Abstract
Actin is the most abundant protein in eukaryotic cells, but its release from cells into blood vessels can be lethal, being associated with clinical situations including hepatic necrosis and septic shock. A homeostatic mechanism, termed the actin-scavenger system, is responsible for the depolymerization and removal of actin from the circulation. During the first phase of this mechanism, gelsolin severs the actin filaments. In the second phase, the vitamin D-binding protein (DBP) traps the actin monomers, which accelerates their clearance. We have determined the crystal structures of DBP by itself and complexed with actin to 2.1 Å resolution. Similar to its homologue serum albumin, DBP consists of three related domains. Yet, in DBP a strikingly different organization of the domains gives rise to a large actin-binding cavity. After complex formation the three domains of DBP move slightly to “clamp” onto actin subdomain 3 and to a lesser extent subdomain 1. Contacts between actin and DBP throughout their extensive 3,454-Å2 intermolecular interface involve a mixture of hydrophobic, electrostatic, and solvent-mediated interactions. The area of actin covered by DBP within the complex approximately equals the sum of those covered by gelsolin and profilin. Moreover, certain interactions of DBP with actin mirror those observed in the actin-gelsolin complex, which may explain how DBP can compete effectively with gelsolin for actin binding. Formation of the strong actin–DBP complex proceeds with limited conformational changes to both proteins, demonstrating how DBP has evolved to become an effective actin-scavenger protein.
Actin is one of the most abundant and highly conserved proteins in eukaryotic cells, being involved in numerous functions including cell motility, control of cell shape, and muscle contraction (1, 2). Central to these functions is the ability of actin to assemble into polymers at physiological salt concentrations. However, this property of actin can lead to lethal consequences when actin is released from the cells to form filaments in the bloodstream under conditions involving cell death or tissue injury. The release of actin is associated with a variety of clinical situations including hepatic necrosis, septic shock, the adult respiratory distress syndrome, and certain disorders of pregnancy (3, 4). In animal experiments, rats given large amounts of actin through i.v. injection die rapidly and show evidence of pulmonary venous obstruction by actin filaments, pulmonary microthrombi, and endothelial injury (5).
A homeostatic mechanism, termed the actin-scavenger system, is responsible for the rapid depolymerization and clearance of actin filaments from the bloodstream (3–5). This system involves the concerted action of gelsolin and the vitamin D-binding protein (DBP), the latter also known as group-specific component (Gc). During the first phase of the scavenger process, severing of the actin filaments results from the binding of gelsolin to the barbed ends of the filaments, which is followed by a second phase during which actin monomers are trapped by DBP into high affinity 1:1 complexes (6–8) (Kd = 10−9 M). Clearance of such a complex, which seems to take place primarily through the liver (4), proceeds much faster (half time, ≈30 min) than that of uncomplexed DBP (half time, ≈24 h). However, in patients with multiple trauma or fulminant hepatic failure, the capacity of this system can be exceeded, leading to immediate death (3, 4, 9, 10).
DBP is a 52-kDa highly expressed and multifunctional serum protein (3). In addition to its role as part of the actin-scavenger system, DBP is the main protein involved in the transport of vitamin D (11), hence its name. It is now well established, however, that less than 5% of circulating DBP is actually complexed with vitamin D metabolites, leaving a considerable amount of the protein available for other functions. There also is good evidence connecting DBP with macrophage activation (12) and chemotaxis (3).
We describe here the crystal structures of DBP by itself and complexed with actin to resolutions of 2.15 and 2.1 Å, respectively. As predicted from sequence analysis, the structures confirm that the three domains of DBP are related to each other and to those of human serum albumin (HSA; ref. 13). A surprising finding, however, is that the overall three-dimensional organization of the domains is different in DBP. The functional meaning of this finding becomes clear after analysis of the structure of the complex of DBP with actin, where actin is found bound within a large cavity formed by the three domains of DBP. Thus, the structures demonstrate that the overall fold of DBP has adapted to become a very effective actin-binding protein.
Materials and Methods
Crystallization.
