Abstract
Expression of retroviral Gag protein in yeast has previously shown Gag targeting to the plasma membrane but little or no production of Gag virus-like particles (VLPs). Here we show that, after removal of the cell wall, the expression of HIV type 1 Gag protein in Saccharomyces cerevisiae spheroplasts allowed simultaneous budding of VLPs from the plasma membrane. Our data show that (i) the VLPs released from yeast spheroplasts were spherical and had morphological features, such as membrane apposed electron-dense layers, characteristic of the immature form of HIV particles; (ii) the VLPs were completely enclosed in the plasma membrane derived from yeast, which is denser than that of higher eukaryotic cells; (iii) the VLP Gag shells remained intact after treatment of nonionic detergent; and (iv) the VLPs were released soon after removal of the cell wall and accumulated up to 300 μg/liter of culture. Our results also show that VLP production was abolished by amino acid substitution of the Gag N-terminal myristoylglycine and impaired when Gag C-terminal deletions were extended beyond the nucleocapsid domain. These results were consistent with those obtained previously in higher eukaryotic expression systems, suggesting that similar Gag domains were used for VLP assembly. We suggest that the system described here offers significant advantages for studying host factors required for VLP budding. The system also may be available for production of vector virus-free VLPs for practical applications such as vaccine development.
Over the last two decades, several lines of evidence have accumulated to suggest that yeast can be a host for full replication of animal and plant viruses. Incubation of yeast spheroplasts with vesicular stomatitis virus has shown internalization of virus particles, suggesting that early stages of a viral life cycle, such as adsorption, penetration, and uncoating, proceed in yeast cells (1, 2). Multiple rounds of transcription and replication of viral genomes have recently been demonstrated by studies in which brome mosaic viral RNA replication genes were expressed or Flock house virus genomic RNAs were transfected in yeast (3, 4). Furthermore, expression in yeast of viral structural components (e.g., hepatitis B virus surface antigen and brome mosaic virus capsid protein) leads to assembly into a particle structure inside the yeast cells, suggesting that late stages of viral replication also occur in yeast (5, 6). These data suggest that yeast supports most of the stages of a virus life cycle. Indeed, transfection of Flock house viral RNAs has resulted in production of infectious virions in yeast cells (4). However, the life cycle of enveloped viruses is completed by a final stage termed budding, which leads to assembly of viral components on the cell membrane and release of viral particles enclosed in the lipid membrane into intracellular vesicles or from the plasma membrane.
Although the viral components essential for particle budding vary for each virus, generally budding of enveloped viruses is driven by either viral capsid protein or membrane protein, or a combination of the two, suggesting that internal enzymatic proteins such as viral DNA/RNA polymerases are not absolutely required (7). Expression of these structural components alone is sufficient for production of virus-like particles (VLPs) in higher eukaryotic cells, indicating that each virus has a minimal assembly machinery for VLP formation. In the case of retroviruses such as HIV, the Gag protein, the main structural component, directs VLP formation. The Gag protein contains the matrix (MA) domain at the N-terminal region, which is essential for association with the plasma membrane (8–11), but Gag assembly itself is driven by the Gag region from the C-terminal part of the central capsid (CA) domain to the nucleocapsid (NC) domain (12–16), presumably through the NC acting to effectively concentrate Gag around RNA. Another Gag region that is responsible for particle release also has been identified and termed the late or L domain (17, 18).
HIV Gag is initially synthesized in the cytosol and then targeted to the plasma membrane where particle assembly and budding occur. A number of studies have shown that expression of HIV Gag in higher eukaryotic cells produce VLP budding (19, 20); in contrast, expression in yeast failed to produce VLPs from the cell surface, despite the fact that Gag is myristoylated and targeted to the plasma membrane (21–25). Similar results have been reported for Rous sarcoma virus (26). However, one report showed structures protruding from the plasma membrane into the cell wall when HIV Gag is expressed in yeast (25). Furthermore, Hayakawa et al. (27) provided initial evidence that a particulate form of human T cell leukemia virus Gag is released from yeast in an N-terminal myristoylation-dependent manner. These observations lead us to hypothesize that formation of VLP is normally arrested at the budding stage in yeast but that, in some conditions, budding of VLP occurs. In this report, we show that removal of the cell wall from yeast expressing HIV Gag protein leads to simultaneous budding and release of HIV VLPs into the culture medium. Because the VLPs are free of any expression vector viruses by virtue of being a plasmid-based expression system and are purified from the culture medium without disruption of yeast cells, we suggest that the budding system described here may be useful for practical applications such as vaccine development.
Materials and Methods
Construction of Yeast Expression Vectors for HIV-1 Gag Proteins.
