Abstract
Phosducin and phosducin-like protein (PhLP) bind G protein βγ subunits and regulate their activity. This report describes a previously uncharacterized binding partner unique to PhLP that was discovered by coimmunoprecipitation coupled with mass spectrometric identification. Chaperonin containing tailless complex polypeptide 1 (CCT), a cytosolic chaperone responsible for the folding of many cellular proteins, binds PhLP with a stoichiometry of one PhLP per CCT complex. Unlike protein-folding substrates of CCT, which interact only in their nonnative conformations, PhLP binds in its native state. Native PhLP competes directly for binding of protein substrates of CCT and thereby inhibits CCT activity. Overexpression of PhLP inhibited the ability of CCT to fold newly synthesized β-actin by 80%. These results suggest that the interaction between PhLP and CCT may be a means to regulate CCT-dependent protein folding or alternatively, to control the availability of PhLP to modulate G protein signaling.
Although all of the necessary information for achieving a specific three-dimensional structure is contained within a protein's primary sequence, many proteins require interaction with chaperones to attain their native conformation in vivo. One class of chaperones, the eukaryotic cytosolic chaperonin CCT [chaperonin containing tailless complex polypeptide 1 (TCP-1)] consists of eight homologous polypeptides that form a ring-shaped structure with a large central cavity (1). Two of these rings are stacked together to make the holo-CCT complex that uses the energy derived from ATP binding and hydrolysis to drive the folding of proteins in the central cavity in an enzyme/substrate fashion (1–5). CCT assists in the folding of actin and tubulin (2, 6, 7) and many other cytosolic proteins (3, 4, 8). In fact, CCT interacts with both nonnative conformations of proteins and nascent polypeptide strands (9). There has been extensive study of the mechanism of protein folding by CCT (1–5), yet little is known about the cellular regulation of this process. In this report, we describe an interaction between CCT and a native polypeptide, phosducin-like protein (PhLP), which blocks the ability of CCT to fold actin and other substrates.
PhLP, so named for its 65% homology in amino acid sequence to phosducin (Pd), was discovered as the product of an ethanol-induced gene in cultured neuronal cells (10). Known functions of Pd and PhLP involve regulation of G protein signaling. Pd binds the Gβγ subunit complex and blocks its interaction with Gα subunits as well as Gβγ effectors (11–15). PhLP also binds Gβγ (16, 17) and prevents Gβγ from binding Gα (18). In addition, both Pd and PhLP interact with the 26S proteasomal subunit, SUG1 (19, 20). However, their cellular expression patterns are very different. Pd is expressed almost exclusively in retinal and pineal cells (21, 22), whereas PhLP mRNA has been found in all tissues examined (10, 23), and immunoblot analyses has confirmed PhLP protein expression in diverse tissues and cell lines (C.D.T., unpublished data). As a result of its broad expression pattern, PhLP has been postulated to be a general regulator of G protein signaling (17). However, a recent study in yeast suggests PhLP involvement in other cellular processes (24). In an effort to better understand the cellular function of PhLP, we sought to identify its in vivo binding partners by coupling coimmunoprecipitation techniques with mass spectrometric identification of coimmunoprecipitating proteins. We report a high-affinity in vivo interaction of PhLP with CCT that inhibits the ability of CCT to catalyze the folding of its protein substrates.
Materials and Methods
cDNA Constructs.
Rat PhLP cDNA was cloned into the vector pcDNA3.1(+)/myc-His B (Invitrogen) to generate a construct with a c-myc epitope tag and a hexahistidine purification tag on the 3′ end. The resulting PhLP-myc-His cDNA was cloned into the vector pET15b (Novagen) for expression and purification. Rat Pd-myc-His was also prepared in pcDNA3.1(+) and pET15b. N-terminally tagged His-PhLP in pET15b was prepared as described (25). The integrity of all constructs was verified by sequence analysis. A β-galactosidase-myc-His vector pcDNA3.1(+)/myc-His/LacZ was obtained from Invitrogen.
Protein Purifications.
PhLP-myc-His was expressed in Escherichia coli and purified by Ni2+ affinity chromatography in the absence of denaturant as described (25). 35S-labeled His-PhLP was expressed and purified in a similar manner with minor modifications. E. coli DE3 cells were grown in M9 minimal medium, and 10 μCi/ml [35S]H2SO4 (NEN-Dupont; 1 Ci = 37 GBq) was added at the time of induction of His-PhLP expression to label the protein. The 35S-His-PhLP was insoluble in cells grown in minimal media, therefore the cell pellets were extracted in buffer with 8 M urea and applied to the Ni2+-chelate column. The 35S-His-PhLP was renatured on the column by washing in buffer containing decreasing amounts of urea and then eluted in its refolded form with imidazole. 35S-labeled Pd was prepared in a similar manner except that urea was not needed to solubilize the protein. Gtα (15), Hsp 70 (26), and CCT (27) were each purified following described protocols. Gtα was labeled with 125I, also as described (28).
Immunoprecipitation.
