Abstract
Maytansinoids are potent antitumor agents found in plants and microorganisms. To elucidate their biosynthesis at the biochemical and genetic level and to set the stage for their structure modification through genetic engineering, we have cloned two gene clusters required for the biosynthesis of the maytansinoid, ansamitocin, from a cosmid library of Actinosynnema pretiosum ssp. auranticum ATCC 31565. This is a rare case in which the genes involved in the formation of a secondary metabolite are dispersed in separate regions in an Actinomycete. A set of genes, asm22–24, asm43–45, and asm47, was identified for the biosynthesis of the starter unit, 3-amino-5-hydroxybenzoic acid (AHBA). Remarkably, there are two AHBA synthase gene homologues, which may have different functions in AHBA formation. Four type I polyketide synthase genes, asmA–D, followed by the downloading asm9, together encode eight homologous sets of enzyme activities (modules), each catalyzing a specific round of chain initiation, elongation, or termination steps, which assemble the ansamitocin polyketide backbone. Another set of genes, asm13–17, encodes the formation of an unusual “methoxymalonate” polyketide chain extension unit that, notably, seems to be synthesized on a dedicated acyl carrier protein rather than as a CoA thioester. Additional ORFs are involved in postsynthetic modifications of the initial polyketide synthase product, which include methylations, an epoxidation, an aromatic chlorination, and the introduction of acyl and carbamoyl groups. Tentative functions of several asm genes were confirmed by inactivation and heterologous expression.
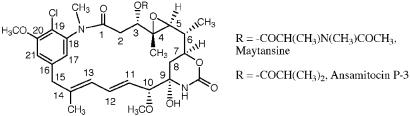
The maytansinoids (Fig. 1) are extraordinarily potent antitumor agents that were originally isolated from members of the higher plant families Celastraceae, Rhamnaceae, and Euphorbiaceae, as well as some mosses, under the auspices of the U.S. National Cancer Institute (1–5). They are 19-membered macrocyclic lactams related to ansamycin antibiotics of microbial origin, such as rifamycin B and geldanamycin (6). The similarity stimulated a search for maytansinoid-producing microorganisms, leading to the isolation of the ansamitocins (Fig. 1) from the Actinomycete Actinosynnema pretiosum ssp. pretiosum and a mutant strain Actinosynnema pretiosum ssp. auranticum (7, 8). Both the structures and antitumor activity of the ansamitocins are similar to those of the maytansinoids from plant sources. A number of structural variations are encountered naturally, including different ester side chains at C-3, and the presence or absence of the 4,5-epoxide, the N-methyl group, the halogen, and oxygens at C-15 and at the C-14 methyl group (9).
Figure 1.
Structures of maytansinoids.
Efforts to develop maytansine into a clinically useful anticancer drug proved disappointing in phase II clinical trials (10), probably because of dose-limiting toxicity in humans, but there is continuing interest in the development of conjugates of maytansinoids for targeted delivery (11, 12). Extensive work by Kupchan et al. (13) and at the Takeda Company (9, 10, 14, 15) to define structure-activity relationships among maytansinoids relied entirely on semisynthesis from the available natural products. For this reason, many backbone alterations of the parent molecular structure could thus far not be explored.
Based on the results of feeding experiments with 13C- and 14C-labeled precursors (16, 17), the biosynthesis of the maytansinoids can be predicted to involve the assembly of the carbon framework on a type I modular polyketide synthase (PKS) from 3-amino-5-hydroxybenzoic acid (AHBA) as the starter unit. Chain extension proceeds by incorporation of three propionates, three acetates, and an unusual hydroxylated two-carbon extender unit (“glycolate” unit) to give a 19-membered macrocyclic lactam. This initial proansamitocin then undergoes a series of post-PKS modifications, which introduce three methyl groups, a halogen, a carbamoyl group, an epoxide, and an ester side chain. In the present article, we report the cloning, sequencing, and characterization of the ansamitocin biosynthetic gene cluster (asm) from A. pretiosum. This work opens the way for a detailed analysis of ansamitocin biosynthesis at the genetic and biochemical level and sets the stage for the genetic engineering of chemically inaccessible, backbone-modified maytansinoid analogs.
Materials and Methods
Materials and General Methods.
A. pretiosum ssp. auranticum 31565 was obtained from the American Type Culture Collection. For ansamitocin production, the strain was cultivated in yeast/malt/glucose (YMG) medium containing 0.4% yeast extract, 1% malt extract, and 0.4% glucose at pH 7.3. Escherichia coli strains XL1-Blue MRF′ (Stratagene) and ET12567/pUZ8002 (18) were used throughout the study as a cloning host and transient host for conjugation sets, respectively. Conjugations between E. coli and A. pretiosum were performed as described by Kieser et al. (19). The freshly cultured A. pretiosum mycelia and the overnight-grown E. coli cells were mixed and plated on YMG agar plates supplemented with 10 mM MgCl2. After incubation at 37°C for 16 h, the plates were overlaid with 1 ml of deionized water containing 1 mg of nalidixic acid (Sigma) and 10 μl of a dimethyl sulfoxide solution of thiostrepton (50 mg/ml; Sigma). A. pretiosum conjugant colonies were selected by further incubation at 28°C for 5–7 days. A. pretiosum protoplasts were prepared in P buffer (19) containing lysozyme (10 mg/ml; Sigma) at 37°C for 1 h. The polyethylene glycol-based transformation (19) was carried out with denatured plasmid DNA. Additional details regarding methods are published as supporting information on the PNAS web site, www.pnas.org.
Vectors, DNA Manipulation, and Cosmid Library Construction.
A. pretiosum ssp. auranticum ATCC 31565 chromosomal DNA was partially digested with Sau3AI, dephosphorylated and ligated into SuperCos 1 (Stratagene), pretreated with XbaI, dephosphorylated, and restricted with BamHI. Packaging the ligated mixture with Gigapack III Gold (Stratagene) and transducing into E. coli SURE (Stratagene) generated the genomic library. The entire asm gene cluster was obtained through sequential chromosome walking to extend the primary cosmid clones containing the rifK homologues.
asm Gene Inactivations.
