Abstract
Voltage-gated K+ channels control excitability in neuronal and various other tissues. We identified three unique α-subunits of voltage-gated K+-channels in the human genome. Analysis of the full-length sequences indicated that one represents a previously uncharacterized member of the Kv6 subfamily, Kv6.3, whereas the others are the first members of two unique subfamilies, Kv10.1 and Kv11.1. Although they have all of the hallmarks of voltage-gated K+ channel subunits, they did not produce K+ currents when expressed in mammalian cells. Confocal microscopy showed that Kv6.3, Kv10.1, and Kv11.1 alone did not reach the plasma membrane, but were retained in the endoplasmic reticulum. Yeast two-hybrid experiments failed to show homotetrameric interactions, but showed interactions with Kv2.1, Kv3.1, and Kv5.1. Co-expression of each of the previously uncharacterized subunits with Kv2.1 resulted in plasma membrane localization with currents that differed from typical Kv2.1 currents. This heteromerization was confirmed by co-immunoprecipitation. The Kv2 subfamily consists of only two members and uses interaction with “silent subunits” to diversify its function. Including the subunits described here, the “silent subunits” represent one-third of all Kv subunits, suggesting that obligatory heterotetramer formation is more widespread than previously thought.
Keywords: electrically silent subunits‖ER retention‖heterotetrameric assembly‖KCNG3
Voltage-gated potassium channels are transmembrane proteins consisting of four α-subunits that form a central permeation pathway. Each subunit contains six transmembrane domains (S1–S6) and a pore loop containing the GYG-motif, the signature sequence for potassium selectivity. The fourth transmembrane domain (S4) contains positively charged residues and is the major part of the voltage sensor. Voltage-gated potassium channels serve a wide range of functions including regulation of the resting membrane potential and control of the shape, duration, and frequency of action potentials (1–3).
At present, 26 genes have been described encoding for different Kv α-subunits. These are divided into subfamilies by sequence similarities: within a subfamily members share ≈70% of sequence identity, whereas between different subfamilies this percentage drops to ≈40%, reflecting the homology in the core section S1–S6 (4). The Kv family of potassium channels consists of nine subfamilies, Kv1 through Kv9, although Kv7 has only been described for Aplysia (5). The subunits of the Kv1 through Kv4 subfamilies all show functional expression in a homotetrameric configuration. Despite having the typical topology of voltage-gated potassium channel subunits, the subunits of the Kv5 through Kv9 families cannot generate current by themselves (6–10). For instance, Kv6.1 fails to form homotetrameric channels, but it is able to form heterotetrameric channels with Kv2.1; expression of these heterotetramers resulted in currents with clearly distinguishable properties (11). All known “electrically silent” subunits have been shown to form heterotetrameric channels with the members of the Kv2 subfamily (8–10). In a sense, these “silent” subunits can be considered regulatory subunits—e.g., the metabolic regulation of the Kv2.1/Kv9.3 heteromultimer might play an important role in hypoxic pulmonary artery vasoconstriction and in the possible development of pulmonary hypertension (8).
In this study we report the cloning and functional properties of three previously uncharacterized subunits that were identified in the early public draft version of the human genome. Based on sequence identity, one of these is a previously uncharacterized member of the Kv6 subfamily (Kv6.3), whereas the others are the first members of two unique subfamilies, Kv10.1 and Kv11.1. Biochemical, microscopic, and functional analysis indicated that these previously uncharacterized subunits are all “silent subunits,” which may explain why they have not been cloned previously. Through obligatory heterotetramerization they exert a function-altering effect on other Kv subunits.
Experimental Procedures
Cloning of Kv2.1, Kv6.3, Kv10.1, and Kv11.1.
The coding sequence of human Kv2.1 was amplified from a human brain library (CLONTECH) and cloned into pEGFP-N1. The channel sequences of Kv6.3, Kv10.1, and Kv11.1 were obtained through a blast search of the high throughput genomic sequence (htgs) database (July 2000). The coding sequences were cloned using PCR amplification from a human brain library (CLONTECH) or a human testis library (TaKaRa, Shiga, Japan) for Kv10.1 and Kv11.1, respectively. Both coding exons of Kv6.3 were amplified from human genomic DNA. The BsaMI restriction site at the start of the second exon was used to join the two coding exons.
Amino Acid Sequence Alignments and Phylogenetic Tree.
Computer analyses were performed using megalign (DNAstar, Madison, WI). The phylogenetic tree and the percentage of identity were obtained by aligning the core S1–S6 sequences (e.g., aa 252–518 in Kv1.5).
Expression Analysis.
A cDNA panel from different tissues was obtained from CLONTECH (cDNA panel I and II). PCR was performed with primer sets that were selected to ensure the amplification of the correct subunit, without amplification of homologous subunits. All reactions were done with 38 cycles and PCR products were analyzed on a 2% agarose gel.
Transfection.
Ltk− cells were cultured and transfected with cDNA as reported (12). Each subunit was coexpressed with Kv2.1 (10:1 ratio). At this ratio, less than 0.01% of the channels will be wild-type Kv2.1. Between 12 and 24 h post-transfection the cells were trypsinized and used for analysis.
