Abstract
IFN-γ induces a number of genes to up-regulate cellular responses by using specific transcription factors and the cognate elements. We recently discovered that CCAAT/enhancer-binding protein-β (C/EBP-β) induces gene transcription through an IFN-response element called γ-IFN-activated transcriptional element (GATE). Using mutant cells, chemical inhibitors, and specific dominant negative inhibitors, we show that induction of GATE-driven gene expression depends on MEK1 (mitogen-activated protein kinase kinase/extracellular signal-regulated protein kinase kinase) and ERKs (extracellular signal-regulated protein kinases) but is independent of Raf-1. Interestingly in cells lacking the MEKK1 gene or expressing the dominant negative MEKK1, ERK activation, and GATE dependent gene expression is inhibited. A dominant negative MEKK1 blocks C/EBP-β-driven gene expression stimulated by IFN-γ. These studies describe an IFN-γ-stimulated pathway that involves MEKK1-MEK1-ERK1/2 kinases to regulate C/EBP-β-dependent gene expression.
Keywords: cytokines‖cell growth‖differentiation‖antiviral‖immune response
The IFN family of cytokines regulates antiviral, antitumor, and immune responses by inducing the transcription of a number of IFN-stimulated genes (ISGs) by activating the Janus tyrosine kinase (JAK) and signal transducing activator of transcription (STAT) pathway (1, 2). IFN-α/β induce the tyrosine phosphorylation of STAT1 and STAT2 by using Tyk2 and JAK1. The STAT2-STAT1 dimer forms a complex with another protein, p48 (ISGF3γ), binds to the IFN-stimulated response elements, and induces gene expression (1, 2). IFN-γ, through JAK1 and JAK2, selectively activates the phosphorylation of STAT1. STAT1 dimers induce the expression of ISGs that contain a γ-IFN-activated site (1, 2). Although IFN-α alone induces several ISGs, pretreatment of cells with IFN-γ leads to a strong augmentation of gene expression (3, 4) and antiviral responses (5). This effect is caused in part by the induction of the ISGF3γ gene (3, 4). Indeed, down-regulation of ISGF3γ expression by viral products provides an escape route against the antiviral action of IFNs (6).
ISGF3γ gene induction is delayed, and its promoter lacks STAT-binding elements (7). Instead, a regulatory element, γ-IFN-activated transcriptional element (GATE), and its cognate transcription factors control ISGF3γ expression. We have shown earlier that transcription factor C/EBP-β (8, 9), a regulator of acute phase responses and cell differentiation (10, 11), can induce gene expression through GATE and that its activity is stimulated by the mitogen-activated protein kinases (MAPKs) extracellular signal-regulated protein kinase (ERK)1/2. These studies, however, did not identify the upstream regulators of IFN-γ-induced signaling to GATE through C/EBP-β. Here we show that MEKK1, an upstream regulator of stress-responsive kinases, feeds into the ERK pathway to stimulate gene expression. Thus, our studies identify a previously uncharacterized IFN-γ signaling pathway.
Materials and Methods
Reagents.
Murine IFN-γ (Roche Molecular Biochemicals); SB202190 (Calbiochem), U0126 (Promega), epidermal growth factor (EGF), eicosatetranoic acid, antibodies specific for phospho-ERK1/2; actin (Sigma); ERK2; ISGF3γ, MEKK1; and C/EBP-β (Santa Cruz Biotechnology) were used.
Cell Culture and Plasmids.
Murine macrophage cell line RAW (RAW264.7) was grown in RPMI medium 1640 with 5% FBS. Isogenic mouse fibroblasts lacking c-Raf (12), MEKK1 (13), and ERK1 (14) were described earlier. Isogenic fibroblasts prepared from wild-type and C/EBP-β−/− mouse embryos (15) were grown in DMEM with 5% FBS. The murine ISGF3γ (p48) reporter construct, P4, was described earlier (7). In this construct a 74-bp element of murine p48 gene promoter, encompassing GATE, was cloned upstream of the simian virus 40 (SV40) early promoter driving luciferase. Mutagenesis of the GATE sequence in this construct caused a loss of IFN-γ response and C/EBP-β binding (8). The GATE-Luc and GATE-Mu constructs contained a single copy of wild-type and mutant GATE, respectively, cloned upstream of simian virus 40 minimal promoter in pGL3 promoter vector (7). The GATE-Mu bore mutations within the C/EBP-β binding site. Wild-type and T235A mutant of human C/EBP-β cloned in a pCMV vector was a gift from S. Akira (16). Catalytically inactive dominant negative (DN) mutants of ERK1 (K71R) and ERK2 (K52R) cloned in pCEP4 vector (17) were provided by M. H. Cobb (University of Texas Southwestern Medical Center, Dallas). A constitutively active (CA) MEK1 and DN-MEK1 cloned in pCMV were described (18, 19). Wild type, CA-MEKK1 (Δ367), and DN-MEKK1 (Δ367 KR) were provided by D. J. Templeton (Case Western Reserve University, Cleveland; refs. 20 and 21).
Gene Expression Analyses.
Western blot analyses, transfection, β-galactosidase and luciferase assays, and SDS-PAGE analyses were performed as described (7, 22). The total amount of transfected DNA (1.0 μg) was kept constant by adding pBluescript SK(+) DNA, if required. In general, 0.4 μg of luciferase and 0.1 μg of C/EBP-β expression vector were used for transfection. A β-actin promoter driven β-galactosidase reporter (0.2 μg) was used as an internal control for normalizing variations in transfection efficiency (7). ERK activation was monitored by immunoblot analysis of cell extracts with a phospho-ERK1/2-specific antibody (19). Total ERK was determined in these samples by using antibodies specific for ERK2.