A 1.1-kbp DNA fragment of pIJ101 carrying the tsr gene for thiostrepton resistance (21) and the 0.7-kbp RK2 replication oriT origin (22) were routinely used as the selection marker and for gene-disruption constructs. The target genes or DNA fragments containing the regions to be deleted (Fig. 2A) were D1 deletion (S16–24)—the cosmid 8C2 was cut with SacI and ligated, followed by insertion of the tsr-oriT cassette from pHGF9027 (L.B., T.-W.Y., and H.G.F., unpublished data) to create pHGF9029; D2 deletion (S22–27)—the 4.6-kbp EcoRI (no. 2)-SacI (no. 22) and 5.2-kbp SacI (no. 27)-EcoRI (no. 3) DNA fragments were ligated with the EcoRI-pretreated pDH5 (23) to create pHGF9011; and D3 deletion (asmB)—to truncate asmB, the 2.5-kbp KpnI-EcoRI and 3.5-kbp EcoRI-HindIII inserts that contain the 5′ and 3′ end sequences of asmB (Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org), which were recovered from a set of subclones of pDDc7, religated to generate a 6.0-kbp KpnI-HindIII fragment carrying the truncated asmB in which the internal 2,923-aa residues were deleted and replaced by five amino acids, P-S-N-S-I, which were introduced with the cloning linker, 5′-CCA-TCG-AAT-TCG-ATC-3′ and cloned into pDH5 pretreated with KpnI/HindIII. The oriT fragment was further added to the resulting clone to create pHGF9001. All of the constructs, pHGF9001, -9027, and -9029 were delivered into A. pretiosum by either polyethylene glycol-based transformation or conjugation with E. coli. The thiostrepton-resistant recombinants resulting from the homologous recombination between the delivered DNA vector and the wild-type A. pretiosum were selected, transferred to TSB broth (Difco) for three more rounds of relaxed cultivation, and screened for thiostrepton-sensitive recombinants derived from a second crossover event. Plasmid integrations into the chromosome as well as the thiostrepton-sensitive recombinants were confirmed by Southern blot analysis of total genomic DNA.
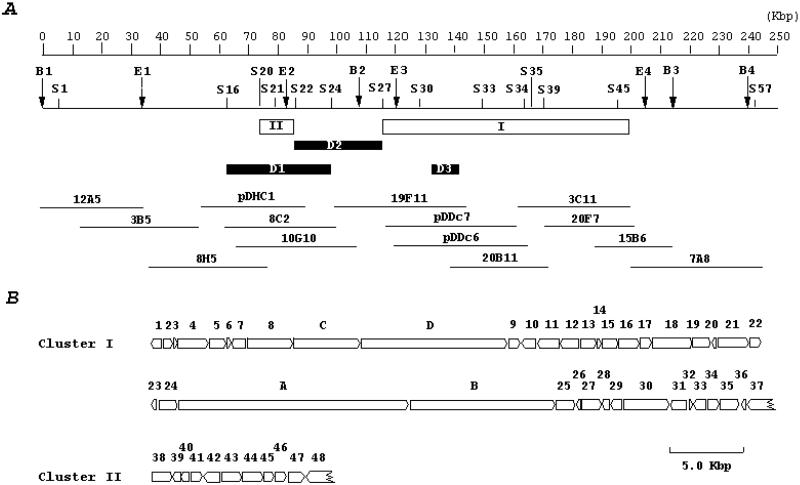
Figure 2.
Organization of the ansamitocin biosynthetic gene cluster. (A) Restriction map of the genomic region and overlapping inserts of 14 cosmid clones used for mapping and sequencing. BglII (B1–4), EcoRI (E1–4), and SacI (S1–57) restriction sites are indicated on the map. Clusters I and II, which have been completely sequenced, are indicated by rectangles. The regions D1, D2, and D3 were deleted in the mutants HGF056, HGF057, and HGF051, respectively. (B) The direction of transcription and the relative sizes of the ORFs deduced from analysis of the nucleotide sequence in clusters I and II. Letters or numbers above the arrows relate to ORFs and asm gene products listed in Table 2.
Constructs for Expression of the asmA and asmAB Genes in Streptomyces coelicolor.
The N-terminal 383 amino acids of the asmA-coding region were PCR-amplified with sense primer MAY003PacI, 5′-CATCGATTAATTAACGGAGAGGCCATATGCTGCGAAGCGACCTGA TCCGTCCC-3′, and antisense primer MAY004KpnI, 5′-TGCGGTACCAGCCGTCGCGCA GC-3′ (restriction sites are underlined in the primers), using pDDc7 as a template. The entire asmA was reassembled by ligating the 1.2-kbp PacI/KpnI-restricted PCR product with the 12.9-kbp KpnI/EcoRI fragment carrying the C-terminal asmA from pDDc7 and cloned downstream of the pactI promoter in PacI/EcoRI-restricted pHGF7505 (24) to yield pHGF7544. The 15.8-kbp HindIII/EcoRI insert carrying the asmA and actII-orf4 regulatory genes from pHGF7544 was relocated and replaced the 9.4-kbp HindIII/EcoRI insert carrying the AHBA synthesis genes in the E. coli-Streptomyces bifunctional plasmid pHGF7543, a pHGF7612 (24) derivative in which the tsr gene has been inactivated by the insertion of an apramycin-resistance aac(3)IV gene (25) to yield pHGF7545. With the same strategy, pHGF7547 was constructed to assemble the entire asmAB by replacing the 12.9-kbp KpnI/EcoRI insert of pHGF7545 with the 22.1-kbp KpnI/EcoRI fragment carrying the C-terminal asmA and asmB from pDDc7.
Ansamitocin Detection and Analysis.
Ansamitocins and related compounds were extracted from the culture broth with ethyl acetate and analyzed by bioassay or isolated by HPLC (Beckman System Gold, C18 reverse-phase column, MeOH/water gradient, diode array detector at λ = 234 and 280 nm) and analyzed by electrospray MS (Bruker Esquire ion trap mass spectrometer, positive ion mode). Inhibitory activity was assayed against the growth of Penicillium avellaneum. Liquid chromatography (LC) MS analyses were carried out with a Shimadzu LC-10AD pump connected to a Micromass (Manchester, U.K.) Quattro II mass spectrometer.