The full-length and the C-terminally truncated HIV-1 (IIIB strain) gag genes were amplified by PCR using a forward primer, 5′-CGCGGGATCCATGGGTGCGAGAGCGTCAGT-3′, and reverse primers, 5′-CCGGGAATTCTTATTGTGACGAGGGGTC-3′ (for the full-length Gag protein), 5′-CGCGGAATTCTCAATTAGCCTGTCTCTCAG-3′ (for the MA-CA-p2-NC protein), and 5′-CGCGGAATTCTACATTATGGTAGCTGTATTT-3′ (for the MA-CA-p2 protein). For the nonmyristoylated Gag protein, replacement of the N-terminal glycine with alanine was done by PCR using 5′-CGCGGGATCCATGGCTGCGAGAGCGTCAG-3′. For the Gag-Pol polyprotein, replacement of the aspartate at amino acid 25 in the protease domain, which is the active-site residue, was carried out by overlapping PCR using 5′-GAAGCTCTATTAAATACAGGAGCA-3′ and its complementary oligonucleotide in the context of the gag-pol gene. The PCR fragments were cloned into yeast expression vector pKT10 (28), which is a 2-μ plasmid containing the URA3 gene as a selective marker and the constitutive promoter for the yeast glyceraldehyde-3-phosphate dehydrogenase gene. To produce the Gag-V3 fusion protein, the gag gene (truncated just before the termination codon) was amplified by PCR using a forward primer, 5′-CGCGGGATCCATGGGTGCGAGAGCGTCAGT-3′, and a reverse primer, 5′-CGCGGGTACCTTGTGACGAGGGGTCGTTGC-3′ (underlined is a KpnI linker), and the HIV-1 env gene encoding the V3 domain and its flanking portions was amplified by using a forward primer, 5′-CGCGGGTACCAACCAATCTGTAGAAATTAATTGT-3′ (underlined is a KpnI linker), and a reverse primer, 5′-CGCGGTCGACTTACCATTTTGCTCTACTAATGTTACA-3′. Both fragments were ligated in-frame at the KpnI site and cloned into vector pKT10. The S. cerevisiae strain RAY3A-D (MATa/α ura3/ura3 his3/his3 leu2/leu2 trp1/trp1) (29) was transformed with the recombinant plasmids.
Growth of Yeast Cells and Preparation of Spheroplasts.
Yeast transformants were grown at 30°C in synthetic defined medium without uracil (0.67% yeast nitrogen base, 2% glucose, and amino acid mixtures without uracil) to OD600 = 2.0–2.5. Spheroplast formation was carried out by the standard procedures. In brief, cells were collected by low-speed centrifugation and washed with wash buffer (50 mM Tris, pH 7.5/5 mM MgCl2/1 M sorbitol). The cells were suspended in wash buffer containing 30 mM DTT and incubated at 30°C for 20 min with gentle shaking. To digest the cell wall, the cells were resuspended in wash buffer containing 3 mM DTT, and Zymolyase-100T (Seikagaku, Tokyo) was added to a final concentration of 0.4 mg/ml. The digestion reaction was carried out at 30°C for 20 min with gentle shaking. After being washed twice with 1 M sorbitol, spheroplasts were cultured at 30°C in yeast extract/peptone/dextrose medium containing 1 M sorbitol for 2 h, unless otherwise indicated.
Purification of Gag VLPs.
The standard procedure for purification of Gag VLPs (30) was slightly modified. In brief, culture media of yeast spheroplasts were clarified and then centrifuged through 30% (wt/vol) sucrose cushions at 4°C at 26,000 rpm for 1.5 h. The VLP pellets were resuspended with PBS and centrifuged on 20–70% (wt/vol) sucrose gradients at 4°C at 35,000 rpm overnight. Purified VLPs were obtained by fractionation of the gradients. For comparison, Spodoptera frugiperda (Sf9) cells were infected with recombinant Autographa californica nuclear polyhedrosis viruses (baculoviruses) containing the full-length HIV-1 gag gene (30), and the VLPs were similarly purified from the culture medium.
Delipidization of Gag VLPs and Digestion with Trypsin.
For removal of the VLP lipid bilayer, Gag VLPs were treated with 1% Triton X-100. In some experiments, Gag VLPs were incubated with 1 μg/ml trypsin (Roche Diagnostics) at 30°C for 30 min in the presence or absence of 1% Triton X-100 (see Fig. 3).
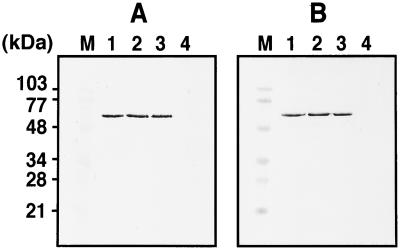
Figure 3.
Trypsin digestion of VLPs in the presence or absence of detergent. Purified VLPs were incubated with 1 μg/ml trypsin at 30°C for 30 min in the presence or absence of 1% Triton X-100, and Gag antigens were detected by Western blotting using anti-HIV-1 CA Ab. (A) VLPs purified from culture medium of yeast spheroplasts expressing Gag protein. (B) VLPs purified from culture medium of Sf9 cells expressing Gag protein. Lanes: M, prestained molecular weight markers; 1, untreated; 2, treated with detergent alone; 3, treated with trypsin alone; and 4, treated by a combination of detergent and trypsin.