Chinese hamster ovary (CHO) cells were grown in 6-well plates to 90% confluency. Cells were transfected with 1 μg DNA per well by using 2 μg of Lipofectamine Plus reagent (GIBCO/BRL) according to the manufacturer's protocols. Two days (48 h) later, cells from each well were washed twice in ice-cold PBS and lysed in 200 μl of immunoprecipitation buffer [20 mM NaH2PO4, pH 7.4/150 mM NaCl/2% IGEPAL (Sigma)/8 μM E-64 (Boehringer Mannheim)/0.6 mM PMSF]. Lysates were passed seven times through a 25-gauge needle and centrifuged at 11,000 × g for 10 min. Tagged PhLP was immunoprecipitated with 4 μl of 0.7 μg/μl anti-c-myc antibody (BioMol, Plymouth Meeting, PA) for 30 min at 4°C and 20 μl of a 50% slurry of Protein A/G PLUS agarose (Santa Cruz Biotechnology) for an additional 30 min at 4°C. Samples were centrifuged, and the beads were washed three times with 300 μl of immunoprecipitation buffer. The pellet was solubilized in Laemmli SDS/PAGE sample buffer, separated on a 12% Laemmli SDS/PAGE gel, and stained with GelCode Coomassie Reagent (Pierce) or immunoblotted by using a 1:50,000 dilution of anti-PhLP antiserum (18), a 1:10,000 dilution of anti-Pd antiserum (18), or a 1:500 dilution of anti-TCP-1α (Calbiochem), β, or ɛ antiserum (Stressgen) as primary antibodies. Blots were developed by using ECL+ (Amersham Pharmacia) and were visualized on a Storm 860 PhosphorImager (Molecular Dynamics) in chemiluminescence mode. Bands were quantified by using ImageQuant software (Molecular Dynamics) and the ratio of PhLP to CCT in the immunoprecipitate was determined by comparing the chemiluminescence from known amounts of purified PhLP or CCT with that from the immunoprecipitates. Coimmunoprecipitation of endogenous PhLP with CCT was performed in a similar manner but with untransfected CHO cells and 4 μl of 1 μg/μl anti-TCP-1α antibody as the precipitating antibody.
For in vitro immunoprecipitations from human cell lines, cell lysates were prepared as described above. After clarifying by centrifugation, 10 μg of purified PhLP-myc-His was added and incubated 30 min at 4°C. Immunoprecipitation was as described above.
For in vitro immunoprecipitations from rabbit reticulocyte lysate, 30 μM 125I-Gtα, 35S-His-PhLP, or 35S-His-Pd was diluted 1:6 in denaturing buffer (7 M urea/10 mM Tris, pH 7.5/2 mM DTT) or binding buffer (10 mM Tris, pH 7.5/100 mM KCl/2 mM MgCl2/1 mM EGTA/1 mM DTT) and incubated at 23°C for 10 min. Samples were diluted 1:100 in binding buffer containing 10% rabbit reticulocyte lysate (Promega) and incubated at 4°C for 30 min. One microliter of 1 μg/μl anti-TCP-1α (Calbiochem) was added and incubated 4°C for 30 min. Immunoprecipitation was as described above. Radioactivity on the gel was quantified by using the PhosphorImager.
Inhibition of 125I-Gtα binding to CCT by PhLP was determined by adding the indicated concentrations of PhLP to 1 μM 125I-Gtα in 10% rabbit reticulocyte lysate and immunoprecipitating as described above. The amount of 125I-Gtα in the SDS/PAGE gel was quantified by using the PhosphorImager.
Native Gels.
Purified 35S-PhLP (50 nM) was incubated with or without 10% rabbit reticulocyte lysate in binding buffer for 30 min on ice. Samples were run on a native polyacrylamide gel with a 3.5% stacking gel in 0.125 M Tris (pH 8.0) and a 4.0% separating gel in 0.375 M Tris (pH 8.7). The running buffer was 66 mM Tris/121 mM glycine, pH 8.7. Gels were analyzed for 35S-PhLP by using the PhosphorImager or immunoblotted for TCP-1α. Purified CCT was also run and stained with Coomassie reagent.
Spontaneous Refolding.
His-PhLP was urea-denatured under the same conditions as the immunoprecipitation described above. Samples were diluted 1:100 into binding buffer, and PhLP inhibition of 125I-Gtα and Gtβγ binding to light-activated rhodopsin was determined as described (25).
Mass Spectrometric Identification.
Bands from human adrenocortical H295R cell extracts were excised, digested in-gel with trypsin (29) and analyzed on a PE Biosystems Voyager DE STR MALDI-TOF (matrix-assisted laser desorption ionization time-of-flight) in reflectron mode, by using α-cyano-4-hydroxy-transcinnamic acid as the matrix. A region of blank gel equivalent in size to the excised bands was used to generate a background spectrum. Peptide masses of peaks not observed in the background spectrum were used to search the nonredundant human NCBI database via the ProFound application (30).
Kinetics of Luciferase Refolding.
The procedure followed described methods for measuring CCT-dependent luciferase refolding (31, 32). Firefly luciferase (Boehringer Mannheim) in refolding buffer (10 mM Mops, pH 7.2/50 mM KCl/3 mM MgCl2/2 mM DDT) was denatured by incubation at 44°C for 10 min in the presence of Hsc 70 (molar ratio of 47:1, Hsc70/Luc). The denatured luciferase was then renatured by incubation at 30°C in refolding buffer containing 5% rabbit reticulocyte lysate and 2 mM ATP. Purified PhLP-myc-His was then added to the reaction at the indicated amounts. Luciferase activity was measured according to the manufacturer's protocol (PharMingen) at increasing times after the addition of reticulocyte lysate. From these data, the initial rates of luciferase refolding were determined. No increase in basal luciferase activity with time was observed in the absence of reticulocyte lysate.
In Vivo β-Actin Folding.