Results
IFN-γ-Induced Gene Expression Through GATE Is Inhibited by MAPK Inhibitors but Is Independent of c-Raf.
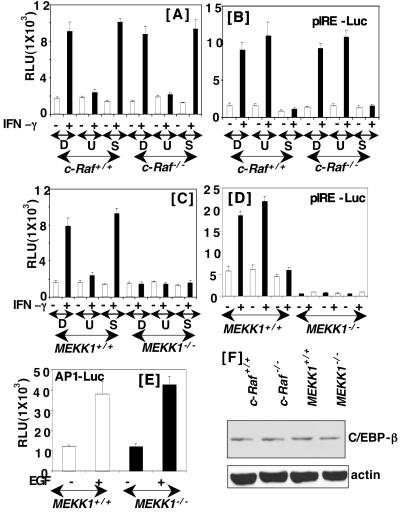
We first studied the effect of two specific inhibitors of MAPK signaling pathways, U0126 (a MEK1/2 inhibitor) and SB202190 (a p38 kinase inhibitor), on IFN-γ-induced gene expression through GATE in c-Raf+/+ mouse embryo fibroblasts. Cells were transfected with the P4 reporter gene, which contains a functional GATE, and then treated with 10 μM of each inhibitor prepared in DMSO before IFN-γ treatment. U0126 but not SB202190 strongly inhibited gene expression (Fig. 1A). Treatment with DMSO (vehicle) had no effect on gene induction by IFN-γ. Under these conditions the MEK1/2 inhibitor had no significant effect on γ-IFN-activated site-dependent gene expression in wild-type or c-Raf−/− cells (Fig. 1B). Thus, MEK1/2 activation seems necessary for IFN-γ-induced gene expression through GATE. Because MEK1 is activated by an upstream regulator, c-Raf, we also repeated these studies in c-Raf−/− mouse embryo fibroblasts (Fig. 1A). Interestingly, IFN-γ was able to induce gene expression in the absence of c-Raf, and it was inhibited by U0126. Thus, c-Raf is not necessary for inducing GATE-dependent gene expression. These results are consistent with our previous observations that DN mutants of either Ras or Raf fail to inhibit IFN-γ-induced expression of the GATE-driven reporter genes in the mouse macrophage cell line RAW. We also examined the effect of IFN-γ on another reporter, palindromic IFN response element (pIRE)-luciferase, the expression of which depends on IFN-γ-activated STAT1 binding to the pIRE (or γ-IFN-activated site). STAT1-dependent gene expression was unaffected in both in the wild-type and c-Raf null cells (Fig. 1B). Furthermore, the induction of γ-IFN-activated site-driven gene expression was inhibited by the p38 kinase inhibitor but not the MEK1/2 inhibitor. This observation is consistent with previous reports indicating the requirement of p38 kinase for STAT1 serine phosphorylation and IFN-inducible gene expression in some cell types (23, 24), although studies in other cell types do not support a requirement for p38 kinase (25).
Figure 1.
Effect of MAPK pathway inhibitors on IFN-γ-induced gene expression. The indicated cells were transfected with the P4-luicferase construct (0.4 μg) and treated with murine IFN-γ (500 units/ml). Transfection was performed and luciferase activity was determined [relative luciferase units (RLU)] after treatment with the indicated agents for 16 h. Before stimulating with IFN-γ, cells were exposed to the DMSO (D), U0126 (U, 10 μM), and SB202190 (S, 25 μM) for 30 min. A plus (+) sign indicates IFN-γ treatment. Bars represent the mean ± SE of triplicates. C is similar to A except different cells were used. (B and D) The effects of MEK1 and p38 kinase inhibitors on the IFN-γ-induced luciferase gene expression from pIRE, a STAT1 binding element. (E) The effect of EGF (10 ng/ml) on AP1-RE-driven luciferase gene expression. (F) Western blots of cell extracts (50 μg) probed with antibodies specific for C/EBP-β and actin.
In light of these observations we have determined whether MEKK1, an upstream of activator of stress-responsive kinases, is necessary for IFN-γ-induced GATE-dependent gene transcription (Fig. 1C). IFN-γ strongly induced the reporter gene in the wild-type cells but not in the MEKK1−/− cells. IFN-γ-stimulated gene expression was inhibited by U0126 in MEKK1+/+ cells. These observations indicate that MEK1 activation by MEKK1 is required for GATE-driven gene induction. Similarly, expression of a reporter, A6-Luc, bearing the native p48 promoter but not its corresponding mutant lacking a functional GATE (GATE pm) was induced in MEKK1+/+ cells after IFN-γ treatment. Such induction was inhibited by U0126. Furthermore, A6-Luc was induced also by IFN-γ in c-Raf−/− cells (data not shown). Thus, the native and minimal enhancer behaved similarly. We next tested whether pIRE-dependent gene expression was affected similarly by the absence of MEKK1 (Fig. 1D). Indeed, pIRE-driven gene expression was inhibited in the MEKK1−/− but not MEKK1+/+ cells. Because the p38 kinase is controlled by MEKK1, these results were expected. As observed in c-Raf+/+ and c-Raf−/− cells, SB202190, but not U0126 inhibited reporter gene expression in MEKK1+/+ cells. Together these results indicate that MEKK1 is critical for IFN-γ-dependent gene expression.