Gradient Analysis.
Gag VLPs and delipidized Gag shells were applied onto 20–70% (wt/vol) sucrose density gradients and centrifuged in an SW55 rotor (Beckman) at 4°C at 35,000 rpm for 16 h, during which time they reached equilibrium. After centrifugation, the gradients were fractionated from the bottom to the top.
Protein Detection and Quantitation.
Protein samples were separated by SDS/PAGE. Western blotting was carried out by using anti-HIV-1 CA (Advanced Biotechnologies, Columbia, MD) or V3 (DuPont/NEN) mAb and anti-mouse IgG alkaline phosphatase conjugate (Cappel).
Purified Gag VLPs were quantitated by Western blotting and Coomassie brilliant blue staining. Test results of serial 3-fold dilutions of the VLP fractions were compared to standard dilutions prepared with purified soluble Gag protein (Innogenetics, Alpharetta, GA) for Western blotting or BSA (Sigma) for Coomassie brilliant blue staining. Expression level of intracellular Gag protein was similarly estimated by Western blotting. Total yeast protein was quantified by Bradford's method (31).
Electron Microscopy.
Yeast spheroplasts were fixed in 2% glutaraldehyde in 100 mM cacodylate buffer (pH 7.2) and postfixed with 1% osmium tetroxide. Purified VLPs were similarly processed. Electron microscopic observation was performed by the standard procedures.
Results
Release of VLPs from Yeast Spheroplasts Expressing HIV Gag Protein.
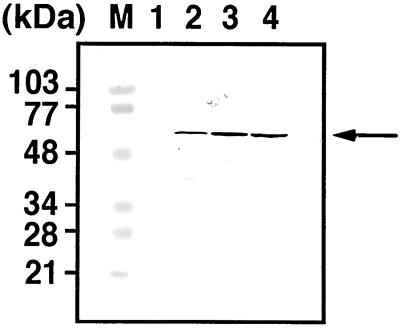
Yeast cells expressing HIV Gag protein were grown in synthetic defined medium without uracil, and after removal of the cell wall, spheroplasts were maintained under isotonic conditions. The cells were harvested at each stage of preparation, and intracellular Gag antigens were examined by Western blotting using anti-HIV-1 CA mAb (Fig. 1). Broadly equivalent levels of intact Gag protein were detected in all cell lysates although each was accompanied by some degradation (Fig. 1, lanes 2–4). In contrast, no reactive Gag antigens were observed in the cells transformed with a parental vector (Fig. 1, lane 1).
Figure 1.
Detection of HIV Gag antigens expressed in yeast cells. Yeast cells were transformed with pKT10 vector containing the full-length HIV-1 gag gene and grown at 30°C in uracil dropout medium. After removal of the cell wall, spheroplasts were cultured at 30°C in yeast extract/peptone/dextrose medium containing 1 M sorbitol for 2 h. Cells were harvested at each stage of preparation, and 0.5 OD of cells were subjected to SDS/PAGE followed by Western blotting using anti-HIV-1 CA Ab. Lanes: M, prestained molecular weight markers; 1, cells transformed with a parental vector; 2, cells with the gag gene-containing vector before removal of the cell wall; 3, spheroplasts with the gag gene-containing vector just after removal of the cell wall; and 4, spheroplasts with the gag gene-containing vector after 2 h of cultivation. The arrow indicates intact Gag proteins.