The procedure followed described methods for measuring CCT-dependent actin folding in cells (3). CHO cells were transiently transfected as before with PhLP or LacZ in the pcDNA3.1(+)B vector. Cells were exposed to 35S-Met (NEN-Dupont) for 10 min. Protein synthesis was stopped by addition of 4 μM cycloheximide (ICN) and the incubation was continued an additional 5 min. Cells were washed three times with PBS to remove free 35S-Met and were lysed in 20 mM Hepes, pH 7.4/50 mM KCl/1 mM DTT/0.2 mM CaCl2/4 mM cycloheximide/1 mM PMSF/0.5 μg/ml leupeptin/0 μM pepstatin/0.4 units/μl hexokinase (Sigma)/40 mM glucose by passing through a 25-gauge needle several times. Cellular debris was removed by centrifugation, and the supernatants were incubated for 1 h in the presence of DNase I-affigel beads prepared as described (33). Beads were washed five times in 400 mM NH4Cl/10 mM Tris, pH 7.8/0.2 mM CaCl2/0.2 mM DTT, and were brought up in Laemmli SDS/PAGE sample buffer. Samples were separated by SDS/PAGE, and the radioactivity in the β-actin band was quantified by using the PhosphorImager. Data were corrected for the 50% transfection efficiency of the CHO cells as determined in the LacZ control by staining transfected cells with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (Fermentas).
Immunoblot Quantification of PhLP and CCT Expression Levels.
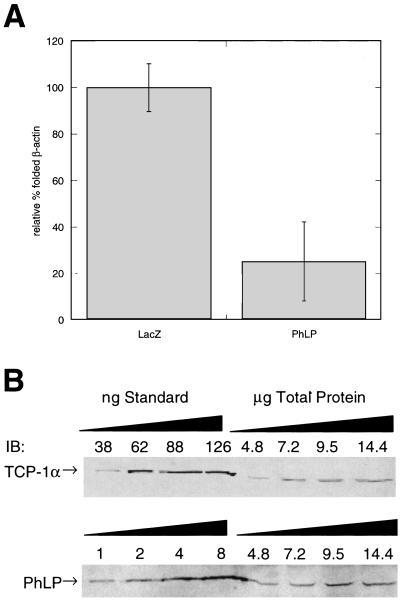
CHO cells were cultured to 90% confluence and then harvested in 1% SDS. Total protein was determined by BCA protein assay (Pierce). Increasing amounts of total protein from cell extracts as well as purified samples of CCT or PhLP-myc-His were subjected to SDS/PAGE followed by immunoblot analysis. Blots were probed with either anti-TCP-1α or anti-PhLP antibodies. Blots were developed by using ECL+ referred to previously and visualized on the PhosphorImager. Bands were quantified by using imagequant software, and curves for both the standards and cell extracts were generated and compared with obtain the ratio of PhLP to CCT in these cells.
Results and Discussion
PhLP Interacts with CCT.
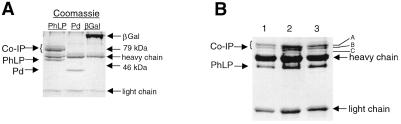
PhLP and Pd, tagged with a c-myc epitope at the carboxy-terminus, were transiently expressed in CHO cells and immunoprecipitated from cell lysates by using a monoclonal antibody to the c-myc tag. A myc-tagged β-galactosidase was also over-expressed as a control. Fig. 1A shows a Coomassie blue-stained gel of the immunoprecipitates, revealing bands with mobilities expected for PhLP-myc, Pd-myc, and β-galactosidase-myc in the appropriate samples. These assignments were confirmed by immunoblotting (data not shown). Interestingly, in the PhLP-myc, samples several intense coimmunoprecipitating bands were observed in the ≈60-kDa mass range. These bands were not found in the Pd-myc or the β-galactosidase-myc samples. The strong coimmunoprecipitation of these polypeptides exclusively with PhLP-myc suggests a specific, high-affinity interaction with PhLP.
Figure 1.
Coimmunoprecipitation of ≈60-kDa polypeptides with PhLP-myc. (A) CHO cells were transiently transfected with c-myc-tagged variants of PhLP, Pd, or LacZ as indicated. Cytosolic extracts of each were prepared and target proteins were immunoprecipitated by using an anti-myc monoclonal antibody. Samples were analyzed by SDS/PAGE and stained with Coomassie blue. (B) A Coomassie blue-stained gel of immunoprecipitates resulting from incubating purified PhLP-myc with lysates from HT29, HCT15, and H295R cells is shown (lanes 1–3, respectively).
