1175–1183. doi: 10.1074/jbc.273.2.1175. [DOI] [PubMed] [Google Scholar]
- 14.Klein-Hitpass L, Cato A C, Henderson D, Ryffel G U. Nucleic Acids Res. 1991;19:1227–1234. doi: 10.1093/nar/19.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spitz I M, Chwalisz K. Steroids. 2000;65:807–815. doi: 10.1016/s0039-128x(00)00194-x. [DOI] [PubMed] [Google Scholar]
- 16.Bocquel M T, Kumar V, Stricker C, Chambon P, Gronemeyer H. Nucleic Acids Res. 1989;17:2581–2595. doi: 10.1093/nar/17.7.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzenellenbogen J A, O'Malley B W, Katzenellenbogen B S. Mol Endocrinol. 1996;10:119–131. doi: 10.1210/mend.10.2.8825552. [DOI] [PubMed] [Google Scholar]
- 18.Beck C A, Weigel N L, Moyer M L, Nordeen S K, Edwards D P. Proc Natl Acad Sci USA. 1993;90:4441–4445. doi: 10.1073/pnas.90.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sartorius C A, Tung L, Takimoto G S, Horwitz K B. J Biol Chem. 1993;268:9262–9266. [PubMed] [Google Scholar]
- 20.Meyer M E, Pornon A, Ji J W, Bocquel M T, Chambon P, Gronemeyer H. EMBO J. 1990;9:3923–3932. doi: 10.1002/j.1460-2075.1990.tb07613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner B L, Pollio G, Leonhardt S, Wani M C, Lee D Y-W, Imhof M O, Edwards D P, Cook C E, McDonnell D P. Proc Natl Acad Sci USA. 1996;93:8739–8744. doi: 10.1073/pnas.93.16.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weigel N L, Beck C A, Estes P A, Prendergast P, Altmann M, Christiensen K, Edwards D P. Mol Endocrinol. 1992;6:1585–1597. doi: 10.1210/mend.6.10.1448113. [DOI] [PubMed] [Google Scholar]
- 23.Benhamou B, Garcia T, Lerouge T, Vergezac A, Gofflo D, Bigogne C, Chambon P, Gronemeyer H. Science. 1992;255:206–209. doi: 10.1126/science.1372753. [DOI] [PubMed] [Google Scholar]
- 24.Vegeto E, Allan G F, Schrader W T, Tsai M J, McDonnell D P, O'Malley B W. Cell. 1992;69:703–713. doi: 10.1016/0092-8674(92)90234-4. [DOI] [PubMed] [Google Scholar]
- 25.Jackson T A, Richer J K, Bain D L, Takimoto G S, Tung L, Horowiz K B. Mol Endocrinol. 1997;11:693–705. doi: 10.1210/mend.11.6.0004. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Jeyakumar M, Petukhov S, Bagchi M K. Mol Endocrinol. 1998;12:513–524. doi: 10.1210/mend.12.4.0089. [DOI] [PubMed] [Google Scholar]
- 27.Wagner B L, Norris J D, Knotts T A, Weigel N L, McDonnell D P. Mol Cell Biol. 1998;18:1369–1378. doi: 10.1128/mcb.18.3.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith C L, Nawaz Z, O'Malley B W. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 29.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, et al. Nature (London) 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 30.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 31.Glass C K, Rosenfeld M G. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 32.Liu Z, Wong J, Tsai S Y, Tsai M-J, O'Malley B W. Proc Natl Acad Sci USA. 1999;96:9485–9490. doi: 10.1073/pnas.96.17.9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna N J, Lanz R B, O'Malley B W. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 34.Shang Y, Brown M. Science. 2002;295:2465–2471. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]