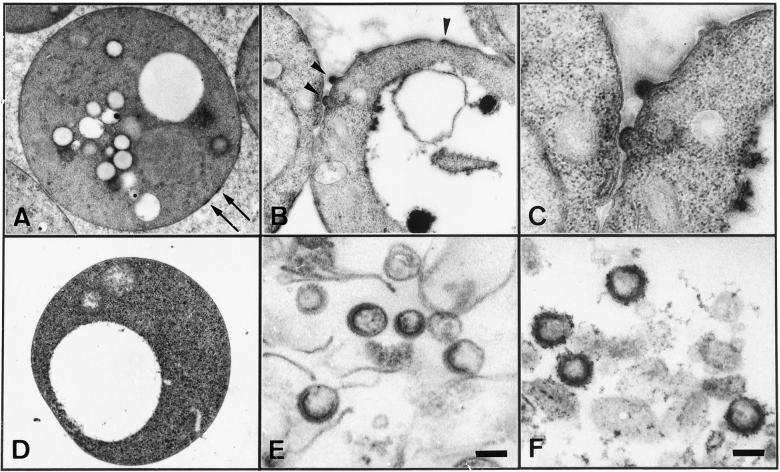
Electron microscopic analysis was carried out to examine whether Gag VLPs budded from the cell surface of the spheroplasts. Immediately after removal of the cell wall, the spheroplasts showed electron-dense patches layered underneath the plasma membrane, likely corresponding to gathered Gag proteins. No budding structures were visible on the cell surfaces observed at this time point (Fig. 2A). However, when spheroplasts were cultured for 2 h in isotonic conditions, one-half spherical structures protruding from the plasma membrane were observed (Fig. 2B). The buds contained electron-dense submembrane layers, suggestive of VLP budding (Fig. 2C). To confirm the release of VLPs, the culture medium of the spheroplasts was harvested and subjected to centrifugation through a sucrose gradient followed by fractionation (see Fig. 4A). When the Gag-rich fractions were subjected to electron microscopic analysis, a number of VLPs were observed (Fig. 2E). The VLPs were spherical (138 ± 20 nm in diameter) with electron-dense submembrane layers, similar to the immature form of authentic HIV particles. The VLPs, however, exhibited electron-dense submembrane layers that were crescent-shaped and not circularly closed (Fig. 2E), although similar features were often observed for control VLPs (121 ± 13 nm in diameter) obtained by Gag expression using recombinant baculoviruses (Fig. 2F). Neither electron-dense layers nor budding structures were observed when spheroplasts containing a parental vector were similarly cultured, showing that the VLPs observed were not derived from yeast Ty retrotransposon elements (Fig. 2D). These observations suggest that removal of the cell wall allows development and release of VLPs from yeast spheroplasts expressing HIV Gag protein.
Figure 2.
Electron microscopic examination of yeast spheroplasts expressing HIV Gag protein and VLPs purified from the culture medium. (A–C) Yeast spheroplasts expressing Gag protein. (D) Yeast spheroplasts transformed with a parental vector (as a control for A–C). (E) VLPs purified from culture medium of yeast spheroplasts expressing Gag protein. (F) VLPs purified from culture medium of Sf9 cells infected with recombinant baculoviruses expressing Gag protein (for comparison to E). Soon after removal of the cell wall (A) and after 2 h of cultivation (B–D). Arrows show Gag layers underneath the plasma membrane and arrowheads a budding VLP. (Scale bars, 100 nm.)
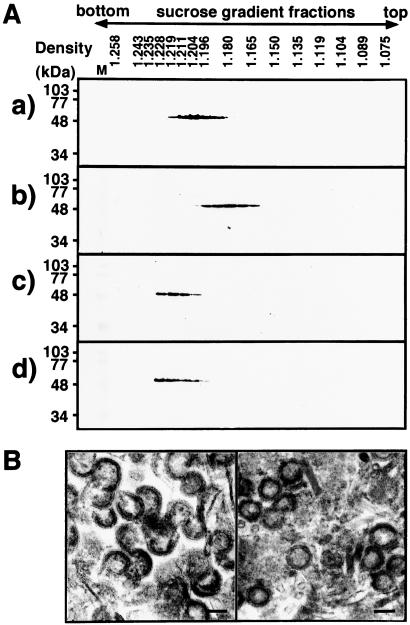
Figure 4.
Gradient analyses of VLPs and delipidized Gag shells and electron microscopic examination of Gag shells. (A) Equilibrium centrifugation of VLPs and delipidized Gag shells. Delipidization was carried out by treatment with 1% Triton X-100. VLPs and delipidized Gag shells were applied onto 20–70% (wt/vol) sucrose gradients and centrifuged at 4°C at 35,000 rpm for 16 h. After centrifugation, gradients were fractionated from the bottom to the top (left to right) and subjected to Western blotting by using anti-HIV-1 CA Ab. (a) VLPs (yeast-produced). (b) VLPs (Sf9 cell-produced). (c) Gag shells (yeast-produced). (d) Gag shells (Sf9 cell-produced). Lane M shows prestained molecular weight markers for SDS/PAGE. (B) Electron micrographs of delipidized Gag shells; yeast-produced (Left) and Sf9 cell-produced (Right). (Scale bars, 100 nm.)
Characterization of VLPs Released from Yeast Spheroplasts.
VLPs completely enveloped by the lipid bilayer are resistant to protease digestion unless delipidized with detergent (32, 33). To examine whether the VLPs released from Gag-expressing yeast spheroplasts are encased in the plasma membrane, purified VLPs were digested with trypsin in the presence or absence of 1% Triton X-100 and analyzed by Western blotting using anti-HIV-1 CA Ab (Fig. 3). The Gag protein was completely digested with trypsin in the presence of the detergent whereas, in contrast, no digestion was observed in the absence of the detergent (Fig. 3A). Similar profiles were observed when VLPs obtained by a baculovirus expression system were used as a control (Fig. 3B). These results indicate that Gag VLPs released from the yeast spheroplasts were completely enveloped by the lipid membrane.