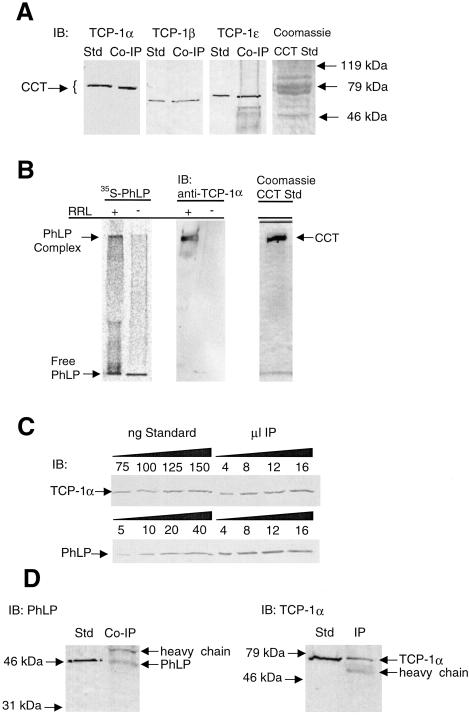
The identity of the PhLP-interacting proteins was determined by mass spectrometric analysis. To improve the potential for successful protein identification, PhLP coimmunoprecipitation experiments were performed in human cell lines including HT29 and HCT15 (colorectal adenocarcinoma) and H295R cells (adrenocortical carcinoma). Because of low transfection efficiency in these lines, purified PhLP-myc was added to cell lysate, incubated, and immunoprecipitated with the anti-myc antibody. Gel analysis of the resulting immunoprecipitates (Fig. 1B) shows the same 60-kDa coimmunoprecipitating bands observed in CHO cells. Three bands from H295R cell lysates were excised and in-gel digested with trypsin. The resulting peptides were analyzed by MALDI-TOF mass spectrometry (Fig. 2). The masses of these peptides were used to search the NCBI nonredundant human protein database, yielding good matches with the α (band A), θ (band B), and β (band C) subunits of the CCT complex. This result was confirmed by immunoblot analysis of the PhLP-myc coimmunoprecipitates from CHO cells (Fig. 3A). Antibodies to the α, β, and ɛ subunits of CCT each recognized appropriate molecular weight bands in the immunoprecipitate, confirming the mass spectrometric identification for α and β subunits and adding the ɛ subunit as part of the complex bound to PhLP-myc.
Figure 2.
Mass spectrometric identification of coimmunoprecipiting proteins. Bands A, B, and C from H295R cell extracts (Fig. 1B, lane 3) were excised, digested in-gel with trypsin and analyzed on a PE Biosystems Voyager DE STR MALDI-TOF. Peptide masses were used to search the nonredundant NCBI database via the ProFound application. (A) Band A matched TCP-1α with a probability of 0.99 with 14% protein coverage. The next best match had a probability 0.004. (B) Band B matched TCP-1θ with probability 0.83 with 21% protein coverage. The next best match had probability 0.11 and was a human cytokeratin. When peaks suspected to result from keratin contamination were deleted from the search data, the probability of the TCP-1θ match rose to 1.0. (C) Band C matched TCP-1β with a probability of 0.98 with 20% protein coverage. The next best match had probability 0.002. (Inset) Tables showing matched peptides, comparing the observed masses (MWobs) with the calculated molecular weights (MWcalc). All masses are monoisotopic and are given as the M+H. Masses at m/z 842, 1045, and 2211 result from trypsin autolysis and were not used in the search.
Figure 3.
Confirmation of the PhLP⋅CCT interaction and determination of the stoichiometry of binding. (A) Immunoprecipitates identical to those from Fig. 1A were immunoblotted with antibodies to TCP-1α, β, and ɛ. Purified CCT was used as a standard in each blot. (B) 35S-PhLP (50 nM) was incubated with or without 10% rabbit reticulocyte lysate (RRL) containing CCT and incubated for 30 min at 30°C. Samples were run on a 4% native gel. (Left) A visualization of the 35S-PhLP from the PhosphorImager. (Center) An immunoblot (IB) for TCP-1α. (Right) A Coomassie blue stain of purified CCT run under the same conditions. The PhLP⋅CCT complex entered the gel and migrated at an Rf value of 0.11, whereas free PhLP migrated with the tracking dye. (C) Increasing amounts of PhLP-myc immunoprecipitates identical to those of Fig. 1A were immunoblotted with antibodies specific to PhLP or TCP-1α, and band intensities were compared with standards for both PhLP and CCT to determine the stoichiometry of the coimmunoprecipitation. The average ratio of TCP-1α to PhLP from three separate immunoprecipitations was 2.2 ± 0.3 to 1. (D) CCT was immunoprecipitated from unmodified CHO cells by using the anti-TCP-1α antibody. The resulting immunoprecipitate was immunoblotted with anti-PhLP or anti-TCP-1α antibodies as indicated.
These results demonstrate that PhLP interacts with at least four subunits of CCT in human cell extracts and in CHO cells under immunoprecipitation conditions that are not expected to disrupt the CCT oligomer (34). Thus, it appears that PhLP is binding to the holo-CCT complex. To further test this possibility, complex formation between PhLP and holo-CCT was determined by native gel analysis. Purified PhLP, metabolically radiolabeled with 35S, was added to rabbit reticulocyte lysate containing CCT and run on a native gel. In the absence of lysate, all of the 35S-PhLP migrated rapidly near the dye front, whereas in the presence of lysate, a significant portion migrated much slower, at the same mobility as TCP-1α and purified holo-CCT, indicating that PhLP was indeed binding to the intact CCT complex (Fig. 3B).
From the relative intensity of the bands in Fig. 1A, there appears to be more TCP-1α than PhLP in the immunoprecipitate. This finding would be expected if one PhLP polypeptide bound per CCT complex because CCT consists of pairs of each of the eight polypeptides that make up the two ring structures (1). To address the question of the stoichiometry of PhLP binding to CCT, the amounts of PhLP and TCP-1α in the immunoprecipitate were determined by immunoblot analysis. By comparing the band intensities from known amounts of purified CCT or PhLP to that from TCP-1α and PhLP in the immunoprecipitate, a ratio of 2.2 ± 0.3 (n = 3) TCP-1α per PhLP was obtained (Fig. 3C). Because there are two TCP-1α subunits in each double-ring CCT complex, it appears that one PhLP polypeptide is binding to one CCT oligomer.