Results
Identification and Cloning of the Ansamitocin Biosynthetic Gene Cluster.
Heterologous hybridization was used to identify genes for AHBA and ansamitocin biosynthesis in A. pretiosum ssp. auranticum ATCC 31565. Initial Southern blot analysis with the rifK gene, encoding the rifamycin AHBA synthase from Amycolatopsis mediterranei S699 (26), revealed two separate rifK-homologous DNA fragments in the genome. A total of 250 kbp of contiguous DNA was cloned and mapped with restriction enzymes BglII, EcoRI, and SacI (S), locating the rifK homologues at two regions, S20–22 and S33–34 respectively (Fig. 2A), 68 kbp apart from each other. Homologues of five other AHBA biosynthesis genes from the rifamycin gene cluster (rif) (24, 27) were also present and mapped at fragments S20–21 (rifG, -L, and -M homologues) and S33–34 (rifJ and -N homologues).
Two gene disruptions were carried out to address the essential role of the AHBA gene homologues and to define the boundary of the biosynthetic gene cluster. The mutant strain HGF056, in which a 35.3-kbp SacI DNA fragment (S16–24) carrying the rifK–M and -G homologues had been deleted from the genome (Fig. 2A), was no longer able to produce any ansamitocins, but production could be restored by supplementation with AHBA (Table 1). There was no significant effect on ansamitocin production in the mutant HGF057, which carried a 30.2-kbp SacI DNA fragment deletion (S22–27) in the region between the two rifK homologues (Fig. 2A). Flanking the second rifK homologue and its associated AHBA synthesis genes (S33–34), two large contiguous regions, spanning S30–35 and S39–45, showed strong signals of hybridization to type I PKS probes. Between the two PKS regions, random sequencing detected homologues of a carbamoyl transferase (29), an acyltransferase (AT) (30), and a halogenase (31).
Table 1.
A. pretiosum mutants
| Strain | Mutant characteristic | Ansamitocin P-3 production | Ansamitocin derivatives accumulated | Ref. |
|---|---|---|---|---|
| HGF051 | asmB deletion | No | No | This study |
| HGF052 | asm19 deletion | No | N-desmethyl-desepoxy-maytansinol | 28 |
| HGF053 | asm15: aac(3)IV | No | 10-Desmethoxyansamitocin P-3 | 44 |
| HGF054 | asm12:aac(3)IV | No | 19-Deschloro-ansamitocins | * |
| HGF056 | Region D1 deletion | No | No | This study |
| HGF057 | Region D2 deletion | Yes | Yes | This study |
L.B., P. Spitelled, S.J.M., S.T., T.-W.Y., and H.G.F., unpublished data.
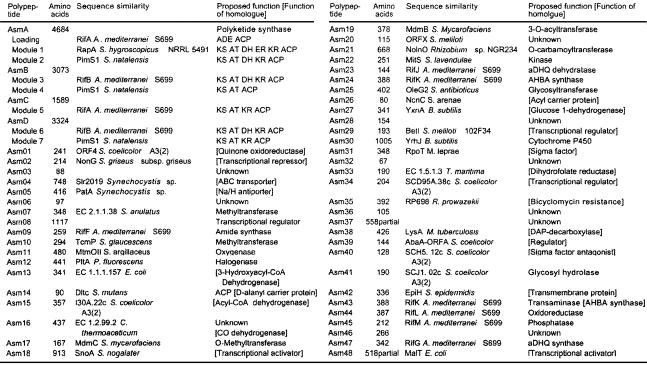
The complete nucleotide sequence of two clusters within the mapped 250-kbp region was then determined and revealed the ansamitocin biosynthetic genes. Clusters I (GenBank accession no. AF453501) and II (GenBank accession no. U33059) were shown to contain between them 50 complete and two partial ORFs that span 96 kbp (Fig. 2 A and B). The genes sequenced and deduced functions of their products are listed in Table 2.
Table 2.
Deduced functions of ORFs in the ansamitocin biosynthetic gene cluster
ADE, carboxylic acid: ACP ligase; KS, β-ketoacyl-ACP synthase; DH, β-hydroxyacyl-thioester dehydratase; ER, enoyl reductase; KR, β-ketoacyl-ACP reductase.
The asm PKS Genes.
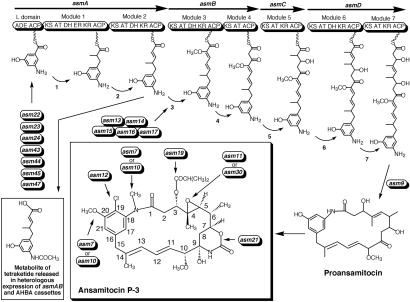
Four large ORFs, asmA–D, encode the loading domain and seven chain-extension modules of a multifunctional PKS. AsmA (483.1 kDa) contains a chain-initiation domain, consisting of an acyl carrier protein (ACP) and an adenyltransferase (32), presumably required for the activation of the starter unit, AHBA, and the first two expected modules for ansamitocin polyketide chain extension. AsmB (313.2 kDa), AsmC (164.6 kDa), and AsmD (339.3 kDa) contain the next five modules required to complete the polyketide portion of ansamitocin. The AT domain of module 3 of AsmB, which is predicted to recognize the “glycolate” extender unit, shows no unusual sequence signature that would distinguish it from other AT domains. Located immediately downstream of asmD is asm9, which encodes a protein with a high degree of similarity to RifF, the chain-terminating, cyclizing enzyme of rifamycin biosynthesis (33, 34). It is proposed that the C terminus of the fully extended polyketide chain is transferred from AsmD to the conserved Cys-73 on the Asm9 protein, followed by intramolecular amide bond formation with the aromatic amino group to release a 19-membered macrocyclic lactam, proansamitocin (Fig. 3).
Figure 3.