To further characterize the VLPs released from yeast spheroplasts, the VLPs were subjected to centrifugation on 20–70% (wt/vol) sucrose gradients and the fractions containing Gag proteins were detected by Western blotting. For the purpose of comparison, VLPs produced from Sf9 insect cells by using recombinant baculoviruses were analyzed in parallel. After equilibrium centrifugation on sucrose gradients, the VLPs produced by yeast were detected at a density of 1.20–1.21 g/ml (Fig. 4Aa). In contrast, the VLPs obtained by using recombinant baculoviruses occurred at a density of 1.18 g/ml (Fig. 4Ab), consistent with the density of authentic HIV. The gradients show that the VLPs released from yeast spheroplasts have a higher buoyant density than those released from higher eukaryotic cells.
To test whether the differences in VLP densities are due to the lipid envelope that is derived from different host cells, the lipid membrane was removed by treatment with 1% Triton X-100. Immature capsids of retroviral Gag precursor (e.g., avian leukosis virus and HIV) have been reported to be stable in nonionic detergents (34, 35). When both sets of delipidized VLPs were analyzed by equilibrium centrifugation on sucrose gradients, Gag antigen was found only at a density of 1.22 g/ml (Fig. 4 Ac and Ad). To rule out the possibility that aggregation of Gag protein might give rise to the density, the delipidized VLPs were subjected to electron microscopic analysis. Both delipidized VLPs showed doughnut-shaped structures, although the Gag shells derived from yeast were often seen to be crescent-shaped, suggesting that both Gag shells were intact after the treatment (Fig. 4B). These data suggest that the VLPs budded from yeast spheroplasts were enveloped by the lipid membrane which was denser than the higher eukaryotic membrane, consistent with previous reports on chemical analysis of yeast plasma membrane (36).
VLP Formation Competence of Gag in Yeast.
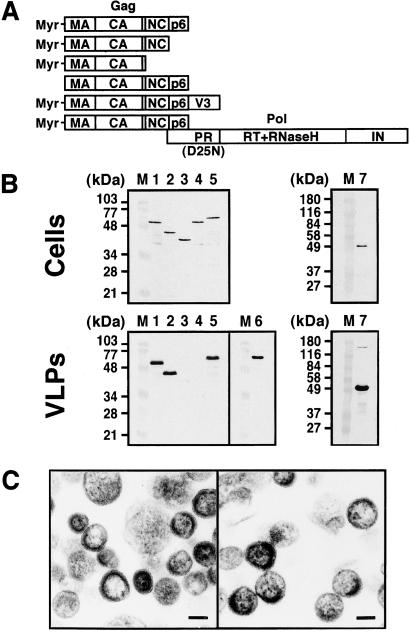
It is known for HIV that nonmyristoylated Gag protein obtained by amino acid substitution at the N-terminal glycine, the acceptor site of the myristoyl moiety, fails to bud VLP (8–10) and that a C-terminal deletion extending beyond the NC domain also abolishes VLP production although the C-terminal p6 domain is dispensable for VLP budding (16, 19). We examined whether these Gag mutants show similar assembly phenotypes when expressed in yeast (Fig. 5). The expression levels of each Gag protein in the yeast cells were broadly equivalent (Fig. 5B Upper). After removal of the cell wall, the culture media were subjected to centrifugation on sucrose gradients and VLPs present were detected by Western blotting (Fig. 5B Lower). Deletion of the C-terminal p6 domain did not affect VLP budding (Fig. 5B Lower, lane 2) whereas a C-terminal deletion that included the NC domain impaired VLP production (Fig. 5B Lower, lane 3). No VLPs were produced by the nonmyristoylated form of Gag (Fig. 5B Lower, lane 4). These data are consistent with those observed in higher eukaryotic cells, suggesting that similar Gag domains may be used for VLP assembly in yeast.
Figure 5.
VLP formation of various Gag constructs. (A) Schematic representation of the Gags constructed. (B) Intracellular Gag expression and purified VLPs. Yeast cells (0.5 OD) expressing each Gag construct were subjected to SDS/PAGE followed by Western blotting (Upper). After removal of the cell wall, the spheroplasts were cultured for 2 h. VLPs were purified from the culture medium by sucrose gradient centrifugation and detected by Western blotting (Lower). Western blotting was carried out using anti-HIV-1 CA (for lanes 1–5 and 7) and V3 (for lane 6) Abs. Lanes: M, prestained molecular weight markers; 1, Myr-MA-CA-p2-NC-p6 (wild-type); 2, Myr-MA-CA-p2-NC; 3, Myr-MA-CA-p2; 4, nonmyristoylated MA-CA-p2-NC-p6; 5 and 6, Myr-MA-CA-p2-NC-p6-V3 fusion; and 7, gag-pol (containing the inactive form of protease). (C) Electron micrographs of VLPs. (Left) VLPs produced by expression of Myr-MA-CA-p2-NC. (Right) VLPs produced by expression of Myr-MA-CA-p2-NC-p6-V3 fusion. (Scale bars, 100 nm.)