Whole-Cell Current Recording.
Current recordings were made with an Axopatch-200B amplifier (Axon instruments, Union City, CA) in the whole cell configuration of the patch-clamp technique (13) as reported (12).
Pulse Protocols and Data Analysis.
The applied pulse protocols are listed in the figure legends. The voltage dependence of channel opening and inactivation (activation and inactivation curves) was fitted with a Boltzmann equation according to y = 1/{1 + exp[−(E − V½)/k]}, where V½ represents the voltage at which 50% of the channels are open or inactivated and k the slope factor. Activation and deactivation kinetics were fitted with a single or double exponential function by using a nonlinear least-squares (Gauss-Newton) algorithm. Results are presented as mean ± SEM; statistical analysis was done using the Student's t test; probability values are presented in the text.
Yeast Two-Hybrid System and Protein Constructs.
The MATCHMAKER Yeast Two-Hybrid System 3 (CLONTECH) was used to assay for protein–protein interactions. The amino termini of Kv1.5, Kv2.1, Kv3.1, Kv4.3, Kv5.1, Kv6.1, Kv6.3, Kv8.1, Kv9.3, Kv10.1, and Kv11.1 were cloned into the vector pGBKT7. The amino termini of Kv2.1, Kv6.3, Kv10.1, and Kv11.1 were also cloned into the vector pGADT7. AH109 cells were transformed with the plasmid constructs of interest (100 ng of each) and plated on −Trp/−Leu/+XαGAL media to select for cells containing both vectors and to test for interaction. The degree of interaction was determined from the speed and intensity of the blue color development.
Coimmunoprecipitation.
Kv2.1 was c-myc-tagged at the C terminus and cotransfected with green fluorescent protein (GFP)-tagged Kv2.1, Kv1.5, Kv6.3, Kv10.1, or Kv11.1 into HEK293 cells. The next day the cells were solubilized on ice with a PBS buffer supplemented with 5 mM EDTA, 1% Triton X-100, and a complete protease inhibitor mixture (Roche Diagnostics). For the immunoprecipitation Protein G Agarose beads and 2 μg of anti-GFP (CLONTECH) were added. The samples were incubated overnight at 4°C with rocking. Beads were then washed with ice-cold solubilization buffer. Proteins were eluted from the beads by boiling in SDS sample buffer and analyzed on 8% SDS/PAGE. Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham Pharmacia Biotech) and the blot was blocked. The blot was incubated with anti-c-myc (CLONTECH); afterwards, anti-mouse IgG (Amersham Pharmacia Biotech) was added, followed by ECL detection (Amersham Pharmacia Biotech).
Confocal Imaging.
Kv6.3, Kv10.1, and Kv11.1 were tagged with GFP at their carboxy terminus. HEK293 cells were cultivated on coverslips. For cotransfections, a 1:10 ratio of channel DNA versus Kv2.1 was added. The endoplasmic reticulum (ER) was visualized with the DsRed ER localization vector. This was constructed starting from the pDsRed vector (CLONTECH). The first 17 aa from calreticulin were amplified from brain cDNA and cloned in frame with the DsRed sequence of pDsRed. The KDEL sequence was inserted behind DsRed by using a mutagenesis PCR. Transfections and cotransfections (ratio 1:10 GFP-labeled channel DNA versus unlabeled Kv2.1 DNA) were done using the lipofectamine method (see above). Confocal images were obtained on a Zeiss CLSM 510, equipped with an argon laser (excitation, 488 nm) for the visualization of GFP and DsRed.
Results
Cloning of Kv6.3, Kv10.1, and Kv11.1.
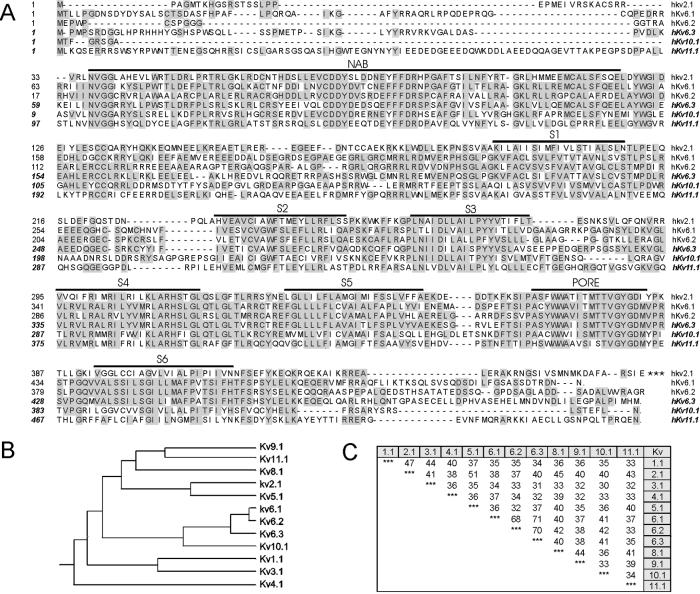
A search of the GenBank high throughput genomic sequence (htgs) database revealed genomic contigs containing exons coding for three previously uncharacterized homologues of Kv channels. The sequences of the genomic contigs were analyzed using genefinder to determine the full coding sequences of the genes. The predicted proteins displayed the typical topology of a Kv subunit: six transmembrane segments (S1–S6), with an array of five to six positive charges in S4, and the potassium selectivity motif “GYG” in the P-loop between S5 and S6 (Fig. 1). Each gene was predicted to consist of two coding exons, without evidence for alternate splicing.