Domain organization of the asm PKS and biosynthetic model for ansamitocin P-3 formation. Each module incorporates the essential KS, AT, and ACP domains, whereas all but one include optional modifying activities (KR, DH, ER). The abbreviations of domain designations are ADE, carboxylic acid: ACP ligase; KS, β-ketoacyl-ACP synthase; DH, β-hydroxyacyl-thioester dehydratase; KR, β-ketoacyl-ACP reductase; ER, enoyl reductase. The putative intermediates in chain-extension cycles and the asm genes involved in the various biosynthetic steps are indicated.
To probe the function of the identified PKS genes in ansamitocin biosynthesis, we constructed the mutant HGF051, in which 2,923 amino acid residues were deleted from the coding region of asmB and replaced by five amino acids, P-S-N-S-I (see Materials and Methods). The mutant HGF051 failed to produce ansamitocin P-3 or any known related compounds based on bioassay, HPLC, and liquid chromatography MS analysis. The functional competence of the asm PKS genes was probed further by heterologous expression under the control of the bidirectional actI/actIII promoters and the actII-ORF4 regulator in S. coelicolor YU105 (24). The cotransformants of pHGF7612 (24), carrying all of the AHBA synthesis genes from the rif cluster, and pHGF7545, carrying asmA, accumulated no polyketide-related intermediate. Notably, however, coexpression of the AHBA genes with the asmAB cassette, pHGF7547, gave a triketide, isolated as its N-acetyl metabolite, with a structure matching that expected for the product assembled by the AsmA protein alone (Fig. 3).
The asm AHBA Biosynthesis Genes.
Seven AHBA biosynthetic genes similar to those cloned from the rifamycin, mitomycin, ansatrienin, and naphthomycin clusters (27, 35, 36) were identified in two groups (Fig. 2B). asm22, asm23, and asm24 were found in close proximity to asmAB. asm24, one of the two rifK homologues, is located upstream of asmA and may be cotranscribed with the asmAB PKS genes. Its product presumably catalyzes the dehydration of 5-deoxy-5-amino-3-dehydroshikimate (aminoDHS) to AHBA (26). asm23, located upstream of asm24 and transcribed in the opposite direction, encodes a type II dehydroquinate dehydratase homologous to RifJ, which catalyzes the dehydration of 5-deoxy-5-amino-3-dehydroquinate (aminoDHQ) to aminoDHS (24). Asm22, related to MitS and RifN (24, 27, 35), shows significant homology with a bacterial glucose kinase involved in glucose repression (37, 38). The genes asm43-47 are apparently organized into a single operon. With the exception of asm46, which shows no sequence similarity with any known gene, their deduced proteins exhibit high homology with known products of AHBA biosynthesis genes. In addition to the second AHBA synthase, Asm43 ∼ RifK, they include Asm44 ∼ RifL, related to a class of sugar oxidoreductases (39), Asm45 ∼ RifM, similar to a phosphoglycolate phosphatase involved in glycolate oxidation (40), and Asm47 ∼ RifG, homologous to dehydroquinate synthases of the shikimate pathway, respectively. Interestingly, both AHBA synthase genes were shown to be functionally competent: asm43 by expression in E. coli and demonstration of AHBA synthase activity and asm24 by complementation of a rifK mutant of A. mediterranei to restore rifamycin production (D.H., T.-W.Y., E.L. and H.G.F., unpublished data.). Surprisingly, despite extensive sequencing and Southern hybridization analysis, the 3,4-dideoxy-4-amino-d-arabino-heptulosonate 7-phosphate (aminoDAHP) synthase gene corresponding to rifH required in the de novo AHBA biosynthesis (24) was not found in the cloned 250-kbp DNA region containing the asm cluster.
Genes Involved in the Biosynthesis of the Unusual “Glycolate” Extender Unit.
Four contiguous genes, asm13–16, were identified 4.3 kbp downstream of asmCD-9. The deduced products of asm13 and asm15 show similarities to an NAD+-dependent 3-hydroxybutyryl-CoA dehydrogenase and an acyl-CoA dehydrogenase, respectively (41, 42). asm13 and asm14 may be translationally coupled as the putative ATG start codon of asm14 overlaps with the TGA stop codon of asm13. The asm14 product (9.7 kDa) resembles acyl carrier proteins, particularly D-alanyl carrier proteins operating in D-alanyl-lipoteichoic acid synthesis (43), and has been shown by gene inactivation to be essential for ansamitocin biosynthesis (44). Asm16 is related only to a protein of unknown function encoded by a gene in the fatty acid biosynthetic gene cluster of Bacillus halodurans (45).
A mutant, HGF053, in which the asm15 gene was interrupted by inserting the aac(3)IV gene, showed no ansamitocin production, even in the presence of the side chain precursor, isobutyrate (44), ruling out the involvement of asm15 in the formation of the C-3 ester side chain. However, HPLC, electrospray MS, and NMR analysis revealed the presence of a small amount of 10-desmethoxyansamitocin P-3 in the fermentation of HGF053 (44). Therefore, the asm13–16 subcluster, possibly additionally including asm17, may be responsible for generating the “glycolate” extender unit, which is incorporated at C-9 and C-10 of ansamitocin. asm17 is probably cotranscribed with asm13–16 and encodes an O-methyltransferase. The biosynthetic gene cluster for FK520, a macrolide also involving “glycolate” extender units, contains a set of five genes, fkbGHIJK, which are homologous to asm13–17 (46). This fact further supports the notion that asm13–17 are involved in the synthesis of this unusual polyketide chain extension unit.
Genes for the Postsynthetic Modification of Proansamitocin.
The asm cluster contains candidate genes for all of the steps required to elaborate the final ansamitocin structure from proansamitocin. asm19 is related to mdmB from Streptomyces mycarofaciens and acyA from Streptomyces thermotolerans, which encode macrolide O-acyltransferases (30, 47). The functional assignment as an acyltransferase attaching the C-3 ester group of ansamitocin was verified by inactivating asm19 through an internal 549-bp in frame deletion. The mutant, HGF052, produced neither ansamitocin P-3 nor the corresponding alcohol, maytansinol. Instead, it synthesized a compound identified as N-desmethyl-desepoxymaytansinol (28), indicating that acylation is not the final step in ansamitocin biosynthesis.