It also has been shown that relatively short C-terminal extensions, such as green fluorescent protein or the HIV envelope V3 region, onto Gag are tolerated for VLP formation (37, 38); in contrast, Gag with larger C-terminal extensions (e.g., β-galactosidase or the authentic Gag-Pol polyprotein), does not form VLP alone but is incorporated into VLP through coassembly with Gag (39, 40). To test the VLP production by Gag constructs with such C-terminal extensions, a Gag-V3 fusion construct and the gag-pol gene with the authentic frameshifting site but the inactive form of protease were similarly expressed in yeast and, after spheroplast formation, the culture media were analyzed. Gradient analysis and subsequent Western blotting revealed that expression of Gag-V3 produced VLPs that are solely composed of 60- to 62-kDa protein reactive with both anti-CA and anti-V3 Abs (Fig. 5B Lower, lanes 5 and 6). Expression of the gag-pol construct produced VLPs containing a trace of 160-kDa protein as well as 55-kDa protein, both of which were reactive with anti-CA Ab (Fig. 5B Lower, lane 7). This result shows that the ribosomal frameshifting from gag to pol occurred in yeast and that the product was incorporated into VLPs. Together, these data suggest that two types of assembly by Gag fusions into VLPs, assembly alone or coassembly, are possible in the yeast spheroplast system.
These data were confirmed by electron microscopic analysis. Both the p6-deleted Gag VLPs and the chimeric Gag-V3 VLPs were spheres with crescent-shaped electron-dense layers, similar to the wild-type Gag VLP observed in this system (Fig. 5C). The chimeric Gag-V3 VLPs, but not the p6-deleted Gag VLPs, were slightly larger (173 ± 35 nm in diameter) than the wild-type Gag VLPs, consistent with previous observations of the chimeric Gag-V3 VLPs released from higher eukaryotic cells (38).
Kinetics of VLP Production from Yeast Spheroplasts.
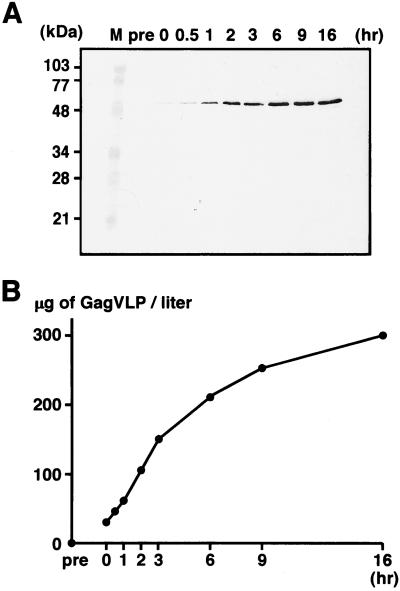
To assess the level of VLP production in this yeast expression system, the culture media of the yeast spheroplasts expressing the full-length Gag protein were harvested at intervals and VLPs were purified by centrifugation on sucrose gradients. When equivalent volumes of the relevant gradient fractions were analyzed by Western blotting, Gag antigens, which were absent in the preparation from the culture medium of the cell wall-bearing yeast (Fig. 6A, lane pre), were detected in the fractions prepared soon after removal of the cell wall (Fig. 6A, lane 0), suggesting that the VLP development and release occur immediately after removal of the cell wall. The level of VLP production increased in a time-dependent manner within the period observed in this study (up to 16 h), suggesting continuous production of VLPs (Fig. 6A). For quantitation of the VLP yield at each time point, each VLP fraction was serially diluted and subjected to Western blotting and Coomassie brilliant blue staining. When compared to standards prepared with purified soluble Gag protein (for Western blotting) or BSA (for staining), the yield of VLPs was estimated to reach ≈300 μg of Gag VLP/liter of spheroplast culture by 16 h (Fig. 6B). This value corresponded to 0.13–0.15% of total yeast protein. The intracellular expression level of Gag in yeast was 0.25–0.33% of total yeast protein when similarly quantitated by Western blotting.
Figure 6.
Time courses of VLP production. After removal of the cell wall, yeast spheroplasts expressing the full-length Gag protein were cultured at 30°C. At each time point, VLPs were purified from the culture medium by sucrose gradient centrifugation. (A) VLP fractions detected by Western blotting using anti-HIV-1 CA Ab. Lanes: M, prestained molecular weight markers; pre, before removal of the cell wall; 0, just after removal of the cell wall; and 0.5–16, hours of cultivation following the removal. (B) VLP yields estimated by quantitative Western blotting and Coomassie brilliant blue staining. The VLP yield at each time point was estimated by comparison to standard dilutions prepared with purified soluble Gag protein (for Western blotting) or BSA (for Coomassie brilliant blue staining).