Figure 1.
Sequence alignment, phylogenetic tree, and percent sequence identity of Kv6.3, Kv10.1, and Kv11.1. (A) The amino acid sequences of Kv2.1, Kv6.1, Kv6.2, Kv6.3, Kv10.1, and Kv11.1 were aligned using megalign. For convenience, only the first 460 aa of Kv2.1 are shown. Gaps (indicated by dashes) were introduced in the sequence to maintain the alignment. Conserved amino acids are shaded in gray. The six putative transmembrane domains and the pore region are indicated by an overline. (B) The phylogenetic tree for the Kv family. (C) The percent sequence similarity based on the S1–S6 core.
One of the predicted proteins consisted of 519 aa and shared more than 70% sequence identity with Kv6.1 and Kv6.2 (Fig. 1). Therefore, this protein has to be regarded as a previously uncharacterized member of the Kv6 subfamily, Kv6.3 or KCNG3. The other two proteins were composed of 436 and 545 aa and shared only ≈40% sequence identity with any of the previously identified Kv subunits. Therefore, we classified them as the first members of two previously uncharacterized subfamilies, Kv10.1 and Kv11.1.
The chromosomal locations of the genomic contigs containing Kv6.3, Kv10.1, and Kv11.1 are 16q24.1, 2p21, and 9p24.2, respectively. The complete cDNA sequences have been submitted to the GenBank database under accession nos. AF348982, AF348983, and AF348984 for Kv10.1, Kv11.1, and Kv6.3, respectively.
Tissue Distribution of Kv6.3, Kv10.1, and Kv11.1.
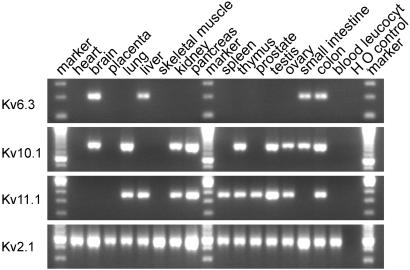
The search of the GenBank EST database yielded several hits for all three sequences, indicating that their mRNAs are indeed expressed. PCR analysis was used to assess the expression of Kv6.3, Kv10.1, and Kv11.1 mRNA in various human tissues (Fig. 2). Kv6.3 showed strong expression in brain and low expression in liver, small intestine, and colon. Kv10.1 was strongly expressed in pancreas and testis and weakly in brain, lung, kidney, thymus, ovary, small intestine, and colon. Kv11.1 gave a strong signal in pancreas and testis and a weaker signal in lung, liver, kidney, spleen, thymus, prostate, and ovary.
Figure 2.
Expression of Kv6.3, Kv10.1, Kv11.1, and Kv2.1 in human tissues. A PCR analysis was performed on a cDNA panel of the indicated human tissues with gene-specific primers for the subunits indicated on the left.
Functional Expression of Kv6.3, Kv10.1, and Kv11.1 in Ltk− Cells.
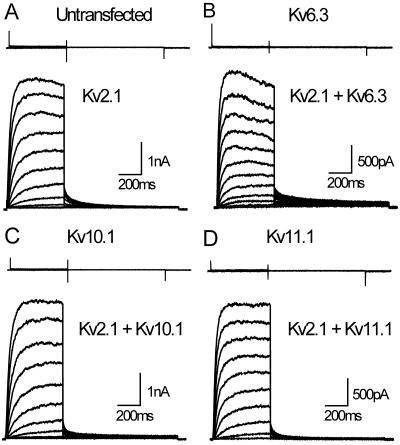
The coding sequences of Kv6.3, Kv10.1, and Kv11.1 were cloned into mammalian expression vectors for transient transfection in Ltk− cells. The subunits Kv6.3, Kv10.1, and Kv11.1 each failed to generate current above background in these cells, as shown in the top sections of each panel in Fig. 3 (n > 10 cells, for at least two independent transfections for each clone). Previously discovered silent subunits can form heterotetrameric channels with the Kv2 subfamily (6–10). To test whether this could also be the case for the previously uncharacterized subunits, we performed coexpressions with Kv2.1.
Figure 3.
Whole-cell current recordings of Kv6.3, Kv10.1, and Kv11.1, and the cotransfections with Kv2.1. The top sections in each panel show typical recordings for untransfected Ltk− cells (A), or for cells expressing Kv6.3 (B), Kv10.1 (C), and Kv11.1 (D). The holding potential was −80 mV and cells were depolarized in 20-mV increments from −80 mV to +60 mV, 500 ms in duration, followed by a repolarizing pulse at −25 mV, 850 ms in duration. The bottom sections of each panel show typical recordings from Ltk− cells expressing Kv2.1 (A), Kv2.1 + Kv6.3 (B), Kv2.1 + Kv10.1 (C), and Kv2.1 + Kv11.1 (D). The holding potential was −80 mV and cells were depolarized by 10-mV increments from −60 mV to +70 mV, 500 ms in duration. Deactivating tails were recorded at −25 mV or −35 mV for 850 ms.