Asm21 is highly similar to the nodulation proteins of Rhizobium sp. (29) and Bradyrhizobium japonicum (48), as well as the products of cmcH of Streptomyces clavuligerus and novN of Streptomyces spheroides, which carry out the O-carbamoylation steps in cephamycin and novobiocin biosynthesis (49, 50). Therefore, asm21 is assigned the putative function of introducing the cyclic carbinolamide group of ansamitocin.
The asm30 gene product strongly resembles the bifunctional P450-NADPH:P450 reductase P450BM-3, a fatty acid monooxygenase from Bradyrhizobium megaterium (51, 52). The heme-P450 and the FMN/FAD-containing reductase domains are linked together on a single 108.6-kDa polypeptide. P450BM-3 performs as a self-sufficient fatty acid hydroxylase, converting lauric, myristic, and palmitic acids to omega-1, -2, and -3 hydroxy analogs. By mechanistic analogy, Asm30 is considered to be responsible for the epoxidation step in ansamitocin formation.
Ansamitocin biosynthesis requires three methylations, of the nitrogen and of the oxygens at C-10 and C-20, all using S-adenosyl-L-methionine (AdoMet) as the methyl donor (17). Three AdoMet-dependent methyltransferase genes, asm7, asm10, and asm17, were identified in the asm cluster. asm17, which is homologous to several O-methyltransferase genes (30, 47, 53), because of its linkage to asm13–16, is assigned to the O-methylation at C-10. Asm7 is related to Streptomyces antibiotic biosynthetic O-methyltransferases (35, 54, 55) and may be responsible for either the N-methylation or the C-20 O-methylation. Asm10 has significant similarity to proteins of unknown function from Mycobacterium tuberculosis and S. coelicolor and shares weak similarity with TcmP, an O-methyltransferase involved in tetracenomycin C synthesis in Streptomyces glaucescens (56).
The asm10 gene seems to be polycistronically transcribed with asm11 and asm12. The deduced products of asm11 and asm12 show similarity to Cts8 and Cts4 of Streptomyces aureofaciens, responsible for tetracycline 6-hydroxylation and ring chlorination, respectively (31). The function of Asm11 is unclear, but it may be an alternative candidate to Asm30 to catalyze the epoxidation at C-4/C-5. The assignment of Asm12 as a halogenase introducing the chlorine at C-19 was confirmed by inactivation of asm12 to give a mutant, HGF054, which no longer produces ansamitocin P-3 (L.B., P. Spiteller, S.J.M., S.T., T.-W.Y., and H.G.F., unpublished work). HGF054 accumulates a series of 19-deschloro-ansamitocin derivatives, rather than a single compound, suggesting that the presence of the halogen is not an absolute requirement for some of the other post-PKS modification reactions.
Discussion
The strategy for cloning the asm gene cluster from A. pretiosum relied on earlier work on rifamycin biosynthesis, using the unique rifK and other AHBA synthesis genes as probes to identify the asm genes. Surprisingly, two rifK homologues, located 68 kbp apart, were found in A. pretiosum, neither one accompanied by a full set of the genes required for AHBA formation (24). The rifK homologue in cluster I is associated with an almost full complement of genes expected for ansamitocin biosynthesis (Fig. 2). These include genes for a complete type I PKS (asmA–D) and its downloading enzyme (asm9) and candidate genes for all of the predicted downstream processing reactions. In addition, there are several possible regulatory and transport genes, and a glycosyltransferase gene (asm25), which together with asm41 may be part of a glycosylation/deglycosylation excretion system (57).
Absent from cluster I are the rifL and rifM homologues, asm44 and asm45, encoding an oxidoreductase and a phosphatase, respectively, which are essential for AHBA formation (24), as well as a 5-deoxy-5-amino-3-dehydroquinate synthase gene (asm47); these are associated with the second rifK homologue in cluster II. It follows that both clusters I and II must be required for ansamitocin formation, and this was confirmed by deletions of the asmB gene from cluster I and of a large DNA fragment carrying cluster II, respectively. Because AHBA restores ansamitocin production in the cluster II deletion mutant, HGF056, cluster II contains genes required only for the formation of the starter unit, AHBA. Apparently, the 30 kbp of DNA located between the two clusters carry no genes essential for either ansamitocin formation or growth, because deletion of this region, including parts of asm37 and asm38, in mutant HGF057 caused no discernible phenotypic changes. It is not clear why the asm genes are split into two separate clusters; possibly they were at one time present as a single cluster and were later separated by a reorganization of the genome.
There are indications that RifK may have two enzymatic activities, the well-characterized dehydratase activity aromatizing 5-deoxy-5-amino-3-dehydroshikimate (26) and an aminotransferase activity introducing the nitrogen into a carbohydrate precursor of aminoDAHP (24). The presence of two rifK homologues in the asm gene cluster is consistent with this notion. Although both gene products are functionally competent as AHBA synthases, they may be optimized for different reactions in AHBA biosynthesis, one for introducing the nitrogen and the other for the later aromatization reaction. Interestingly, no rifH homologue is present in the asm gene cluster. The absence of an aminoDAHP synthase gene has also been reported for the mitomycin C biosynthetic gene cluster from Streptomyces lavendulae (35). These findings suggest that the corresponding shikimate pathway enzyme, DAHP synthase, may participate in the formation of aminoDAHP. Indeed, one plant-type DAHP synthase gene has been identified in A. pretiosum and shown to not be linked to the asm cluster (T.-W.Y. and H.G.F., unpublished data).
The formation of the polyketide backbone of the ansamitocins is catalyzed by four genes of 38 kbp encoding a type I PKS with an aromatic loading domain closely resembling that of the rif cluster (27, 58) and seven chain-extension modules. Each module contains the predicted functional domains for chain extension and chain modification, in an arrangement matching that in other type I PKSs analyzed (59), and the modules are arranged colinear with their function in the biosynthetic assembly process (Fig. 3). The PKS genes asmAB are separated from asmCD by a set of other asm genes involved in substrate supply for chain initiation and extension and in downstream processing. The last PKS gene is followed by a gene encoding an amide synthase that is recognized from work on rifamycin as the “downloading” enzyme (33, 36), catalyzing the release of the completed polyketide chain from the PKS and cyclizing it to a macrocyclic lactam. Therefore, AsmABCD + 9 constitute the minimal enzymatic machinery required to catalyze the formation of a hypothetical proansamitocin, the first complete, cyclic polyketide precursor of ansamitocin.