Discussion
Particle assembly of nonenveloped viruses takes place in the cytosol and progeny viruses are released concomitant with lysis of the host cells. Consistent with these morphogenetic features observed in higher eukaryotic cells, expression in yeast of these viral major components reveals intracellular assembly of VLPs, which are purified after deliberate disruption of yeast cells (5, 6, 41, 42). However, this preparation procedure for VLPs is inappropriate for enveloped viruses because the particles form only concomitant with budding from the host lipid membrane at which time they adopt the lipid envelope. In this report, we chose HIV, an enveloped virus, and described a system of yeast spheroplasts in which the expression of Gag protein resulted in release of VLPs into the culture medium. These data demonstrate that yeast supports the viral budding process, characteristic of the life cycle of enveloped viruses. The data also indicate that the presence of the cell wall may serve to inhibit VLP budding from the yeast cell surface, likely due to either a physical blockage or reduction of membrane fluidity.
VLPs formed by HIV Gag and budded from yeast spheroplasts were encased in the lipid membrane, which was denser than higher eukaryotic membranes. This finding is consistent with studies on previous mass analyses of plasma membranes in which the protein-to-lipid ratio was estimated to be ≈2.2 for yeast but 1.0–1.5 for higher eukaryotes (36, 43, 44). The high ratio of protein to lipid, characteristic of the plasma membrane of wall-bearing cells, has also been observed for the cytoplasmic membrane of Escherichia coli (45). In contrast, intracellular vesicles of yeast show a significantly lower ratio of protein to lipid (0.6–0.7) (36). These data support our electron microscopic observations that the VLPs budded directly from the plasma membrane and were not released along the secretory pathway after intracellular budding.
Current vaccine development strategies use a number of expression systems, and noninfectious particles represent new candidates as safe and efficacious vaccine components. Although a high yield of VLPs for enveloped viruses has been achieved by use of viral expression vectors such as recombinant baculoviruses and vaccinia viruses (19, 46), the VLP fractions obtained by these expression systems are contaminated with the vector viruses themselves, many of which are infectious. In contrast, the yeast system described here produces VLPs not only at a level feasible for practical use but also without contamination of infectious viruses. We suggest therefore that a yeast spheroplast system offers production of VLPs with guaranteed safety, and that HIV VLPs produced by our system constitute a new candidate for a safe and efficacious anti-HIV vaccine. We also suggest that application of yeast genetics to the VLP budding system described here will lead to identification of the host factor(s) essential for VLP budding and a fuller understanding of the budding mechanisms of enveloped viruses.
Acknowledgments
We thank K. Mizumoto and Y. Shibagaki (Kitasato University, Tokyo) for supply of a yeast expression vector. Y.M. thanks A. Nomoto, M. Masuda (University of Tokyo), and I. M. Jones (University of Reading, U.K.) for review of the manuscript. This work was supported by an AIDS grant from the Ministry of Health, Labour, and Welfare of Japan.
Abbreviations
- VLP
virus-like particle
- MA
matrix
- CA
capsid
- NC
nucleocapsid
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Makarow M. EMBO J. 1985;4:1855–1860. doi: 10.1002/j.1460-2075.1985.tb03860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makarow M, Sareneva H, von Bonsdorff C H. J Biol Chem. 1987;262:1836–1841. [PubMed] [Google Scholar]
- 3.Janda M, Ahlquist P. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 4.Price B D, Rueckert R R, Ahlquist P. Proc Natl Acad Sci USA. 1996;93:9465–9470. doi: 10.1073/pnas.93.18.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valenzuela P, Medina A, Rutter W J, Ammerer G, Hall B D. Nature (London) 1982;298:347–350. doi: 10.1038/298347a0. [DOI] [PubMed] [Google Scholar]