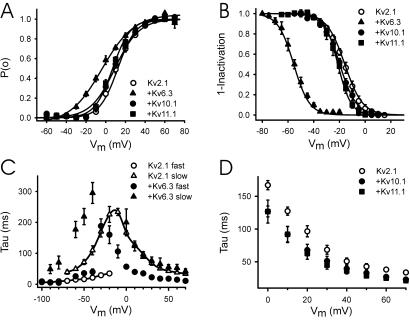
Expression of the human Kv2.1 subunit alone resulted in a typical rapidly activating delayed outward rectifier K+ current with functional properties as described (14, 15). The bottom sections of each panel in Fig. 3 show that coexpression with either previously uncharacterized subunit resulted in currents with distinct properties. For the cotransfection of Kv2.1 with Kv6.3, the threshold for activation was shifted by approximately 20 mV in hyperpolarizing direction compared with Kv2.1 alone (Fig. 4A). In addition, V½ (Table 1) was significantly (P < 0.001) shifted toward hyperpolarizing voltages, and the slope decreased as well (P < 0.001). Cotransfection of Kv2.1 with Kv10.1 had no significant (P > 0.05) effect on the activation curve, whereas with Kv11.1 a small but consistent (P < 0.05) −5 mV shift was observed.
Figure 4.
(A) Voltage dependence of activation. The activation curves of Kv2.1, Kv2.1 + Kv6.3, Kv2.1 + Kv10.1, and Kv2.1 + Kv11.1 were obtained from the normalized initial tail amplitude recorded at −25 mV for Kv2.1, Kv2.1 + Kv10.1, and Kv2.1 + Kv11.1 or at −50 mV for Kv2.1 + Kv6.3 after 500-ms prepulses ranging from −60 mV to 70 mV in 10-mV steps. The solid line represents the Boltzmann function fitted to the experimental data (see Experimental Procedures). (B) Voltage dependence of inactivation. The inactivation curves of Kv2.1, Kv2.1 + Kv6.3, Kv2.1 + Kv10.1, and Kv2.1 + Kv11.1 were obtained from the normalized peak currents recorded during a 250-ms test pulse to 50 mV as a function of the 5-s prepulse ranging from −50 mV to 10 mV for Kv2.1, Kv2.1 + Kv10.1, and Kv2.1 + Kv11.1 and from −80 mV to −20 mV for Kv2.1 + Kv6.3. Experimental data were fitted with a Boltzmann function (solid lines). (C) Kinetics of activation and deactivation of Kv2.1 and Kv2.1 + Kv6.3. Mean time constants ± SEM of activation and deactivation are plotted as a function of the test potential. To obtain the time constants for activation, test pulses were applied ranging from −10 mV to 70 mV for Kv2.1 and −30 mV to 70 mV for Kv2.1 + Kv6.3 in 10-mV steps, 500 ms in duration. To obtain the time constants for deactivation, a 200-ms prepulse to 50 mV was followed by test pulses ranging from −20 mV to −100 mV in 10-mV steps, 850 ms in duration. The experimental data were fitted with mono- or double-exponential functions, as appropriate. The slow component of activation and deactivation are shown as triangles, whereas the fast components are shown as circles. WT Kv2.1 gating kinetics are connected with a solid line. (D) Kinetics of activation of Kv2.1, Kv2.1 + Kv10.1, and Kv2.1 + Kv11.1. Mean time constants ± SEM of activation are shown as a function of the step potentials (−10 mV to 70 mV). The pulse protocol for Kv2.1 + Kv10.1 and Kv2.1 + Kv11.1 is the same as for Kv2.1 alone in C.
Table 1.
Electrophysiological parameters
| Voltage dependence
|
Time constants, ms
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Activation
|
Inactivation
|
Activation at 0 mV
|
Deactivation at −40 mV
|
|||||||||
| V1/2, mV | k, mV | n | V1/2, mV | k, mV | n | Fast | Slow | n | Fast | Slow | n | |
| Kv2.1 | 12.2 ± 1.4 | 9.5 ± 0.6 | 11 | −15.9 ± 1.2 | 7.2 ± 0.6 | 5 | 167 ± 12 | N.A. | 9 | 21.4 ± 2.1 | 110 ± 15 | 5 |
| + Kv6.3 | −4.2 ± 0.7 | 15.1 ± 0.7 | 5 | −55.6 ± 1.1 | 6.6 ± 0.8 | 6 | 58.2 ± 4.9 | 200 ± 38 | 8 | 40.1 ± 9.2 | 295 ± 34 | 5 |
| + Kv10.1 | 9.3 ± 2.0 | 9.8 ± 0.7 | 9 | −19.8 ± 1.2 | 6.4 ± 0.2 | 5 | 127 ± 8 | N.A. | 9 | 18.0 ± 1.2 | 115 ± 6 | 6 |
| + Kv11.1 | 7.0 ± 1.3 | 9.8 ± 0.7 | 11 | −21.2 ± 1.7 | 5.2 ± 0.2 | 7 | 127 ± 18 | N.A. | 10 | 18.8 ± 1.2 | 89.6 ± 3.1 | 7 |
Values are given as mean ± SEM; n = number of experiments. V1/2 and k obtained from Boltzmann fit (see Experimental Procedures). N.A., not applicable.