The polyketide synthesized by the asm PKS shows two unusual features. One is the incorporation of a rare hydroxylated 2-carbon extender unit in the third chain-extension step. Such “glycolate” units are found in a number of other antibiotics, such as geldanamycin (60), leucomycin (61), soraphen (62), FK520 and FK506 (63), and concanamycin A (64). Their origin is not clear from the various feeding experiments reported (17, 60–65). The C-2 hydroxy group of this extender unit is usually but not always (65) methylated. A subcluster of five genes, asm13–17, which form an operon, is evidently responsible for the formation of the substrate for this particular chain-extension step, as has been proposed earlier for the homologous genes fkbG–K in the FK520 cluster (46). In recent work, coexpression in Streptomyces lividans of asm13–17 with a cassette of eryABC, encoding the erythromycin PKS, in which the AT6 had been replaced with the hydroxymalonate-specifying AT8 from the FK520 PKS, resulted in the formation of 2-desmethyl-2-methoxy-6-deoxyerythronolide B by incorporation of a methoxymalonate instead of a methylmalonate unit, whereas asm13-16 in place of asm13–17 gave no new product (66). Based on the available evidence, it is proposed that the substrate for this unusual chain-extension reaction is 2-methoxymalonyl-ACP, which is synthesized by the asm13,15–17 gene products on the activated (20) ACP, Asm14. This ACP, which is essential for ansamitocin formation (44), shows greater sequence similarity with ACPs in nonribosomal peptide synthases than in PKSs.
A second unusual feature of the ansamitocin structure is the position of the double bonds in the ansa ring, at the C-11 and C-13 positions rather than at C-10 and C-12 where normal PKS-processing would place them. This double-bond shift may result from an isomerization of the completed polyketide or it may be a part of the polyketide assembly process. The modifying domains in modules 2 and 3 of the asm PKS, or in any module, show no evidence for unusual catalytic functions. In particular, DH3 has the sequence signature of a normal dehydratase domain. There are also no obvious candidate genes for such a double-bond isomerization outside the PKS region. The possibility that the double-bond migration is related to the presence of the extra oxygen function at C-10 is ruled out by the formation of 10-desmethoxyansamitocin, with the double bonds in the “abnormal” position, in the asm15 mutant (44).
The precise structure of the first cyclic product of the asm PKS, the hypothetical proansamitocin (Fig. 3), is not certain. It depends on the timing of the double-bond migration and on the substrate for the “glycolate” chain-extension step. We have embarked on a strategy to express the asm PKS genes, together with gene cassettes ensuring precursor supply, in the heterologous host S. coelicolor. A first step is the synthesis of a triketide by a transformant expressing an AHBA synthesis cassette together with asmAB, but not with asmA. The requirement for the presence of the AsmB protein suggests that the product of AsmA can be released from its cognate ACP2 only when it can be transferred to the next β-ketoacyl-ACP synthase but not be processed further because of a lack of substrate (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Linking AsmA to a thioesterase domain eliminates the need for AsmB for product release (T.-W.Y. and H.G.F., unpublished data).
The work reported here sets the stage for the detailed biochemical and genetic analysis of the ansamitocin biosynthetic pathway as a step toward genetic engineering of structurally modified maytansinoids. It also provides tools for the study of the intriguing question of the biosynthetic source of the maytansinoids found in higher plants, i.e., whether the polyketide backbone is synthesized by the plant itself, possibly because of a lateral gene transfer from a microorganism, or whether the plants acquire it from an associated microorganism.
Supplementary Material
Acknowledgments
We are grateful to Takeda Chemical Industries, Ltd. (Osaka), for providing samples of ansamitocin P-2, P-3, P-4, and PND-3. This work was supported by National Institutes of Health Research Grant CA 76461 (to H.G.F. and T.-W.Y.), Deutsche Forschungsgemeinschaft Research Grant Le260/15-2, a grant from the Fonds der Chemischen Industrie (to E.L.), and by North Atlantic Treaty Organization Collaborative Research Grant Sa 5-2-05 9 (to E.L. and H.F.G.).
Abbreviations
- PKS
polyketide synthase
- AHBA
3-amino-5-hydroxybenzoic acid
- ACP
acyl carrier protein
- AT
acyltransferase
- DAHP
3,4-dideoxy-4-amino-d-arabino-heptulosonate 7-phosphates
Footnotes
References
- 1.Kupchan S M, Komoda Y, Court W A, Thomas G J, Smith R M, Karim A, Gilmore C J, Haltiwanger R C, Bryan R F. J Am Chem Soc. 1972;94:1354–1356. doi: 10.1021/ja00759a054. [DOI] [PubMed] [Google Scholar]
- 2. Wani, M. C., Taylor, H. L. & Wall, M. E. (1973) J. Chem. Soc. Chem. Commun., 390.