- 6.Krol M A, Olson N H, Tate J, Johnson J E, Baker T S, Ahlquist P. Proc Natl Acad Sci USA. 1999;96:13650–13655. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garoff H, Hewson R, Opstelten D-J E. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rein A, McClure M R, Rice N R, Luftig R B, Schultz A M. Proc Natl Acad Sci USA. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlinger H G, Sodroski J G, Haseltine W A. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant M, Ratner L. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelle T D, Wills J W. J Virol. 1996;70:2269–2276. doi: 10.1128/jvi.70.4.2269-2276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wills J W, Craven R C. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Weldon R A, Jr, Wills J W. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borsetti A, Ohagen A, Gottlinger H G. J Virol. 1998;72:9313–9317. doi: 10.1128/jvi.72.11.9313-9317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C T, Lai H Y, Li J J. J Virol. 1998;72:7950–7959. doi: 10.1128/jvi.72.10.7950-7959.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson L, Yu X F. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 17.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, de Wilde M. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 20.Smith A J, Srinivasakumar N, Hammarskjold M L, Rekosh D. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer R A, Schaber M D, Skalka A M, Ganguly K, Wong-Staal F, Reddy E P. Science. 1986;231:1580–1584. doi: 10.1126/science.2420008. [DOI] [PubMed] [Google Scholar]
- 22.Bathurst I C, Chester N, Gibson H L, Dennis A F, Steimer K S, Barr P J. J Virol. 1989;63:3176–3179. doi: 10.1128/jvi.63.7.3176-3179.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobs E, Gheysen D, Thines D, Francotte M, de Wilde M. Gene. 1989;79:71–81. doi: 10.1016/0378-1119(89)90093-0. [DOI] [PubMed] [Google Scholar]
- 24.Vlasuk G P, Waxman L, Davis L J, Dixon R A, Schultz L D, Hofmann K J, Tung J S, Schulman C A, Ellis R W, Bencen G H, et al. J Biol Chem. 1989;264:12106–12112. [PubMed] [Google Scholar]
- 25.Biemans R, Thines D, Gheysen D, Rutgers T, Cabezon T. AIDS. 1992;6:541–546. doi: 10.1097/00002030-199206000-00003. [DOI] [PubMed] [Google Scholar]
- 26.Bonnet D, Spahr P F. J Virol. 1990;64:5628–5632. doi: 10.1128/jvi.64.11.5628-5632.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayakawa T, Miyazaki T, Misumi Y, Kobayashi M, Fujisawa Y. Gene. 1992;119:273–277. doi: 10.1016/0378-1119(92)90282-t. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka K, Nakafuku M, Tamanoi F, Kaziro Y, Matsumoto K, Toh-e A. Mol Cell Biol. 1990;10:4303–4313. doi: 10.1128/mcb.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruggieri R, Tanaka K, Nakafuku M, Kaziro Y, Toh-e A, Matsumoto K. Proc Natl Acad Sci USA. 1989;86:8778–8782. doi: 10.1073/pnas.86.22.8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morikawa Y, Hinata S, Tomoda H, Goto T, Nakai M, Aizawa C, Tanaka H, Omura S. J Biol Chem. 1996;271:2868–2873. doi: 10.1074/jbc.271.5.2868. [DOI] [PubMed] [Google Scholar]
- 31.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 32.Konvalinka J, Heuser A M, Hruskova-Heidingsfeldova O, Vogt V M, Sedlacek J, Strop P, Krausslich H G. Eur J Biochem. 1995;228:191–198. doi: 10.1111/j.1432-1033.1995.tb20249.x. [DOI] [PubMed] [Google Scholar]
- 33.Spearman P, Ratner L. J Virol. 1996;70:8187–8194. doi: 10.1128/jvi.70.11.8187-8194.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart L, Schatz G, Vogt V M. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose J R, Babe L M, Craik C S. J Virol. 1995;69:2751–2758. doi: 10.1128/jvi.69.5.2751-2758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kramer R, Kopp F, Niedermeyer W, Fuhrmann G F. Biochim Biophys Acta. 1978;507:369–380. [Google Scholar]
- 37.Perrin-Tricaud C, Davoust J, Jones I M. Virology. 1999;255:20–25. doi: 10.1006/viro.1998.9573. [DOI] [PubMed] [Google Scholar]
- 38.Luo L, Li Y, Cannon P M, Kim S, Kang C Y. Proc Natl Acad Sci USA. 1992;89:10527–10531. doi: 10.1073/pnas.89.21.10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C T, Stegeman-Olsen J, Zhang Y, Barklis E. Virology. 1994;200:524–534. doi: 10.1006/viro.1994.1215. [DOI] [PubMed] [Google Scholar]
- 40.Park J, Morrow C D. J Virol. 1992;66:6304–6313. doi: 10.1128/jvi.66.11.6304-6313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jore J P, Veldhuisen G, Kottenhagen M, Pouwels P H, Foriers A, Rombaut B, Boeye A. Yeast. 1994;10:907–922. doi: 10.1002/yea.320100706. [DOI] [PubMed] [Google Scholar]
- 42.Cook J C, Joyce J G, George H A, Schultz L D, Hurni W M, Jansen K U, Hepler R W, Ip C, Lowe R S, Keller P M, et al. Protein Expression Purif. 1999;17:477–484. doi: 10.1006/prep.1999.1155. [DOI] [PubMed] [Google Scholar]
- 43.Eylar E H, Hagopian A. Methods Enzymol. 1971;22:123–130. [Google Scholar]
- 44.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. The Molecular Biology of the Cell. New York: Garland; 1995. pp. 255–318. [Google Scholar]
- 45.Kaback H R. Methods Enzymol. 1971;22:99–120. [Google Scholar]
- 46.Karacostas V, Nagashima K, Gonda M A, Moss B. Proc Natl Acad Sci USA. 1989;86:8964–8967. doi: 10.1073/pnas.86.22.8964. [DOI] [PMC free article] [PubMed] [Google Scholar]