To determine whether PhLP interacts with CCT at concentrations found endogenously in CHO cells, coimmunoprecipitation was investigated in untransfected CHO cells by using the anti-TCP-1α antibody. PhLP was found in the TCP-1α immunoprecipitate (Fig. 3D), indicating that the interaction did indeed occur at concentrations normally found in these cells.
PhLP Interacts with CCT as a Binding Partner and Not as a Substrate.
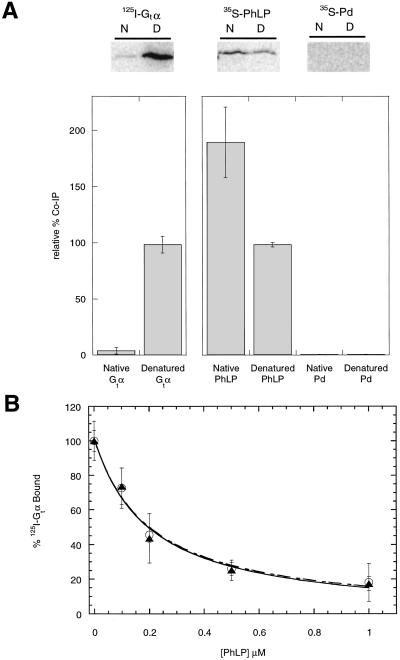
CCT substrates such as actin and tubulin bind with high affinity only in their unfolded states and with little or no affinity in their native state (2–7). This property allows the release of folded proteins from the cavity after they have reached their native conformation. To address the question of whether PhLP interacts with CCT as a substrate in a misfolded state or as a binding partner in its native state, the binding of native versus denatured PhLP to CCT was compared. Purified 35S-PhLP and 35S-Pd were either denatured in 6 M urea or left in their native form, diluted into rabbit reticulocyte lysate containing CCT (31), and incubated for 30 min on ice. The CCT was immunoprecipitated with the anti-TCP-1α antibody, coimmunoprecipitating proteins were separated by SDS/PAGE, and radioactivity in the gels was measured by PhosphorImager analysis. A known substrate of CCT, the α subunit of the photoreceptor G protein transducin (Gtα) (3), was used as a control for comparison. Gtα was radiolabeled with 125I by using Bolton–Hunter reagent, which does not disturb its Gtβγ binding activity (28). Under native conditions, little Gtα coimmunoprecipitated with CCT, whereas under denaturing conditions, a significant amount of the Gtα was found in the immunoprecipitate (Fig. 4A). In contrast, PhLP coimmunoprecipitated with CCT under both native and denaturing conditions (Fig. 4A), with ≈2-fold more PhLP bound to CCT under native conditions, suggesting that the native form of PhLP binds CCT with higher affinity than the denatured form. Pd was also tested in this experimental format and did not coimmunoprecipitate with CCT under native or denaturing conditions despite its homology to PhLP.
Figure 4.
Native PhLP binds CCT. (A) 125I-Gtα, 35S-PhLP, and 35S-Pd at a final concentration of 5 μM were denatured in 6 M urea or maintained in their native state in buffer without urea and then diluted 100-fold in the presence of CCT from rabbit reticulocyte lysates. Samples were immunoprecipitated by using the anti-TCP-1α antibody and subjected to SDS/PAGE and phosphorimaging. N and D indicate respectively native or urea-denatured samples. The value of urea-treated Gtα was set at 100% for the 125I-labeled samples and the value of urea-treated PhLP was set at 100% for the 35S-labeled samples. Error bars represent the standard deviation from three separate experiments. (B) PhLP was denatured under the same conditions as above. Urea-treated (○) or nontreated (▴) PhLP samples were then incubated with 0.2 μM 125I-Gtα, 0.2 μM Gtβγ, and urea-stripped retinal rod outer segment membranes (1.0 μM rhodopsin). The binding of Gt to light-activated rhodopsin was then assayed as described (15). The final dilution of urea had no effect on the total binding. Error bars represent the standard deviation from three separate experiments.
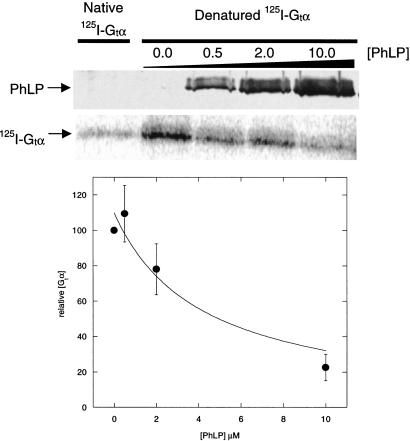
Two explanations for the observed binding of PhLP under denaturing conditions are first that PhLP binds to CCT both in its native and denatured states, representing dual modes of interaction, or second that PhLP could spontaneously refold after dilution from the 6 M urea into rabbit reticulocyte lysate. To test these possibilities, the native state of PhLP was probed by measuring its ability to bind the βγ complex of transducin (Gtβγ) and block interaction with Gtα. Without Gtβγ, the Gtαβγ heterotrimer cannot form and bind light-activated rhodopsin, thus PhLP inhibits light-induced binding of Gtα to rhodopsin-containing membranes (15, 18, 25). When urea-treated PhLP was tested in this assay, it was equally effective as untreated PhLP at inhibiting association of Gtβγ with Gtα (Fig. 4B). The IC50 values for both were 0.2 μM, as would be predicted given the Kd of 0.1 μM for PhLP binding to Gtβγ (25) and the Kd of 0.15 μM for Gtα binding to Gtβγ measured under similar conditions (35), indicating that nearly 100% of the PhLP was active whether treated with urea or not. Therefore, the urea-treated PhLP must have spontaneously refolded under these conditions into a conformation that bound Gtβγ and inhibited Gtα binding. Increasing the urea to 8 M gave the same result (data not shown). Thus, the observed binding of PhLP to CCT after dilution from urea was a result of spontaneous refolding of PhLP and not of an interaction of denatured PhLP with CCT. Together, the data in Fig. 4 indicate that PhLP does not require CCT to refold and that it binds CCT as a native binding partner and not as a misfolded substrate.