- 3.Powell R G, Smith C R, Jr, Plattner R D, Jones B E. J Nat Prod. 1983;46:660–666. [Google Scholar]
- 4.Sakai K, Ichikawa T, Yamada K, Yamashita M, Tanimoto M, Hikita A, Ijuin H, Kondo K. J Nat Prod. 1988;51:845–850. doi: 10.1021/np50059a005. [DOI] [PubMed] [Google Scholar]
- 5.Suwanborirux K, Chang C J, Spjut R W, Cassady J M. Experientia. 1990;46:117–120. doi: 10.1007/BF01955435. [DOI] [PubMed] [Google Scholar]
- 6.Rinehart K L, Jr, Shield L S. Fortsch Chem Org Naturst. 1976;33:231–307. doi: 10.1007/978-3-7091-3262-3_3. [DOI] [PubMed] [Google Scholar]
- 7.Higashide E, Asai M, Ootsu K, Tanida S, Kozai Y, Hasegawa T, Kishi T, Sugino Y, Yoneda M. Nature (London) 1977;270:721–722. doi: 10.1038/270721a0. [DOI] [PubMed] [Google Scholar]
- 8.Asai M, Mizuta E, Izawa M, Haibara K, Kishi T. Tetrahedron. 1978;35:1079–1085. [Google Scholar]
- 9.Smith C R, Jr, Powell R G. In: Alkaloids. Pelletier S W, editor. Vol. 2. NY: Wiley; 1984. pp. 149–204. [Google Scholar]
- 10.Reider P J, Roland D M. In: The Alkaloids. Brossi A, editor. Vol. 23. New York: Academic; 1984. pp. 71–156. [Google Scholar]
- 11.Ladino C A, Chari R V, Bourret L A, Kedersha N L, Goldmacher V S. Int J Cancer. 1997;73:859–864. doi: 10.1002/(sici)1097-0215(19971210)73:6<859::aid-ijc16>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Tadayoni B M, Bourret L A, Mattocks K M, Derr S M, Widdison W C, Kedersha N L, Ariniello P D, Goldmacher V S, Lambert J M, et al. Proc Natl Acad Sci USA. 1996;93:8618–8623. doi: 10.1073/pnas.93.16.8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kupchan S M, Sneden A T, Branfman A R, Howie G A, Rebhun L I, McIvor W E, Wang R W, Schnaitman T C. J Med Chem. 1978;21:31–37. doi: 10.1021/jm00199a006. [DOI] [PubMed] [Google Scholar]
- 14.Izawa M, Wada Y, Kasahara F, Asai M, Kishi T. J Antibiot. 1981;34:1591–1595. doi: 10.7164/antibiotics.34.1591. [DOI] [PubMed] [Google Scholar]
- 15.Kawai A, Akimoto H, Kozai Y, Ootsu K, Tanida S, Hashimoto N, Nomura H. Chem Pharm Bull. 1984;32:3341–3351. [PubMed] [Google Scholar]
- 16.Hatano K, Akiyama S-I, Asai M, Rickards R W. J Antibiot. 1982;35:1415–1417. doi: 10.7164/antibiotics.35.1415. [DOI] [PubMed] [Google Scholar]
- 17.Hatano K, Mizuta E, Akiyama S-I, Higashide E, Nakao Y. Agric Biol Chem. 1985;49:327–333. [Google Scholar]
- 18.MacNeil D J, Gewain K M, Ruby C L, Dezeny G, Gibbons P H, MacNeil T. Gene. 1992;111:61–68. doi: 10.1016/0378-1119(92)90603-m. [DOI] [PubMed] [Google Scholar]
- 19.Kieser T, Bibb M J, Buttner M J, Chater K F, Hopwood D A. Practical Streptomyces Genetics. Norwich, U.K.: The John Innes Foundation; 2000. [Google Scholar]
- 20.Lambalot R H, Gehring A M, Flugel R S, Zuber P, LaCelle M, Marahiel M A, Khosla C, Walsh C T. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 21.Kendall K J, Cohen S N. J Bacteriol. 1988;170:4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labigne-Roussel A, Harel J, Tompkins L. J Bacteriol. 1987;169:5320–5323. doi: 10.1128/jb.169.11.5320-5323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hillemann D, Pühler A, Wohlleben W. Nucleic Acids Res. 1991;19:727–731. doi: 10.1093/nar/19.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu T-W, Müller R, Müller M, Zhang X, Draeger G, Kim C-G, Leistner E, Floss H G. J Biol Chem. 2001;276:12546–12555. doi: 10.1074/jbc.M009667200. [DOI] [PubMed] [Google Scholar]
- 25.Blondelet-Rouault M H, Weiser J, Lebrihi A, Branny P, Pernodet J L. Gene. 1997;190:315–317. doi: 10.1016/s0378-1119(97)00014-0. [DOI] [PubMed] [Google Scholar]
- 26.Kim C-G, Yu T-W, Fryhle C B, Handa S, Floss H G. J Biol Chem. 1998;273:6030–6040. doi: 10.1074/jbc.273.11.6030. [DOI] [PubMed] [Google Scholar]
- 27.August P R, Tang L, Yoon Y J, Ning S, Müller R, Yu T-W, Taylor M, Hoffmann D, Kim C-G, Zhang X, et al. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 28. Moss, S. J., Bai, L., Toelzer, S., Carroll, B. J., Mahmud, T., Yu, T.-W. & Floss, H. G. (2002) J. Am. Chem. Soc., in press. [DOI] [PubMed]
- 29.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Nature (London) 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 30.Hara O, Hutchinson C R. J Bacteriol. 1992;174:5141–5144. doi: 10.1128/jb.174.15.5141-5144.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dairi T, Nakano T, Aisaka K, Katsumata R, Hasegawa M. Biosci Biotechnol Biochem. 1995;59:1099–1106. doi: 10.1271/bbb.59.1099. [DOI] [PubMed] [Google Scholar]
- 32.Admiraal S J, Walsh C T, Khosla C. Biochemistry. 2001;40:6116–6123. doi: 10.1021/bi010080z. [DOI] [PubMed] [Google Scholar]
- 33.Yu T-W, Shen Y, Doi-Katayama Y, Tang L, Park C, Moore B S, Hutchinson C R, Floss H G. Proc Natl Acad Sci USA. 1999;96:9051–9056. doi: 10.1073/pnas.96.16.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stratmann A, Toupet C, Schilling W, Traber R, Oberer L, Schupp T. Microbiology. 1999;145:3365–3375. doi: 10.1099/00221287-145-12-3365. [DOI] [PubMed] [Google Scholar]