To test whether MEKK1 was required for other ligand-induced pathways, we examined EGF-inducible gene expression through an AP1-responsive element (Fig. 1E). In contrast to IFN-γ, EGF-induced luciferase expression equivalently in MEKK1+/+ and MEKK1−/− cells. Because our previous studies have shown that C/EBP-β is a regulator of GATE-dependent gene expression, we examined whether the differential response in c-Raf−/− and MEKK1−/− cells was caused by a difference in the levels of C/EBP-β protein. Western blot analysis of the cell extracts from wild-type, c-Raf−/− and MEKK1−/− cells revealed no significant difference in the levels of C/EBP-β protein in these cells (Fig. 1F).
A DN-MEKK1 Inhibits IFN-γ-Induced Gene Expression from GATE.
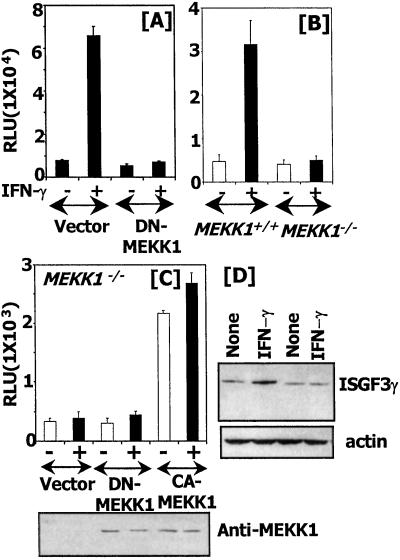
The importance of MEKK1 for GATE-dependent transcription was assessed further in another cell type. Mouse macrophage RAW cells were transfected with GATE-Luc (which bears the minimal GATE) in the presence of an empty expression vector or a DN-MEKK1 mutant. After IFN-γ treatment, the luciferase gene was induced robustly in the presence of the vector but not in the presence of DN-MEKK1 (Fig. 2A). Transfection analyses with the same reporter into MEKK1−/− cells yielded similar results. The reporter was induced strongly by IFN-γ in MEKK1+/+ but not in MEKK1−/− cells (Fig. 2B). These observations show the importance of MEKK1 for regulating GATE-driven gene expression in different cell types. A converse experiment was conducted in MEKK1−/− cells to test whether a CA-MEKK1 would restore GATE-driven responses. Cells were transfected with P4 reporter along with empty expression vector, a DN-MEKK1, or a CA-MEKK1, and luciferase activity was measured. Neither the vector nor DN-MEKK1 restored gene induction in these cells. As expected, CA-MEKK1 restored the gene expression, and it was not affected significantly by IFN-γ treatment (Fig. 2C). Thus, active MEKK1 is required for GATE-dependent gene expression. A comparable expression of the transfected MEKK1 was confirmed by performing a Western blot. These mutants lack the N-terminal 367 aa of the full-length MEKK1, and proteins of the expected size were expressed (Fig. 2C Bottom). The importance of MEKK1 for the induction of endogenous ISGF3γ was determined by a Western blot analysis (Fig. 2D). IFN-γ readily induced ISGF3 in the wild-type but not MEKK1 null cells. A reprobing of this blot with actin antibodies showed that it was a specific effect on ISGF3γ. Reverse transcription–PCR analysis for p48 mRNA yielded similar results (data not shown).
Figure 2.
MEKK1 is required for GATE-dependent gene expression. GATE-Luc plasmid was transfected into the indicated cells, and gene expression was analyzed as described in Fig. 1. (A) Cells were transfected with the indicated effector plasmids in addition to GATE-Luc. (B) Effect of MEKK1 mutation on GATE-Luc. (C) A CA-MEKK1 rescues GATE-dependent gene expression in MEKK1−/− cells. Cells were transfected with expression vector (pCMV), DN-MEKK1, or CA-MEKK1 along with the P4 reporter, and gene expression was analyzed. A Western blot (Bottom) shows the expression of MEKK1 mutants as analyzed with specific antibodies. (D) Expression of endogenous ISGF3γ. Cell extracts (60 μg) Western blotted by using the indicated antibodies. RLU, relative luciferase units.
MEK1 Is Necessary for MEKK1-Dependent IFN-γ-Induced Transcription Through GATE.
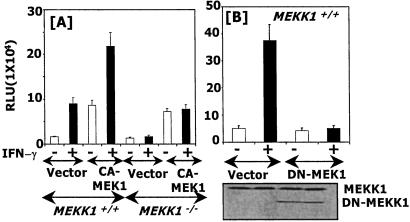
Because MEK1 is essential for ERK1/2 activation in many signaling pathways (26, 27) and GATE-dependent gene expression is inhibited by U0126 (Fig. 1), we examined whether overexpression of a CA-MEK1 would restore GATE-dependent gene expression in MEKK1−/− cells. CA-MEK1 elevated basal expression and further augmented IFN-γ-inducible expression significantly higher than that observed in vector-transfected MEKK1+/+ cells (Fig. 3A). There was very low basal expression of the reporter in MEKK1−/− cells, which was enhanced significantly in the presence of the CA-MEK1. However, no further augmentation of IFN-induced gene expression occurred, indicating that CA-MEK1 partially restores GATE-dependent gene expression in the absence of MEKK1. In a converse experiment, we determined whether coexpression of a DN-MEK1 mutant would block GATE-controlled gene expression in the MEKK1+/+ cells. As observed with U0126, DN-MEK1 strongly inhibited reporter expression in these cells (Fig. 3B). Expression of the DN mutant was shown by a Western blot. A comparable level of endogenous MEKK1 (195 kDa) was present in all lanes. These observations further support the notion that MEKK1 exerts its effects through MEK1.
Figure 3.