PhLP Is a Competitive Inhibitor of CCT.
The fact that native PhLP binds CCT suggests a role for PhLP in the protein folding function of CCT. To begin to test this possibility, the effect of PhLP on the binding of the CCT substrate Gtα (3) was assessed. Urea-treated Gtα was incubated with rabbit reticulocyte lysate containing CCT and increasing concentrations of native PhLP. The CCT was immunoprecipitated with the anti-TCP-1α antibody, and the resulting immunoprecipitates were subjected to SDS/PAGE and PhosphorImager analysis. Fig. 5 Upper shows a representative phosphorimage. As the PhLP concentration increased, the amount of Gtα coimmunoprecipitating with CCT decreased. Fig. 5 Lower shows a graphical representation of the data from three similar experiments. Approximately 80% inhibition of binding of 1 μM urea-treated Gtα to CCT in the lysate was achieved at 10 μM PhLP and half-maximal inhibition occurred at 4 μM PhLP. Thus, PhLP was an effective inhibitor of denatured Gtα binding to CCT, with a binding affinity ≈4-fold less than denatured Gtα. This result demonstrates that the interaction of PhLP with CCT blocks the binding of CCT substrates, suggesting that PhLP can inhibit protein folding by CCT.
Figure 5.
Competition for CCT substrate binding by native PhLP. Urea-denatured 125I-Gtα was diluted 100-fold into rabbit reticulocyte lysate to a final concentration of 1.0 μM. Native PhLP was added to a final concentration of 0.5, 2.0, and 10.0 μM. CCT was immunoprecipitated by using the anti-TCP-1α antibody, and the immunoprecipitates were subjected to SDS/PAGE and phosphorimager analysis or immunoblotting. (Upper) An immunoblot for PhLP. The middle panel is the 125I-Gtα phosphorimage of one gel. (Lower) A graphical representation of the data from three separate experiments. Error bars represent the standard deviation between experiments.
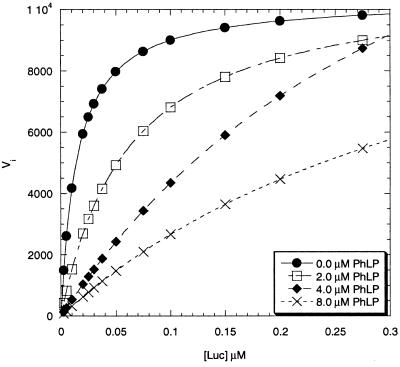
To investigate this possibility, the effects of PhLP on the kinetics of refolding of firefly luciferase were examined. Luciferase is a well-characterized CCT substrate whose recovery of activity in rabbit reticulocyte lysates after heat denaturation has been used to monitor CCT-mediated protein folding (31, 32). Steady-state kinetic analyses (Fig. 6) show that PhLP decreased the rate of luciferase refolding in vitro through a competitive mechanism. This result is consistent with the mutually exclusive binding of PhLP or Gtα to CCT observed in Fig. 5. Thus, it appears that PhLP binding to CCT competitively interferes with the binding of substrate proteins in the folding cavity. Further analysis of the kinetic data in a Dixon plot (data not shown) yielded a Ki of 190 nM for PhLP binding to CCT. This binding affinity falls within the estimated concentration range for cellular expression of PhLP (50–200 nM, ref. 36). Moreover, the in vivo binding affinity may be even higher given the strength of the observed coimmunoprecipitation.
Figure 6.
Inhibition of CCT-dependent luciferase refolding by PhLP. Initial rates of luciferase refolding in rabbit reticulocyte lysate at increasing concentrations of PhLP were measured as described in Materials and Methods. A Michaelis–Menten plot of the kinetic data showing PhLP inhibition of luciferase folding is shown. The Km for luciferase refolding by CCT was 7.3 ± 0.7 nM (n = 4). All four curves converged on similar Vmax values at high luciferase concentration, characteristic of a competitive inhibitor.
PhLP Inhibits CCT Activity in Cells.
The ability of PhLP to inhibit CCT-dependent protein folding was further tested by transiently over-expressing PhLP in CHO cells and assessing the effect on CCT-dependent protein folding. CCT activity was measured by the binding of newly synthesized 35S-β-actin to DNase I-linked beads (3, 33). This method measures CCT activity in vivo by exploiting the fact that DNase I specifically binds native β-actin but not misfolded β-actin. Cells were pulsed with [35S]methionine to label newly synthesized proteins, after which further protein synthesis was blocked with cycloheximide. 35S-labeled proteins were allowed to fold, then the cells were lysed, and native actin was precipitated with DNase I beads. 35S-β-actin that coprecipitates with the DNase I beads represents the fraction of actin that was synthesized during the pulse and was subsequently folded by CCT. PhLP overexpression suppressed 80% of the binding of 35S-β-actin to DNase I beads compared with controls expressing β-galactosidase (Fig. 7A). This result demonstrates the ability of PhLP to inhibit CCT activity in living cells and suggests that PhLP may be a physiologically significant regulator of CCT-dependent protein folding.