Co-transfection of Kv2.1 with Kv6.3 also markedly changed the C-type inactivation (Fig. 4B): the cotransfection resulted in a 40-mV hyperpolarizing shift in the voltage dependence of inactivation (P < 0.001). Cotransfection of Kv10.1 had no significant effect on the voltage dependence of inactivation (P > 0.05), whereas Kv11.1 gave a small −5 mV shift (P < 0.05).
The time-course of activation of Kv2.1 was fitted with a monoexponential function and resulted in time constants shown in Fig. 4 C and D. Upon cotransfection with Kv6.3, activation was accelerated (P values at all voltages <0.001) and the time course of activation was approximated better with a double exponential function (Fig. 4C). The acceleration of activation was less pronounced for the cotransfection of Kv10.1 or Kv11.1 (Fig. 4D), but still statistically significant (P < 0.05 at all voltages). Deactivation was fitted with a mono- or double-exponential function as appropriate. Cotransfection of Kv6.3 slowed deactivation significantly (P value at all voltages <0.001), whereas Kv10.1 and Kv11.1 had no significant effect (P value at all voltages >0.05). A summary of the electrophysiological parameters is given in Table 1.
Tissue Distribution of Kv2.1.
To test whether the previously uncharacterized subunits could be regulatory subunits for Kv2.1 in vivo, we determined the expression of Kv2.1 with the same cDNA panels as we did for Kv6.3, Kv10.1, and Kv11.1 (Fig. 2). Kv2.1 showed very high expression in brain, skeletal muscle, pancreas, and small intestine and moderate to high expression in heart, placenta, lung, liver, kidney, spleen, thymus, prostate, testis, ovary, and colon, consistent with previous reports (6, 16, 17). These results show that Kv6.3, Kv10.1, and Kv11.1 are expressed in several tissues in which Kv2.1 is also expressed, indicating that at least in some tissues these subunits could indeed interact with Kv2.1 to form heterotetrameric channels.
Biochemical Evidence for Selective Interaction with Kv Subunits.
To explore in a more unbiased manner the potential interactions of the three previously uncharacterized subunits with subunits of the known subfamilies, we used a yeast two-hybrid approach. Given the limitations of this method, we screened with the intracellular amino terminal segment, which contains the NAB domain that regulates coassembly (18–21). Kv6.3, Kv10.1, and Kv11.1 each did not show interactions with themselves, nor with each other, Kv1.5, Kv4.3, Kv8.1, and Kv9.1 (Fig. 5). In contrast, a strong interaction with Kv2.1, Kv3.1, and Kv5.1 was seen. For each of the previously uncharacterized subunits this interaction was as strong as the interaction of Kv2.1 with itself (positive control). Kv2.1 failed to interact with Kv1.5, consistent with the known lack of heterotetramerization between Kv1.5 and Kv2.1 (18, 22). The interaction of Kv2.1 with the previously uncharacterized subunits was confirmed with coimmunoprecipitation, using the full-length proteins (Fig. 6).
Figure 5.
Interaction of Kv6.3, Kv10.1, and Kv11.1 with representative subunits of all Kv subfamilies. The intracellular N-terminal segment that contains the subfamily-specific NAB domain was used as bait (B) and/or target (T) in a yeast two-hybrid analysis.
Figure 6.
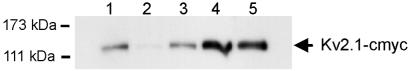
Co-immunoprecipitation of Kv6.3GFP, Kv10.1GFP, and Kv11.1GFP with Kv2.1c-myc. Immunoprecipitation was done with anti-GFP antibodies, Western blot was performed with anti-c-myc. Lanes 3–5 show that Kv2.1c-myc was coprecipitated with Kv6.3GFP, Kv10.1GFP, and Kv11.1GFP. GFP-tagged Kv2.1 (lane 1) and Kv1.5 (lane2) were used as positive and negative controls, respectively.
Subcellular Localization of Kv6.3, Kv10.1, and Kv11.1.