MEK1 is critical for MEKK1-initiated GATE-dependent gene expression. (A) The effects of a CA-MEK1 on GATE-dependent gene expression in MEKK1+/+ and MEKK1−/− cells. After transfection with the indicated plasmids, cells were treated with IFN-γ and analyzed for P4-driven luciferase activity. RLU, relative luciferase units. (B) A catalytically inactive MEK1 inhibits GATE-dependent gene induction in MEKK1+/+ cells. A Western blot showing expression of endogenous and DN-MEKK1 is shown also.
IFN-γ Fails to Activate ERK1/2 in the Absence of MEKK1.
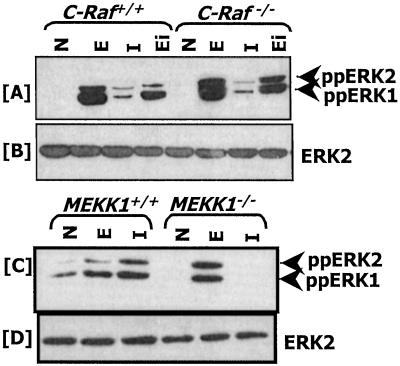
Earlier studies using hematopoietic and HeLa cell lines have shown that IFN-γ activates Ras and Raf (28, 29). Ras and Raf activation by IFN-γ has been suggested to activate ERK1/2. The surprising activation of GATE-dependent gene expression in c-Raf−/− cells (Fig. 1A) compelled us to examine whether Raf-independent ERK1/2 activation occurred. Western blot analysis with antibodies specific for diphosphorylated forms of ERK1/2 (ppERK1/2) showed that ERK1/2 are activated not only by IFN-γ but also EGF and 11,15,17-eicosatetranoic acid, an analogue of arachidonic acid (Fig. 4A). Thus, ERK1/2 activation independent of c-Raf occurs in response to multiple ligands. IFN-γ-induced activation of ERK1/2 was relatively weak in the wild-type and mutant cells compared with the other ligands. Because MEKK1 is essential for the GATE-dependent gene expression and activated MEK1 inhibitor U0126 blocks it, we examined whether ERK1/2 were activated by IFN-γ in cells lacking MEKK1. Western blot analyses revealed that ERK1/2 are activated in MEKK1+/+ but not MEKK1−/− in response to IFN-γ (Fig. 4C). All lanes had a comparable amount of ERK2 (Fig. 4 B and D). Loss of ERK1/2 activation was ligand-specific in MEKK1−/− cells, because EGF activated them normally in both cell types.
Figure 4.
ERK1/2 activation in c-Raf−/− and MEKK1−/− cells. N, no treatment; E, EGF (10 ng/ml); I, IFN-γ (500 units/ml); Ei, 11,15,17-eicosatetranoic acid (10 μM). (A and C) The blots were probed with phospho-ERK-specific antibodies. (B and D) The blots shown in A and C were stripped and probed with ERK2-specific antibodies to demonstrate the presence of an equal amount of ERK in each lane. Whole-cell protein (50 μg) was used from each treatment.
DN-ERK1/2 Mutants Block IFN-γ-Induced GATE-Driven Gene Expression in MEKK+/+ Cells.
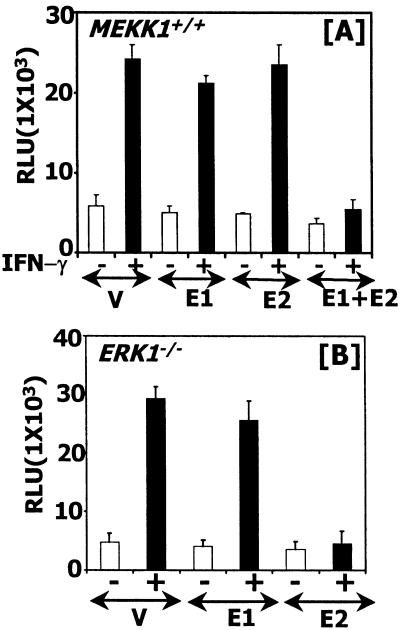
Because MEKK1 exerts its effects through MEK1, and ERK1/2 activation by IFN-γ depended on MEKK1 (Figs. 1, 3, and 4), we next determined whether the downstream effectors ERK1/2 were critical in this pathway to activate GATE-dependent gene expression. To test this possibility, two DN mutants of ERK1 and ERK2 were coexpressed along with the P4 reporter gene into MEKK1+/+ cells. Cotransfection of vector, DN-ERK1, or DN-ERK2 alone with the reporter did not inhibit IFN-γ-induced transcription significantly. However, in the presence of both DN-ERK mutants luciferase gene induction was inhibited strongly (Fig. 5A). To demonstrate further a role of ERK1/2 in regulating GATE-dependent gene expression, this experiment was repeated in ERK1−/− mouse fibroblasts (Fig. 5B). Vector alone had no effect on IFN-γ-induced luciferase expression, nor did DN-ERK1. However, the DN-ERK2 mutant ablated IFN-γ-inducible gene expression. These data suggest that MEKK1-activated signals are routed through ERK1/2 to GATE.
Figure 5.
ERK1/2 are critical for GATE-driven gene expression in MEKK1+/+ cells. (A) Mutant ERKs block IFN-γ-stimulated gene expression through GATE. The P4 reporter was transfected with expression vector carrying DN-ERK1, ERK2 mutants, or both. The total amount of DNA transfected was kept constant by adding pCEP4 vector where necessary. V, vector, pCEP4; E1, ERK1 (K71R) mutant; E2, ERK2 (K52R) mutant. Luciferase activity was determined as described above. A similar experiment was repeated in ERK1−/− cells (B). RLU, relative luciferase units.
MEKK1 Is Necessary for the IFN-γ Activation of Transcription Factor C/EBP-β.