- 35.Mao Y, Varoglu M, Sherman D H. Chem Biol. 1999;6:251–263. doi: 10.1016/S1074-5521(99)80040-4. [DOI] [PubMed] [Google Scholar]
- 36.Chen S, von Bamberg D, Hale V, Breuer M, Hardt B, Müller R, Floss H G, Reynolds K A, Leistner E. Eur J Biochem. 1999;261:98–107. doi: 10.1046/j.1432-1327.1999.00244.x. [DOI] [PubMed] [Google Scholar]
- 37.Angell S, Schwarz E, Bibb M J. Mol Microbiol. 1992;6:2833–2844. doi: 10.1111/j.1365-2958.1992.tb01463.x. [DOI] [PubMed] [Google Scholar]
- 38.Spath C, Kraus A, Hillen W. J Bacteriol. 1997;179:7603–7605. doi: 10.1128/jb.179.23.7603-7605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loos H, Sahm H, Sprenger G A. FEMS Microbiol Lett. 1993;107:293–298. doi: 10.1111/j.1574-6968.1993.tb06045.x. [DOI] [PubMed] [Google Scholar]
- 40.Schaferjohann J, Yoo J G, Kusian B, Bowlen B. J Bacteriol. 1993;175:7329–7340. doi: 10.1128/jb.175.22.7329-7340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Youngleson J S, Jones W A, Jones D T, Woods D R. Gene. 1989;78:355–364. doi: 10.1016/0378-1119(89)90238-2. [DOI] [PubMed] [Google Scholar]
- 42.Matsubara Y, Indo Y, Naito E, Ozasa H, Glassberg R, Vockley J, Ikeda Y, Kraus J, Tanaka K. J Biol Chem. 1989;264:16321–16331. [PubMed] [Google Scholar]
- 43.Boyd D A, Cvitkovitch D G, Bleiweis A S, Kiriukhin M Y, Debabov D V, Neuhaus F C, Hamilton I R. J Bacteriol. 2000;182:6055–6065. doi: 10.1128/jb.182.21.6055-6065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carroll B J, Moss S J, Bai L, Kato Y, Toelzer S, Yu T-W, Floss H G. J Am Chem Soc. 2002;124:4176–4177. doi: 10.1021/ja0124764. [DOI] [PubMed] [Google Scholar]
- 45.Takami H, Takaki Y, Nakasone K, Hirama C, Inoue A, Horikoshi K. Biosci Biotechnol Biochem. 1999;63:452–455. doi: 10.1271/bbb.63.452. [DOI] [PubMed] [Google Scholar]
- 46.Wu K, Chung L, Revill W P, Katz L, Reeves C D. Gene. 2000;251:81–90. doi: 10.1016/s0378-1119(00)00171-2. [DOI] [PubMed] [Google Scholar]
- 47.Epp J K, Huber M L B, Turner J R, Goodson T, Schoner B E. Gene. 1989;85:293–301. doi: 10.1016/0378-1119(89)90421-6. [DOI] [PubMed] [Google Scholar]
- 48.Luka S, Sanjuan J, Carlson R W, Stacey G. J Biol Chem. 1993;268:27053–27059. [PubMed] [Google Scholar]
- 49.Coque J J R, Pérez-Llarena F J, Enguita F J, Fuente J L, Martin J F, Liras P. Gene. 1995;162:21–27. doi: 10.1016/0378-1119(95)00308-s. [DOI] [PubMed] [Google Scholar]
- 50.Steffensky M, Mühlenweg A, Wang Z-X, Li S-M, Heide L. Antimicrob Agents Chemother. 2000;44:1214–1222. doi: 10.1128/aac.44.5.1214-1222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruettinger R T, Wen L P, Fulco A J. J Biol Chem. 1989;264:10987–10995. [PubMed] [Google Scholar]
- 52.Sevrioukova I F, Li H, Zhang H, Peterson J A, Poulos T. Proc Natl Acad Sci USA. 1999;96:1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martz F, Maury S, Pincon G, Legrand M. Plant Mol Biol. 1998;36:427–437. doi: 10.1023/a:1005969825070. [DOI] [PubMed] [Google Scholar]
- 54.Lacalle R A, Ruiz D, Jimenez A. Gene. 1991;109:55–61. doi: 10.1016/0378-1119(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 55.Madduri K, Torti F, Colombo A L, Hutchinson C R. J Bacteriol. 1993;175:3900–3904. doi: 10.1128/jb.175.12.3900-3904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Decker H, Motamedi H, Hutchinson C R. J Bacteriol. 1993;175:3876–3886. doi: 10.1128/jb.175.12.3876-3886.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vilches C, Hernandez C, Salas J A. J Bacteriol. 1992;174:161–165. doi: 10.1128/jb.174.1.161-165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schupp T, Toupet C, Engel N, Goff S. FEMS Microbiol Lett. 1998;159:201–207. doi: 10.1111/j.1574-6968.1998.tb12861.x. [DOI] [PubMed] [Google Scholar]
- 59.Hopwood D A. Chem Rev (Washington, DC) 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 60.Haber A, Johnson R D, Rinehart K L., Jr J Am Chem Soc. 1977;99:3541–3544. doi: 10.1021/ja00452a079. [DOI] [PubMed] [Google Scholar]
- 61.Omura S, Tsuzuki K, Nakagawa A, Lukacs G. J Antibiot. 1983;36:611–613. doi: 10.7164/antibiotics.36.611. [DOI] [PubMed] [Google Scholar]
- 62. Hill, A. M., Harris, J. P. & Siskos, A. P. (1998) J. Chem. Soc. Chem. Commun., 2361–2362.
- 63.Byrne K M, Shafiee A, Nielsen J B, Arison B, Monaghan R L, Kaplan L. Dev Ind Microbiol. 1993;32:29–45. [Google Scholar]
- 64. Bindseil, K. U. & Zeeck, A. (1994) Liebigs Ann. Chem., 305–312.
- 65.Ono M, Sakuda S, Ikeda H, Furihata K, Nakayama J, Suzuki A, Isogai A. J Antibiot. 1998;51:1019–1028. doi: 10.7164/antibiotics.51.1019. [DOI] [PubMed] [Google Scholar]
- 66.Kato Y, Bai L, Xue Q, Revill W P, Yu T-W, Floss H G. J Am Chem Soc. 2002;124:5268–5269. doi: 10.1021/ja0127483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.