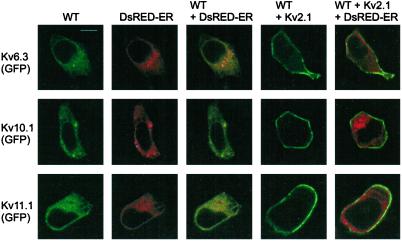
Although the lack of N-terminal tetramerization might explain the lack of current, it is known that Kv2.1 can generate current when the NAB domain is removed (18). Therefore, we also determined the subcellular localization of the previously uncharacterized subunits by using confocal microscopy. To visualize the subcellular protein distribution, GFP was fused to their carboxy termini. Transfected cells expressing only Kv6.3, Kv10.1, or Kv11.1 showed a punctated intracellular appearance without staining of the plasma membrane (Fig. 7, column 1). This indicates that the full-length protein was made, because GFP was added on the C-terminal end. To test whether this pattern reflected retention in the ER, we performed cotransfections with a vector (DsRed-ER) containing the cDNA from the red fluorescent protein DsRed, fused with the ER targeting signal from calreticulin and the ER retention signal, KDEL (Fig. 7, column 2). The localization of the red and green fluorescence overlapped completely, resulting in a yellow-orange color indicating that each of the three subunits were retained in the ER when they were expressed alone (Fig. 7, column 3). When Kv2.1 was coexpressed with these subunits, a redistribution of the green fluorescence was observed. In each case, prominent GFP staining was evident at the plasma membrane with minimal intracellular staining (Fig. 7, column 4). Potassium currents obtained with the GFP-tagged subunits were similar to those shown in Figs. 3 and 4. The intracellular staining was a nearly pure DsRed-ER fluorescence, showing hardly any overlap (Fig. 7, column 5). These results indicate that Kv2.1 promotes trafficking of Kv6.3, Kv10.1, and Kv11.1 to the cell surface membrane, presumably by forming heterotetrameric channels with these subunits.
Figure 7.
Subcellular localization of the channel-GFP fusion proteins assessed by confocal imaging. The rows show images with GFP fusion proteins of Kv6.3, Kv10.1, and Kv11.1, respectively. The first three columns show the fluorescence of the channel subunits, the DsRed-ER localization vector, and the overlay of both, respectively. The last two columns show cells cotransfected with Kv2.1, DsRed-ER, and each of the subunits; the surface staining of the GFP-tagged subunits (fourth column) is obvious with minimal overlap with the DsRed-ER fluorescence (overlay of both in the fifth column). (Scale bar, 10 μm.)
Discussion
This study reports the cloning and characterization of three previously uncharacterized α-subunits of voltage-gated potassium channels: Kv6.3, Kv10.1, and Kv11.1. The conventional methods to clone potassium channels include homology and expression cloning (14, 23). The disadvantage of both techniques is their dependence on expression level or on a functional signature: genes with very low expression levels or lacking a functional signature are not (easily) picked up by these techniques. The human genome project does allow to detect and clone such genes, as is demonstrated here for Kv6.3, Kv10.1, and Kv11.1.
When expressed in mammalian Ltk− cells each of the three subunits was unable to elicit any current, indicating that they belong to the “silent” subunits (6–10). The lack of functional current can be explained by retention in the ER, as was demonstrated with confocal microscopy, comparable with observations for Kv8 and Kv9 subunits (9, 16, 24). Such retention can have various causes such as ER retention signals or improper folding and/or assembly. Investigation of the sequences of Kv6.3, Kv10.1, and Kv11.1 did not reveal known ER retention or export signals, suggesting an assembly problem. For the confocal imaging we used a C-terminal GFP tag, which could interfere with trafficking. Indeed, C-terminal sequences can control efficient cell surface expression and clustering (25, 26). However, the three subunits reported here do not display such sequences and the GFP tag did not effect the currents recorded after coexpression. Inefficient assembly of channel subunits might originate from the aminoterminal “NAB” domain that directs and restricts subunit assembly within Kv subfamilies (18–21). Indeed, the aminotermini of Kv6.3, Kv10.1, and Kv11.1 did not interact with themselves, as was demonstrated with a yeast two-hybrid analysis. Therefore, to the extent that the NAB domain facilitates homotetrameric assembly, these subunits would appear incapable of efficient homotetramerization, which might explain ER retention.
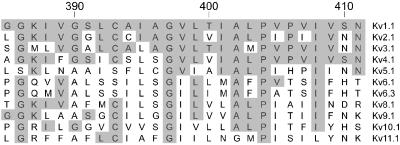
However, these incompatible amino termini may not be the only reason for the lack of functionality for these and other silent Kv channels. Indeed, distinct currents were observed for a chimera between the N terminus of Kv8.1 in a Kv1.3 background (7). However, a chimera with S6 from Kv8.1 in a Kv1.3 background (and vice versa) was not functional, which indicates that part of the nonfunctionality resides in the S6 segment. The alignment of this segment (Fig. 8) demonstrates that the three subunits reported here, as well as previously cloned silent subunits, all lack the second proline of the conserved P–X–P motif of the Kv1–Kv4 subunits. This points to a major structural difference in the S6 segments between the functional and silent subunits. However, when P406 and V409 of Kv2.1 were mutated to the corresponding residues of Kv8.1, altered but functional currents were observed, indicating that these residues alone do not explain the nonfunctional S6 chimera (24). However, P406 is the second proline in the highly conserved P–X–P motif from the functional Kv channels and might be responsible for a sharp bend in the S6 helical structure involved in gating (27). All of the silent subunits lack the second proline of the P–X–P motif, indicating a structural difference of the S6 segment between the functional and the silent subunits, which is apparently compensated in the heterotetrameric configuration.
Figure 8.
Alignment of the S6 segment of the Kv potassium channels. One member of each subfamily is represented. Conserved amino acids are shaded in gray. Sequence numbering of Kv2.1 is shown on top.