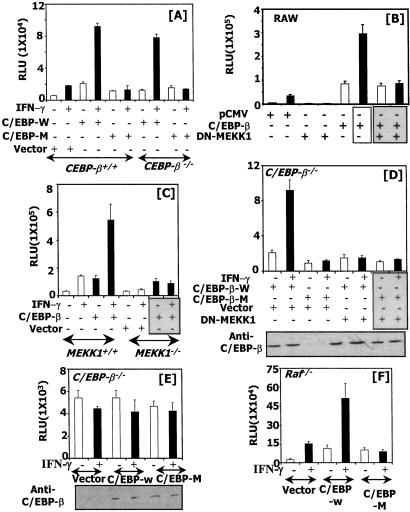
Our previous studies have indicated that C/EBP-β binds to GATE and stimulates transcription in response to IFN-γ (8). These studies have used RAW cells in which endogenous C/EBP-β also activated the IFN-γ response. To confirm these observations we used the isogenic C/EBP-β+/+ and C/EBP-β−/− fibroblasts. These cells were transfected with either wild-type C/EBP-β or a mutant that lacked the critical threonine in the regulatory domain. This threonine is located in a conserved GTPS motif of the regulatory domain and is a substrate for ERK phosphorylation (16). Therefore, we tested the effect of this C/EBP-β mutant on IFN-γ-induced GATE-dependent gene expression. Transfection analyses revealed that wild-type C/EBP-β was able to further stimulate luciferase gene expression in C/EBP-β+/+ cells as compared with the control vector transfection (Fig. 6A). However, the mutant C/EBP-β lacking the critical threonine residue inhibited transcription. As expected, wild-type C/EBP-β but not the mutant promoted luciferase expression in C/EBP-β−/− cells in response to IFN-γ. However, the mutant elevated basal gene expression similar to the wild-type C/EBP-β.
Figure 6.
MEKK1 is necessary for C/EBP-β-dependent gene expression. The indicated cell lines were transfected with the specific plasmids along with the P4 reporter, and luciferase activity was measured. (A) A mutant C/EBP-β lacking ERK consensus motif blocks IFN-γ-stimulated gene expression. C/EBP-W, wild-type C/EBP-β; C/EBP-M, a mutant C/EBP-β with a T-A conversion of the ERK consensus GTPS motif. (B) DN-MEKK1 blocks IFN-γ-stimulated C/EBP-β-dependent gene expression in RAW macrophage cells. (C) IFN-γ-stimulated C/EBP-β-dependent gene expression through GATE is blocked in MEKK1−/− cells. (D) DN-MEKK1 blocks gene induction by the transfected C/EBP-β in response to IFN-γ in C/EBP-β−/− cells. (E) Effect of C/EBP-β on GATE mutant-driven expression. Expression of the transfected C/EBP-β is shown for the luciferase data of D and E under the respective panels. (F) Effect of C/EBP-β on GATE-driven gene expression in Raf−/− cells. RLU, relative luciferase units.
We next determined the effect MEKK1 on C/EBP-β-dependent transcription in RAW macrophages (Fig. 6B). In these cells, coexpression of C/EBP-β with the P4 reporter strongly elevated basal transcription (5-fold), which was stimulated further (15-fold) by IFN-γ. DN-MEKK1 blocked the induction of the reporter gene by IFN-γ as expected. In the presence of DN-MEKK1, C/EBP-β elevated basal transcription but did not stimulate it further after treatment with IFN-γ. The validity of these experiments was tested further in MEKK1+/+ and MEKK1−/− cells by coexpression of C/EBP-β with the P4 reporter gene (Fig. 6C). C/EBP-β up-regulated basal transcription, and IFN-γ significantly stimulated it further in the MEKK1+/+ cells. However, C/EBP-β was unable to enhance IFN-γ-stimulated expression in the MEKK1−/− cells. These results are similar to those shown in Fig. 6B.
To establish a functional relationship between MEKK1 and C/EBP-β+/+, C/EBP-β−/− cells were transfected with an expression vector for DN-MEKK1 in the presence of wild-type or the threonine mutant of C/EBP-β (Fig. 6D). A control transfection with empty expression vector was used for a comparison. As expected, introduction of wild-type C/EBP-β but not the mutant resulted in a robust stimulation of the IFN-γ-induced transcription. However, the wild-type C/EBPβ sustained only basal transcription and failed to support IFN-γ-induced transcription in the presence of DN-MEKK1. DN-MEKK1 had no effect on mutant C/EBP-β. A Western analysis showed that C/EBP mutants expressed comparably (Fig. 6D). Conversely, neither wild-type nor mutant CEBP-β induced a reporter driven by mutant GATE (Fig. 6E) in C/EBP-β−/− cells. Lastly, a wild-type but not mutant C/EBP-β induced GATE-driven gene expression in Raf−/− cells (Fig. 6F). Thus, MEKK1-initiated signals via ERKs cause the phosphorylation of C/EBP-β and regulate GATE-dependent gene expression.