Figure 7.
In vivo inhibition of actin folding by PhLP. (A) CHO cells were transiently transfected with PhLP or LacZ and the folding of nascent β-actin was measured by the amount of 35S-labeled actin bound to DNase I beads. Error bars represent the standard deviation between six separate experiments. (B) The amount of PhLP in untransfected CHO cells was determined by quantitative immunoblot analysis. Increasing amounts of total protein from cell extracts as well as purified samples of CCT or PhLP were immunoblotted (IB) with anti-TCP-1α or PhLP antibodies and band intensities between standards and cell extracts were compared with determine the amount of CCT and PhLP in the CHO cells. From these data a ratio of holo-CCT to PhLP of 3.5 ± 0.5 to 1 was calculated.
Physiological Implications.
To address whether cellular levels of PhLP are sufficient to bind a significant portion of the CCT complexes, CCT and PhLP were quantified in CHO cells by immunoblotting. This yielded a ratio of 3.5 ± 0.5 mol of holo-CCT complex per mol PhLP (n = 4; Fig. 7B), which suggests that PhLP could maximally inhibit folding ≈30% under resting conditions. However, these basal PhLP levels are enough that moderate induction of PhLP expression would be sufficient to stoichiometrically sequester CCT and substantially influence protein folding. For example, treatment of NG108–15 cells with 100 mM ethanol increases the level of PhLP mRNA by three-fold (10), suggesting that cellular stimuli such as ethanol may regulate CCT-dependent protein folding through PhLP expression.
An alternative hypothesis for the physiological function of the PhLP⋅CCT interaction is that CCT regulates the activity of PhLP. There are several possible ways that this could occur. First, CCT may facilitate the formation of the PhLP⋅Gβγ complex as has been reported for other protein complexes (8). Second, CCT may sequester PhLP from Gβγ, thereby blocking the ability of PhLP to regulate Gβγ signaling. Third, CCT may protect PhLP from proteolytic degradation. Biophysical measurements of the N-terminal domain of Pd (37, 38) suggest that this domain is relatively unstructured until it binds Gtβγ. PhLP's homology to Pd suggests a similar structure. An unstructured PhLP N-terminal domain would be a target for proteolytic degradation in the cell, but if it were associated with CCT, it could be protected from proteolysis. Indeed, the PhLP N-terminal domain may be a member of the recently recognized class of proteins and protein domains that are unstructured in their native, functional state. These proteins generally have multiple binding partners, adopting different structures depending on which partner is bound (39).
CCT is the third binding partner of PhLP to be identified, in addition to G protein βγ subunits (17) and the SUG1 subunit of the 26S proteosome (20). It is not known how PhLP binding is distributed between its various partners, but the coimmunoprecipitation data in Fig. 1 indicate a strong preference for PhLP binding to CCT. Bands corresponding to Gβγ (38 kDa and 8 kDa) or SUG-1 (45 kDa) were absent in Coomassie blue-stained immunoprecipitates, and Gβγ bands were only revealed after immunoblotting (data not shown), indicating a predominance of CCT complexes over Gβγ or proteasome interactions. It will be important in the future to determine the factors that control which of its partners PhLP binds under various cellular conditions.
In summary, the data presented here demonstrate a unique interaction of PhLP with CCT. Coimmunoprecipitation was observed in cells at both over-expressed and endogenous levels, and PhLP binding competitively blocked CCT-dependent protein folding both in vitro and in intact cells. These results suggest novel mechanisms for the regulation of PhLP and CCT.