The profound effects of Kv6.3 on Kv2.1 gating properties suggest an important role for these heterotetramers: the latter would be inactivated at potentials close to resting potential (V½ for inactivation is −56 mV) in contrast to the homotetrameric Kv2.1 channels (V½ = −16 mV). Because both subunits are expressed in the brain (Fig. 2) functional heterotetramers could exist (6, 17). Previous studies on the sustained delayed rectifier component of hippocampal neurons showed properties that are comparable with those of Kv2.1 and Kv6.3 heteromultimers (28, 29). At −5 mV the two time constants for activation for the current in those neurons were 53 ms and 190 ms, which is comparable with heterotetrameric channels of Kv2.1 and Kv6.3 (Table 1). In addition, the midpoint of inactivation was more negative (−96 mV), which is at least closer to −56 mV for Kv2.1 and Kv6.3 compared with −16 mV for Kv2.1 alone. Furthermore, the pharmacological profile for homomeric Kv2.1 channels did not correspond completely with that of the sustained delayed rectifier component: the TEA sensitivity depended on the cell type under investigation (29) and differed from Kv2.1. Thus native channels that are considered to contain Kv2 subunits may well be heterotetramers, although it will be a challenge to assign the proper heterotetrameric combination.
In neurons, Kv2.1 is thought to have a role in controlling the membrane potential and in the electrical signaling of cells (30, 31). Using antisense oligonucleotides it was demonstrated that somato-dendritic excitability was regulated by Kv2.1 in hippocampal neurons (32). The down-regulation of the Kv2.1 protein (>90%) was associated with action potential broadening and an increase in intracellular calcium at high-frequency stimulation. The gating properties were not reported in this study but the 90% down regulation of the Kv2.1 protein was associated with only a 50% reduction of the sustained delayed rectifier component. Although the molecular nature of the sustained current remains to be elucidated, a heterotetrameric subunit composition could be compatible with such dominant negative results.
Within the Kv1, Kv3, and Kv4 families, functional diversity is achieved by the different properties of each subunit, and by the heteromeric (intrafamily) assembly of α-subunits resulting in channels with distinct biophysical properties. The Kv2 subfamily contains only two members that have very similar biophysical properties. While Kv2.1 and Kv2.2 are capable of heteromultimerization, the resulting currents are functionally similar to those of their homotetramers (33). It has been suggested that the functional diversity within this family is achieved through heteromeric assembly with other subfamilies of silent subunits (34). Our results now add three more subunits that can expand further the functional diversity in different types of tissue or during development.
Thus far, 19 functional Kv α-subunits had been discovered and only 7 silent subunits. Our results enlarge this last group to 10 subunits. Despite the large number of these subunits, their exact physiological role is still poorly understood mainly because of the difficulty in recognizing a silent subunit in isolated cells or in tissue. Thus far, heterologous expression studies have led to the hypothesis that the silent subunits must interact with other Kv subunits from the Kv2 and Kv3 subfamilies to regulate their function. If each of the silent subunits can interact with the two members of the Kv2 subfamily and the four members of the Kv3 subfamily, then at least 60 different heterotetramers are possible (each with one to three silent subunits). Thus, this growing group of silent subunits considerably expands the potential for molecular diversity of the native K+ channels. Thus, future experiments will be necessary to reveal the true interaction partners and the physiological importance of the silent subunits.
Acknowledgments
We thank Dr. Jean-Pierre Timmermans for the use of the confocal microscope. This work was supported by Flanders Institute for Biotechnology Grant PRJ05 and National Institutes of Health/National Heart, Lung, and Blood Institute Grant HL59689.
Abbreviations
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
Note Added in Proof.
While this paper was under review, another group (35) reported cloning of “Kv6.3,” which corresponds to Kv10.1 in our analysis (see Fig. 1).