Discussion
IFN-γ regulates innate and specific immune responses by inducing a variety of cellular genes (30). Consistent with the diversity of these processes and the genes involved, a number of transcription factors mediate IFN-γ action in addition to STAT1. These factors include IRF-1 (31), ICSBP (32, 33), RF-X (34, 35), CIITA (36), BRCA1 (37), etc. Recent studies have shown that IFN-γ induces several genes in a STAT1-independent manner (38, 39). Previously we reported the identification of GATE, an IFN-γ response element (7), and its regulation in response to IFN-γ (8) by transcription factor C/EBP-β (10, 11, 40). C/EBP-β (NF-IL-6, LAP, CRP2, and NF-M; refs. 10, 11, and 40) responds to a number of extracellular stimuli including IL-6, IL-1, tumor necrosis factor-α, and lipopolysaccharide (10, 11) and is necessary for regulating several processes including carbohydrate metabolism, lipid storage, Th1 immune responses, macrophage-mediated antibacterial and antitumor defenses, and female fertility (40, 41). IFN-β suppresses HIV-1 replication by using a naturally occurring truncated form of C/EBP-β (42). C/EBP-β is required also for B-lymphopoiesis (43), chemical carcinogen-induced Ras-dependent tumor formation in skin keratinocytes (44), and C/EBP-β deficiency causes a lymphoproliferative disorder (45). These and other studies (46, 47) indicate that the functional diversity of C/EBP-β is regulated in part by specific protein kinases in response to disparate extracellular stimuli. Because C/EBP-β enhanced basal and IFN-stimulated gene expression, an involvement of specific protein kinase(s) in this pathway was suspected. We showed earlier that inhibitors of ERK activation block IFN-γ-stimulated GATE-driven gene expression (48). Although ERK1/2 were required, surprisingly DN mutants of Ras and Raf-1 (the upstream activators of this pathway) did not block IFN-stimulated gene expression.
In this study we showed that MEKK1 is required for both STAT1- and C/EBP-β-driven gene expression in response to IFN-γ. Although MEKK1 is required for both of these pathways, only a mutant MEK1 or its inhibitor ablated GATE-driven gene expression. The p38-kinase is required only for STAT1-dependent gene expression, because SB202190 blocks it in the wild-type cells (Fig. 1). Indeed, experimental evidence indicates a role for p38 kinase in IFN-induced responses through STAT1 (23, 24). Thus, MEKK1 controls both the ERK and p38 kinase pathways in response to IFN-γ. MEKK1 activates MEK1, which in turn seems to activate the ERKs. Activated ERK1/2 in turn phosphorylate C/EBP-β to stimulate gene transcription. This suggestion is consistent with the observation that a C/EBP-β lacking its ERK phosphorylation site does not drive IFN-γ-induced transcription. Although neither the ERK1 nor ERK2 mutants alone blocked gene expression, together they strongly repressed IFN-γ-induced gene expression in MEKK1+/+ cells (Fig. 5). Because both ERKs are phosphorylated in response to IFN-γ (Fig. 4), if one of them is blocked by the corresponding mutant, the other will remain active. This point was illustrated further in ERK1−/− cells, where blockade of ERK2 alone with its DN mutant was sufficient to block IFN-γ signaling (Fig. 5B). Although the basis for such redundancy is unclear, it is likely that IFN-γ-activated ERK1/2 may differentially control specific sets of genes by using C/EBP-β and/or other transcription factors. We show a critical role of MEKK1 in IFN-γ-induced and C/EBP-β-dependent pathways. Previously this kinase has been implicated in the regulation of IκB-kinase-dependent NF-κB activation (49, 50) including the activation of the IFN-β gene enhancer (51). Thus, MEKK1 plays a critical role in IFN synthesis and IFN action.
The MAPK pathways are controlled by a sequential activation of specific kinases that up-regulate specific transcriptional targets (26, 27). It is well established that Ras activation of Raf causes the stimulation of ERKs via an intermediate enzyme MEK1. This pathway is stimulated primarily by growth factors. In contrast, stress-inductive stimuli and some cytokines stimulate MEKK1, which activates the p38 kinases and c-Jun N-terminal kinase (JNK; refs. 21 and 52). Recent studies have shown that EGF does not require c-Raf for ERK activation (12, 53). In a similar manner, IFN-γ was independent of c-Raf for ERK activation (Fig. 4A). This scenario is plausible because IFN-γ is a growth inhibitor, and Ras and Raf are growth promoters. Unexpectedly, MEKK1 was required for ERK1/2 activation in response to IFN-γ as revealed by a total lack of their phosphorylation in MEKK1−/− cells. Under the same conditions EGF was able to activate ERK1/2 in these cells and stimulate AP1 responsive element-driven reporter gene expression. ERK activation by lipopolysaccharide and sorbitol was diminished but not abolished totally in MEKK1−/− cells (52). Thus, MEKK1-dependent ERK1/2 activation seems ligand-specific. The MEKK1-dependent gene-stimulatory effect can be blocked by a DN-MEK1 in wild-type cells (Fig. 3A). Conversely, expression of a CA-MEK1 partially relieves this requirement. Although no physiologic ligands have been identified, overexpression studies in 293 and Jurkat cells suggest MEKK1 acts as a scaffold for Raf, MEK1, and ERK2 (54). The noncatalytic large N-terminus of this kinase appears to form a scaffold, leading to the juxtaposition of the kinases and their activation. Furthermore, MEKK1 can phosphorylate MEK1 on the same residue that Raf does (55), which in turn can phosphorylate ERKs. Because ERK activation occurs in the absence of Raf-1, MEKK1 may mediate a direct effect on MEK1 to up-regulate ERK1. Indeed, a CA-MEKK1 activates MEK1. However, no ERK activation was reported (55) in those studies. It is likely that another molecule couples MEK1 and ERK1/2 in vivo for full activation. IFN-γ probably activates such a molecule, the identity of which is unclear at this stage. Thus, IFN-γ seems to be one ligand that requires MEKK1-MEK1-ERK1/2 in a physiologic setting. Previous studies did not identify the functional significance of this pathway (55). More importantly, unlike the present study, the above-mentioned studies did not identify the transcription factors and the regulatory elements influenced by this pathway.