Actin was prepared and purified from rabbit muscle (14), and DBP was purchased from Calbiochem. The conditions used for the crystallization of uncomplexed DBP are similar to those reported by Vogelaar et al. (ref. 15, hanging drops of DBP at ≈20 mg⋅ml−1 were stabilized at 20°C against a reservoir solution containing 28% polyethylene glycol 200 and 100 mM sodium acetate, pH 4.6). Actin and DBP mixed at a 1.2:1.0 molar ratio and purified on a Superose 12 HR column (running buffer: 100 mM Tris, pH 7.6/0.6 mM ATP/0.5 mM β-mercaptoethanol/0.2 mM CaCl2) were concentrated to ≈20 mg⋅ml−1 and crystallized at 4°C in 11% polyethylene glycol 12,000/200 mM magnesium acetate/100 mM sodium cacodylate pH 6.6/20% glycerol. Although these conditions are similar to those of Bogaerts et al. (8), our crystals belong to a different space group (P212121).
Data Collection, Structure Determination, and Refinement.
Complete and highly redundant x-ray data sets to 2.1 and 2.15 Å resolution were collected at 100 K at BioCARS beamline 14-BM-C (Advanced Photon Source, Argonne, IL) from crystals of uncomplexed DBP and DBP complexed with actin. Note that although in both cases diffraction was observed to 1.9 Å resolution or better, the data were incomplete to that resolution. All the diffraction data were indexed and scaled with the programs DENZO and SCALEPACK (ref. 16; Table 1). The structure of the actin–DBP complex was determined first. A molecular replacement solution was found with the program AMORE (17) for the actin portion of the asymmetric unit, using the structure of actin from the DNase I complex (18) as a search model (PDB ID code 1ATN). Note that actin accounts for only ≈45% of the total amino acid content of the asymmetric unit. After the first cycle of refinement with the program CNS (19) with all the diffraction data available (Rfactor = 46.2%, Rfree = 50.1%), electron density maps were calculated by using partial model phases and phases refined with the program DM (20). These maps revealed very limited information about the DBP part of the model. However, we were able to add a few poly(Ala) and/or poly(Ser) fragments taken from the HSA structure (13), which we knew had some structural homology with DBP. Repeated cycles of model building and refinement allowed for the incremental improvement of the electron density maps up to a point where we recognized that a dramatic change had occurred in the domain organization of DBP (compared with that of HSA). At this point we were able to assign a considerable part of the DBP sequence, which was facilitated by the identification of some of the disulfide bridges that characterize DBP and HSA. Refinement of this model resulted in an Rfactor of 29.5% and Rfree of 36.3%. At this stage of the refinement the incomplete structure of DBP from the complex was used to determine the structure of uncomplexed DBP by molecular replacement with the program AMORE (17). Rotation and translation solutions were found for the two molecules of DBP in the asymmetric unit of the P43 crystals of uncomplexed DBP. Although from this point on the two structures were refined independently, they were frequently cross-checked to help with model building and validation of the structures. The quality of the final refined structures was validated with the programs PROCHECK (21) and CNS (19). Table 1 summarizes the data collection and refinement statistics.
Table 1.
Data collection and refinement statistics
| Uncomplexed DBP | Actin–DBP | |
|---|---|---|
| Data collection | ||
| Space group | P43 | P212121 |
| Unit cell parameters | ||
| a, b, c, Å | 133.93, 133.93, 73.58 | 80.76, 87.36, 162.69 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 |
| Molecules per asymmetric unit | 2 | 1 |
| Resolution range, Å | 60.0–2.15 | 40.0–2.10 |
| Crystal mosaicity, ° | 0.745 | 0.465 |
| Completeness, % | 94.9 (81.2)* | 99.8 (98.8) |
| Observations, total/unique | 1,041,011/67,175 | 1,064,049/67,458 |
| Redundancy | 15.5 (8.1) | 15.8 (11.0) |
| Rmerge, %† | 5.9 (21.4) | 9.3 (28.6) |
| Rmerge, %†‡ | 6.9 (22.1) | 9.8 (28.9) |
| Average I/σ | 27.7 (10.2) | 23.4 (9.8) |
| Refinement | ||
| Resolution range, Å | 60.0–2.15 | 40.0–2.10 |
| Completeness, % | 94.9 (81.2) | 99.8 (98.8) |
| σ-cutoff | None | None |
| Rfactor, % (all reflections)§ | 21.8 (25.1) | 21.7 (24.2) |
| Free Rfactor, %‖ | 24.8 (28.7) | 23.6 (24.9) |
| Average B factors, Å2 | ||
| Protein atoms | 49.8 | 37.2 |
| Water molecules | 50.0 | 41.7 |
Highest resolution shell (2.32–2.15 Å for uncomplexed DBP and 2.18–2.1 Å for actin–DBP) values are given in parentheses.