Footnotes
References
- 1.Hille B. Ionic Channels of Excitable Membranes. Sunderland, MA: Sinauer; 1991. [Google Scholar]
- 2.Barry D M, Nerbonne J M. Annu Rev Physiol. 1996;58:363–394. doi: 10.1146/annurev.ph.58.030196.002051. [DOI] [PubMed] [Google Scholar]
- 3.Pongs O. FEBS Lett. 1999;452:31–35. doi: 10.1016/s0014-5793(99)00535-9. [DOI] [PubMed] [Google Scholar]
- 4.Pongs O. Physiol Rev. 1992;72:S69–S88. doi: 10.1152/physrev.1992.72.suppl_4.S69. [DOI] [PubMed] [Google Scholar]
- 5.Zhao B, Rassendren F, Kaang B K, Furukawa Y, Kubo T, Kandel E R. Neuron. 1994;13:1205–1213. doi: 10.1016/0896-6273(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 6.Drewe J A, Verma S, Frech G, Joho R H. J Neurosci. 1992;12:538–548. doi: 10.1523/JNEUROSCI.12-02-00538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hugnot J P, Salinas M, Lesage F, Guillemare E, de Weille J, Heurteaux C, Mattei M G, Lazdunski M. EMBO J. 1996;15:3322–3331. [PMC free article] [PubMed] [Google Scholar]
- 8.Patel A J, Lazdunski M, Honore E. EMBO J. 1997;16:6615–6625. doi: 10.1093/emboj/16.22.6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salinas M, Duprat F, Heurteaux C, Hugnot J P, Lazdunski M. J Biol Chem. 1997;272:24371–24379. doi: 10.1074/jbc.272.39.24371. [DOI] [PubMed] [Google Scholar]
- 10.Zhu X R, Netzer R, Bohlke K, Liu Q, Pongs O. Receptors Channels. 1999;6:337–350. [PubMed] [Google Scholar]
- 11.Post M A, Kirsch G E, Brown A M. FEBS Lett. 1996;399:177–182. doi: 10.1016/s0014-5793(96)01316-6. [DOI] [PubMed] [Google Scholar]
- 12.Snyders D J, Chaudhary A C. Mol Pharmacol. 1996;49:949–955. [PubMed] [Google Scholar]
- 13.Hamill O P, Marty A, Neher E, Sakmann B, Sigworth F J. Pflügers Arch Eur J Physiol. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 14.Frech G C, VanDongen A M, Schuster G, Brown A M, Joho R H. Nature (London) 1989;340:642–645. doi: 10.1038/340642a0. [DOI] [PubMed] [Google Scholar]
- 15.Benndorf K, Koopmann R, Lorra C, Pongs O. J Physiol (London) 1994;477:1–14. doi: 10.1113/jphysiol.1994.sp020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shepard A R, Rae J L. Am J Physiol. 1999;277:C412–C424. doi: 10.1152/ajpcell.1999.277.3.C412. [DOI] [PubMed] [Google Scholar]
- 17.Trimmer J S. Proc Natl Acad Sci USA. 1991;88:10764–10768. doi: 10.1073/pnas.88.23.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Jan Y N, Jan L Y. Science. 1992;257:1225–1230. doi: 10.1126/science.1519059. [DOI] [PubMed] [Google Scholar]
- 19.Xu J, Yu W, Jan Y N, Jan L Y, Li M. J Biol Chem. 1995;270:24761–24768. doi: 10.1074/jbc.270.42.24761. [DOI] [PubMed] [Google Scholar]
- 20.Papazian D M. Neuron. 1999;23:7–10. doi: 10.1016/s0896-6273(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 21.Shen N V, Pfaffinger P J. Neuron. 1995;14:625–633. doi: 10.1016/0896-6273(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 22.Covarrubias M, Wei A A, Salkoff L. Neuron. 1991;7:763–773. doi: 10.1016/0896-6273(91)90279-9. [DOI] [PubMed] [Google Scholar]
- 23.Tamkun M M, Knoth K M, Walbridge J A, Kroemer H, Roden D M, Glover D M. FASEB J. 1991;5:331–337. doi: 10.1096/fasebj.5.3.2001794. [DOI] [PubMed] [Google Scholar]
- 24.Salinas M, de Weille J, Guillemare E, Lazdunski M, Hugnot J P. J Biol Chem. 1997;272:8774–8780. doi: 10.1074/jbc.272.13.8774. [DOI] [PubMed] [Google Scholar]
- 25.Burke N A, Takimoto K, Li D, Han W, Watkins S C, Levitan E S. J Gen Physiol. 1999;113:71–80. doi: 10.1085/jgp.113.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D, Takimoto K, Levitan E S. J Biol Chem. 2000;275:11597–11602. doi: 10.1074/jbc.275.16.11597. [DOI] [PubMed] [Google Scholar]
- 27.del Camino D, Holmgren M, Liu Y, Yellen G. Nature (London) 2000;403:321–325. doi: 10.1038/35002099. [DOI] [PubMed] [Google Scholar]
- 28.Numann R E, Wadman W J, Wong R K. J Physiol (London) 1987;393:331–353. doi: 10.1113/jphysiol.1987.sp016826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, McBain C J. J Physiol (London) 1995;488:647–660. doi: 10.1113/jphysiol.1995.sp020997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murakoshi H, Trimmer J S. J Neurosci. 1999;19:1728–1735. doi: 10.1523/JNEUROSCI.19-05-01728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baranauskas G, Tkatch T, Surmeier D J. J Neurosci. 1999;19:6394–6404. doi: 10.1523/JNEUROSCI.19-15-06394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du J, Haak L L, Phillips T E, Russell J T, McBain C J. J Physiol (London) 2000;522:19–31. doi: 10.1111/j.1469-7793.2000.t01-2-00019.xm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blaine J T, Ribera A B. J Neurosci. 1998;18:9585–9593. doi: 10.1523/JNEUROSCI.18-23-09585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kramer J W, Post M A, Brown A M, Kirsch G E. Am J Physiol. 1998;274:C1501–C1510. doi: 10.1152/ajpcell.1998.274.6.C1501. [DOI] [PubMed] [Google Scholar]
- 35.Sano Y, Mochizuki S, Miyake A, Kitada C, Inamura K, Yokoi H, Nozawa K, Matsushime H, Furuichi K. FEBS Lett. 2002;512:230–234. doi: 10.1016/s0014-5793(02)02267-6. [DOI] [PubMed] [Google Scholar]