The relation between the IFN-γ-dependent MEKK1-MEK1-ERK pathway and C/EBP-β activation was supported by several observations. Wild-type C/EBP-β strongly drove gene expression through GATE in the presence of IFN-γ, and this effect was blocked by DN-MEKK1 in RAW and MEKK1−/− cells (Fig. 6 B and C). In C/EBP-β−/− cells, transfection of wild-type C/EBP-β but not a mutant lacking the ERK phosphorylation site restored IFN-γ responsiveness (Fig. 6A). This observation indicates that without a terminal target, i.e., C/EBP-β, ERKs fail to induce gene expression through GATE. IFN-γ activation of C/EBP-β was blocked by DN-MEKK1; MEK1 inhibitor U0126 blocked IFN-γ-stimulated GATE-dependent gene expression in MEKK1+/+ cells. Neither a DN-p38 kinase (data not shown) nor its inhibitor blocked GATE-dependent gene expression induced by IFN-γ (Fig. 1C) in MEKK1+/+ cells. Together, the evidence presented in this study indicated the operation of MEKK1-MEK1-ERK for up-regulating gene expression through GATE.
Acknowledgments
These studies are supported by National Institutes of Health Grants CA78282 and CA71401 (to D.V.K.), CA73381 and CA77816 (to L.C.P.), and HL58122 (to S.P.M.R.).
Abbreviations
- ISG
IFN-stimulated gene
- STAT
signal transducing activator of transcription
- GATE
γ-IFN-activated transcriptional element
- C/EBP-β
CCAAT/enhancer-binding protein-β
- MAPK
mitogen-activated protein kinase
- ERK
extracellular signal-regulated protein kinase
- EGF
epidermal growth factor
- CA
constitutively active
- DN
dominant negative
- pIRE
palindromic IFN response element
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Stark G R, Kerr I M, Williams B R G, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Levy D E, Lew D J, Decker T, Kessler D S, Darnell J E., Jr EMBO J. 1990;9:1105–1111. doi: 10.1002/j.1460-2075.1990.tb08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandyopadhyay S K, Kalvakolanu D V, Sen G C. Mol Cell Biol. 1990;10:5055–5063. doi: 10.1128/mcb.10.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis J A, Huq A, Shan B. J Virol. 1989;63:4569–4578. doi: 10.1128/jvi.63.11.4569-4578.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalvakolanu D V. Trends Microbiol. 1999;7:166–171. doi: 10.1016/s0966-842x(99)01476-6. [DOI] [PubMed] [Google Scholar]
- 7.Weihua X, Kolla V, Kalvakolanu D V. Proc Natl Acad Sci USA. 1997;94:103–108. doi: 10.1073/pnas.94.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy S K, Wachira S J, Weihua X, Hu J, Kalvakolanu D V. J Biol Chem. 2000;275:12626–12632. doi: 10.1074/jbc.275.17.12626. [DOI] [PubMed] [Google Scholar]
- 9.Xiao W, Wang L, Yang X, Chen T, Hodge D, Johnson P F, Farrar W. J Biol Chem. 2001;276:23275–23281. doi: 10.1074/jbc.M010047200. [DOI] [PubMed] [Google Scholar]
- 10.Akira S, Kishimoto T. Adv Immunol. 1997;65:1–46. [PubMed] [Google Scholar]
- 11.Poli V. J Biol Chem. 1998;273:29279–29282. doi: 10.1074/jbc.273.45.29279. [DOI] [PubMed] [Google Scholar]
- 12.Huser M, Luckett J, Chiloeches A, Mercer K, Iwobi M, Giblett S, Sun X M, Brown J, Marais R, Pritchard C. EMBO J. 2001;20:1940–1951. doi: 10.1093/emboj/20.8.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia Y, Makris C, Su B, Li E, Yang J, Nemerow G R, Karin M. Proc Natl Acad Sci USA. 2000;97:5243–5248. doi: 10.1073/pnas.97.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pages G, Guerin S, Grall D, Bonino F, Smith A, Anjuere F, Auberger P, Pouyssegur J. Science. 1999;286:1374–1377. doi: 10.1126/science.286.5443.1374. [DOI] [PubMed] [Google Scholar]
- 15.Sterneck E, Tessarollo L, Johnson P F. Genes Dev. 1997;11:2153–2162. doi: 10.1101/gad.11.17.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins D J, Zhen E, Owaki H, Vanderbilt C A, Ebert D, Geppert T D, Cobb M H. J Biol Chem. 1993;268:5097–5106. [PubMed] [Google Scholar]
- 18.Tolwinski N S, Shapiro P S, Goueli S, Ahn N G. J Biol Chem. 1999;274:6168–6174. doi: 10.1074/jbc.274.10.6168. [DOI] [PubMed] [Google Scholar]
- 19.Shapiro P S, Ahn N G. J Biol Chem. 1998;273:1788–1793. doi: 10.1074/jbc.273.3.1788. [DOI] [PubMed] [Google Scholar]
- 20.Deak J C, Templeton D J. Biochem J. 1997;322:185–192. doi: 10.1042/bj3220185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan M, Dai T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R, Templeton D J. Nature (London) 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 22.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 23.Goh K C, Haque S J, Williams B R. EMBO J. 1999;18:5601–5608. doi: 10.1093/emboj/18.20.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uddin S, Majchrzak B, Woodson J, Arunkumar P, Alsayed Y, Pine R, Young P R, Fish E N, Platanias L C. J Biol Chem. 1999;274:30127–30131. doi: 10.1074/jbc.274.42.30127. [DOI] [PubMed] [Google Scholar]