Rmerge = Σ |I − 〈I〉|/Σ I, where I is the observed intensity of an individual reflection, and 〈I〉 is the mean intensity of that reflection.
Rmerge = Σ (I − 〈I〉)2/Σ I2, where I is the observed intensity of an individual reflection, and 〈I〉 is the mean intensity of that reflection.
Rfactor = Σ |Fo − Fc|/Σ |Fo|, where Fo and Fc are observed and calculated structure factors, respectively.
Free Rfactor, Rfactor calculated for a subset of the reflections (5%), which were omitted during the refinement and used to monitor its convergence.
Results and Discussion
Structure of Uncomplexed DBP.
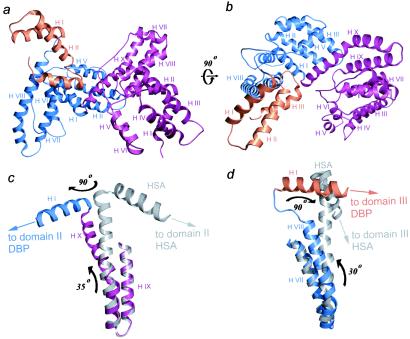
The 2.15-Å resolution structure of uncomplexed DBP (Fig. 1 a and b) was determined by molecular replacement from the structure of its complex with actin (see Materials and Methods). Only the first three amino acids from the N terminus, Leu-17–Arg-19, are missing in the structure, whereas the rest of the sequence is well defined in the electron density maps (note that amino acids Met-1–Ala-16 constitute the signal sequence, which is missing in the mature protein). Similar to HSA (13), DBP comprises three homologous α-helical domains referred to, from N to C terminus, as domains I, II, and III. Domains I and II can be subdivided further into two structurally related subdomains. However, domain III of DBP is shorter than that of HSA, lacking its C-terminal subdomain. Small rotations at the junctions between subdomains account for most of the differences between domains I and II of DBP (see the legend to Fig. 1 for a more detailed comparison of the various domains). There are two independent molecules of DBP within the asymmetric unit of the crystals. Superimposition of these molecules reveals an ≈6° rotation between the N- and C-terminal halves of the molecules relative to a fulcrum located in a loop around residue Pro-316. This difference between the two DBP molecules seems to arise from differences in their crystal-packing environments and suggests that significant rotational freedom among the various subdomains of DBP exists.
Figure 1.
Structure of uncomplexed DBP. (a and b) Two ribbon representations of the DBP structure are shown rotated by 90°. The three domains of DBP are colored burgundy (domain I), blue (domain II), and orange (domain III). Notice how the α-helix X of domain I (best seen in b) and α-helix VIII of domain II (best seen in a) are bent. The structural changes occurring within these two α-helices, which connect domains I and II and II and III of DBP, respectively, account for an entirely different domain organization in DBP as compared with HSA. To better illustrate this point, superimpositions of the two α-helices with their HSA counterparts are shown in c and d (where the corresponding regions of HSA are shown in gray). The arrows in c and d show how the differences between the two proteins can be separated into motions in two nearly perpendicular directions. Note, however, that when analyzed separately the domains of DBP and albumin clearly are related. Thus, whereas domain III of DBP is significantly shorter than that of HSA, domains I and II of the two proteins are related more closely (rms deviations of 2.4 and 2.08 Å for 150 and 145 equivalent Cα positions, respectively).
An unexpected feature of the structure is that the overall three-dimensional organization of the three domains of DBP is strikingly different from that of its homologue HSA. The differences in orientation and position of the domains of DBP, compared with HSA, arise from bends within the C-terminal α-helices of domains I and II and rotations centered at the loops that connect these two domains (Fig. 1 c and d). The C-terminal α-helix of domain I is bent by ≈35° at Leu-206 and twisted at Gly-227 (within the interdomain loop) by ≈90° (Fig. 1c). Similarly, the C-terminal α-helix of domain II is slightly bent over its entire length, which together with a rotation of ≈90° centered at the loop accounts for a markedly different positioning of domain III (Fig. 1d). We cannot identify a unique reason explaining the bending of the α-helices and twists of the loops. Instead, it seems to be the combination of interactions along the α-helices, including interactions with parts of the structure which in HSA are located farther apart, that are optimally responsible for their bending. As discussed below, this unique interdomain organization of DBP is central to its ability to interact with actin.
Limited Structural Changes After Formation of the Actin–DBP Complex.