Abbreviations
- TCP-1
tailless complex polypeptide 1
- CCT
chaperonin containing TCP-1
- PhLP
phosducin-like protein
- Pd
phosducin
- CHO
Chinese hamster ovary
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Llorca O, McCormack E A, Hynes G, Grantham J, Cordell J, Carrascosa J L, Willison K R, Fernandez J J, Valpuesta J M. Nature (London) 1999;402:693–696. doi: 10.1038/45294. [DOI] [PubMed] [Google Scholar]
- 2.Gao Y, Thomas J O, Chow R L, Lee G H, Cowan N J. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- 3.Farr G W, Scharl E C, Schumacher R J, Sondek S, Horwich A L. Cell. 1997;89:927–937. doi: 10.1016/s0092-8674(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 4.Melki R, Batelier G, Soulie S, Williams R C., Jr Biochemistry. 1997;36:5817–26. doi: 10.1021/bi962830o. [DOI] [PubMed] [Google Scholar]
- 5.Llorca O, Martin-Benito J, Grantham J, Ritco-Vonsovici M, Willison K R, Carrascosa J L, Valpuesta J M. EMBO J. 2001;20:4065–4075. doi: 10.1093/emboj/20.15.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall J, Tempst P, Hartl F. EMBO J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yaffe M B, Farr G W, Miklos D, Horwich A L, Sternlicht M L, Sternlicht H. Nature (London) 1992;358:245–248. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]
- 8.Feldman D E, Thulasiraman V, Ferreyra R G, Frydman J. Mol Cell. 1999;4:1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 9.Thulasiraman V, Yang C F, Frydman J. EMBO J. 1999;18:85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miles M F, Barhite S, Sganga M, Elliott M. Proc Natl Acad Sci USA. 1993;90:10831–10835. doi: 10.1073/pnas.90.22.10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer P H, Muller S, Puzicha M, Pippig S, Obermaier B, Helmreich E J, Lohse M J. Nature (London) 1992;358:73–76. doi: 10.1038/358073a0. [DOI] [PubMed] [Google Scholar]
- 12.Lee R H, Ting T D, Lieberman B S, Tobias D E, Lolley R N, Ho Y K. J Biol Chem. 1992;267:25104–25112. [PubMed] [Google Scholar]
- 13.Hawes B E, Touhara K, Kurose H, Lefkowitz R J, Inglese J. J Biol Chem. 1994;269:29825–29830. [PubMed] [Google Scholar]
- 14.Hekman M, Bauer P H, Sohlemann P, Lohse M J. FEBS Lett. 1994;343:120–124. doi: 10.1016/0014-5793(94)80302-1. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida T, Willardson B M, Wilkins J F, Jensen G J, Thornton B D, Bitensky M W. J Biol Chem. 1994;269:24050–24057. [PubMed] [Google Scholar]
- 16.Schroder S, Lohse M J. Proc Natl Acad Sci USA. 1996;93:2100–2104. doi: 10.1073/pnas.93.5.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thibault C, Sganga M W, Miles M F. J Biol Chem. 1997;272:12253–12256. doi: 10.1074/jbc.272.19.12253. [DOI] [PubMed] [Google Scholar]
- 18.Thulin C D, Howes K, Driscoll C D, Savage J R, Rand T A, Baehr W, Willardson B M. Mol Vis. 1999;5:40. [PubMed] [Google Scholar]
- 19.Zhu X, Craft C M. Mol Vis. 1998;4:13. [PubMed] [Google Scholar]
- 20.Barhite S, Thibault C, Miles M F. Biochim Biophys Acta. 1998;1402:95–101. doi: 10.1016/s0167-4889(97)00141-9. [DOI] [PubMed] [Google Scholar]
- 21.Craft C M, Lolley R N, Seldin M F, Lee R H. Genomics. 1991;10:400–409. doi: 10.1016/0888-7543(91)90325-9. [DOI] [PubMed] [Google Scholar]
- 22.Reig J A, Yu L, Klein D C. J Biol Chem. 1990;265:5816–5824. [PubMed] [Google Scholar]
- 23.Lazarov M E, Martin M M, Willardson B M, Elton T S. Biochim Biophys Acta. 1999;1446:253–264. doi: 10.1016/s0167-4781(99)00098-6. [DOI] [PubMed] [Google Scholar]
- 24.Flanary P L, DiBello P R, Estrada P, Dohlman H G. J Biol Chem. 2000;275:18462–18469. doi: 10.1074/jbc.M002163200. [DOI] [PubMed] [Google Scholar]
- 25.Savage J R, McLaughlin J N, Skiba N P, Hamm H E, Willardson B M. J Biol Chem. 2000;275:30399–30407. doi: 10.1074/jbc.M005120200. [DOI] [PubMed] [Google Scholar]
- 26.Chappell T G, Konforti B B, Schmid S L, Rothman J E. J Biol Chem. 1987;262:746–751. [PubMed] [Google Scholar]
- 27.Melki R, Cowan N J. Mol Cell Biol. 1994;14:2895–2904. doi: 10.1128/mcb.14.5.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willardson B M, Pou B, Yoshida T, Bitensky M W. J Biol Chem. 1993;268:6371–6382. [PubMed] [Google Scholar]
- 29.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Chait B T. Anal Chem. 2000;72:2482–2489. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- 31.Gebauer M, Melki R, Gehring U. J Biol Chem. 1998;273:29475–29480. doi: 10.1074/jbc.273.45.29475. [DOI] [PubMed] [Google Scholar]
- 32.Frydman J, Erdjument-Bromage H, Tempst P, Hartl F U. Nat Struct Biol. 1999;6:697–705. doi: 10.1038/10754. [DOI] [PubMed] [Google Scholar]
- 33.Rosenblatt J, Peluso P, Mitchison T J. Mol Biol Cell. 1995;6:227–236. doi: 10.1091/mbc.6.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roobol A, Carden M J. Eur J Cell Biol. 1999;78:21–32. doi: 10.1016/S0171-9335(99)80004-1. [DOI] [PubMed] [Google Scholar]
- 35.Mittal R, Cerione R A, Erickson J W. Biochemistry. 1994;33:10178–10184. doi: 10.1021/bi00199a046. [DOI] [PubMed] [Google Scholar]
- 36.Schroder S, Lohse M J. Naunyn Schmiedebergs Arch Pharmacol. 2000;362:435–439. doi: 10.1007/s002100000298. [DOI] [PubMed] [Google Scholar]
- 37.Gaudet R, Savage J R, McLaughlin J N, Willardson B M, Sigler P B. Mol Cell. 1999;3:649–660. doi: 10.1016/s1097-2765(00)80358-5. [DOI] [PubMed] [Google Scholar]
- 38.Gaudet R, Bohm A, Sigler P B. Cell. 1996;87:577–588. doi: 10.1016/s0092-8674(00)81376-8. [DOI] [PubMed] [Google Scholar]
- 39.Dyson H J, Wright P E. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]