- 25.Kovarik P, Stoiber D, Eyers P A, Menghini R, Neininger A, Gaestel M, Cohen P, Decker T. Proc Natl Acad Sci USA. 1999;96:13956–13961. doi: 10.1073/pnas.96.24.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cobb M H. Prog Biophys Mol Biol. 1999;71:479–500. doi: 10.1016/s0079-6107(98)00056-x. [DOI] [PubMed] [Google Scholar]
- 27.Lewis T L, Shapiro P S, Ahn N G. Adv Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 28.Sakatsume M, Stancato L F, David M, Silvennoinen O, Saharinen P, Pierce J, Larner A C, Finbloom D S. J Biol Chem. 1998;273:3021–3026. doi: 10.1074/jbc.273.5.3021. [DOI] [PubMed] [Google Scholar]
- 29.Stancato L F, Yu C R, Petricoin E F, 3rd, Larner A C. J Biol Chem. 1998;273:18701–18704. doi: 10.1074/jbc.273.30.18701. [DOI] [PubMed] [Google Scholar]
- 30.Boehm U, Klamp T, Groot M, Howard J C. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 31.Briken V, Ruffner H, Schultz U, Schwarz A, Reis L F, Strehlow I, Decker T, Staeheli P. Mol Cell Biol. 1995;15:975–982. doi: 10.1128/mcb.15.2.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Contursi C, Wang I M, Gabriele L, Gadina M, O'Shea J, Morse H C, 3rd, Ozato K. Proc Natl Acad Sci USA. 2000;97:91–96. doi: 10.1073/pnas.97.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang I M, Contursi C, Masumi A, Ma X, Trinchieri G, Ozato K. J Immunol. 2000;165:271–279. doi: 10.4049/jimmunol.165.1.271. [DOI] [PubMed] [Google Scholar]
- 34.Chin K C, Mao C, Skinner C, Riley J L, Wright K L, Moreno C S, Stark G R, Boss J M, Ting J P. Immunity. 1994;1:687–697. doi: 10.1016/1074-7613(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 35.Brickey W J, Wright K L, Zhu X S, Ting J P. J Immunol. 1999;163:6622–6630. [PubMed] [Google Scholar]
- 36.Steimle V, Siegrist C A, Mottet A, Lisowska-Grospierre B, Mach B. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 37.Ouchi T, Lee S W, Ouchi M, Aaronson S A, Horvath C M. Proc Natl Acad Sci USA. 2000;97:5208–5213. doi: 10.1073/pnas.080469697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gil M P, Bohn E, O'Guin A K, Ramana C V, Levine B, Stark G R, Virgin H W, Schreiber R D. Proc Natl Acad Sci USA. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramana C V, Gil M P, Han Y, Ransohoff R M, Schreiber R D, Stark G R. Proc Natl Acad Sci USA. 2001;98:6674–6679. doi: 10.1073/pnas.111164198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lekstrom-Himes J, Xanthopoulos K G. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 42.Honda Y, Rogers L, Nakata K, Zhao B Y, Pine R, Nakai Y, Kurosu K, Rom W N, Weiden M. J Exp Med. 1998;188:1255–1265. doi: 10.1084/jem.188.7.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, Liu W, Ambrosino C, Ruocco M R, Poli V, Romani L, Quinto I, Barbieri S, Holmes K L, Venuta S, Scala G. Blood. 1997;90:156–164. [PubMed] [Google Scholar]
- 44.Zhu S, Yoon K, Sterneck E, Johnson P F, Smart R C. Proc Natl Acad Sci USA. 2002;99:207–212. doi: 10.1073/pnas.012437299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, Sellitto C, Scarpa S, Bellavia D, Lattanzio G, et al. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trautwein C, van der Geer P, Karin M, Hunter T, Chojkier M. J Clin Invest. 1994;93:2554–2561. doi: 10.1172/JCI117266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buck M, Poli V, van der Geer P, Chojkier M, Hunter T. Mol Cell. 1999;4:1087–1092. doi: 10.1016/s1097-2765(00)80237-3. [DOI] [PubMed] [Google Scholar]
- 48.Hu J, Roy S K, Shapiro P S, Rodig S R, Reddy S P, Platanias L C, Schreiber R D, Kalvakolanu D V. J Biol Chem. 2001;276:287–297. doi: 10.1074/jbc.M004885200. [DOI] [PubMed] [Google Scholar]
- 49.Lee F S, Peters R T, Dang L C, Maniatis T. Proc Natl Acad Sci USA. 1998;95:9319–9324. doi: 10.1073/pnas.95.16.9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baud V, Liu Z G, Bennett B, Suzuki N, Xia Y, Karin M. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim T, Kim T Y, Lee W G, Yim J, Kim T K. J Biol Chem. 2000;275:16910–16917. doi: 10.1074/jbc.M000524200. [DOI] [PubMed] [Google Scholar]
- 52.Yujiri T, Sather S, Fanger G R, Johnson G L. Science. 1998;282:1911–1914. doi: 10.1126/science.282.5395.1911. [DOI] [PubMed] [Google Scholar]
- 53.Mikula M, Schreiber M, Husak Z, Kucerova L, Ruth J, Wieser R, Zatloukal K, Beug H, Wagner E F, Baccarini M. EMBO J. 2001;20:1952–1962. doi: 10.1093/emboj/20.8.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karandikar M, Xu S, Cobb M H. J Biol Chem. 2000;275:40120–40127. doi: 10.1074/jbc.M005926200. [DOI] [PubMed] [Google Scholar]
- 55.Xu S, Robbins D, Frost J, Dang A, Lange-Carter C, Cobb M H. Proc Natl Acad Sci USA. 1995;92:6808–6812. doi: 10.1073/pnas.92.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]