The structure of the actin–DBP complex (Fig. 2) was determined and refined to 2.1 Å resolution starting from the molecular replacement solution of the actin portion of the structure (see Materials and Methods). Not included in the final model are a few flexible regions that are poorly defined in the electron density maps, which include actin residues Asp-1–Asp-3, His-40–Asp-51, and Ala-365–Ala-375, DBP residues Gly-107–Ala-125, and the C-terminal Leu-474.
Figure 2.
Structure of the actin–DBP complex. (a) Stereo view of the structure of the complex. The three domains of DBP are colored as described for Fig. 1, and actin is colored green. Gly-227 and Thr-414 of DBP constitute the first amino acids of domains II and III, respectively. (b) Sequence of DBP. The amino acids from the three domains are colored as described for a. Superimposed on the sequence is a representation of the secondary structure assignment. Regions of DBP that interact with actin are underlined in black. Under these underlined sequences of DBP we show the interacting segments in the primary structure of actin (green). (c) Stereo representation of a characteristic section of the 2Fo − Fc electron density map (contoured at 1.5σ) at the interface between actin (yellow trace) and DBP (gray trace). Notice how numerous well defined water molecules mediate many of the interactions between actin and DBP.
The structure of DBP shows only minor changes after formation of the actin–DBP complex. Superimpositions of the two DBP molecules in the asymmetric unit of the crystals of uncomplexed DBP with that of DBP from the complex result in rms deviations of 1.5 and 1.8 Å for 427 and 410 equivalent Cα (differing by ≤ 4.0 Å), respectively. Regardless of which of the two molecules of uncomplexed DBP is considered, the conformational change after actin binding is nearly the same. DBP seems to “clamp” onto actin by narrowing the angle between domain I on one side and domains II and III on the other side by ≈5° relative to a hinge at Gly-227.
The structure of actin does not change significantly after binding to DBP. Comparison of the Cα coordinates of the actins from the DNase I (18) and DBP complexes shows an rms deviation of 0.68 Å for 343 equivalent Cα positions (differing by ≤ 4.0 Å). However, we cannot rule out the possibility of a conformational change within the C terminus of actin, which is not seen clearly in this structure. In previous actin structures (18, 22–25) the C-terminal amino acids Gly-366–Phe-375 form a short α-helix after a kink at Gly-366. If a similar conformation of the actin C terminus exists in the actin–DBP complex, the C terminus of actin would have to interact with DBP directly.
In the complex with DBP, actin has Mg2+⋅ATP at its nucleotide-binding site. It is striking that in this complex, as well as in the other three complexes of actin studied by protein crystallography [DNase I (18), gelsolin (22), and profilin (25)], the hydrolysis of ATP is either blocked or slowed down dramatically. This observation may be an indication that during ATP hydrolysis actin undergoes certain conformational transitions that cannot take place when it is bound to the above proteins. Note also that the presence of the intact nucleotide in the actin–DBP structure results in the side chain of the catalytic Ser-14, and hence subdomain 2, being oriented as in the other three ATP–actin structures (18, 22, 25) but different from that of ADP-bound actin (23).
It is well known that the polymerization of Mg2+⋅ATP–actin occurs faster than that of Ca2+⋅ATP–actin (26), which is believed to result from distinctive structural differences brought about by these ions (26, 27). However, in this structure, which contains Mg2+ bound in the nucleotide site (as compared to Ca2+ in previous actin structures), we do not observe any structural difference that would support this idea. The only change concerns the water coordination of the Mg2+, which in this structure can be described as a classical nearly octahedral coordination with four water molecules and two O atoms from the nucleotide β- and γ-phosphates at the vertices (as compared with the pentagonal bipyramidal coordination of Ca2+, which involves an additional water molecule). Therefore, other factors such as differences in ATPase activity (26) may account for the different polymerization rates of Mg2+⋅ATP–actin and Ca2+⋅ATP–actin.
Intermolecular Interface of the Actin–DBP Complex.
Actin binds into a large cavity formed by the three domains of DBP (Figs. 2–4). Contacts with DBP involve actin subdomain 3 and to a lesser extent subdomain 1 (Fig. 2 a and b). The three domains of DBP engulf approximately one half of the surface area of actin subdomain 3, making extensive contacts on both sides of this subdomain (Fig. 4). The area covered by DBP on actin approximately corresponds to the sum of the areas covered by gelsolin domain 1 (22) and profilin (25) within their respective complexes with actin. Indeed, gelsolin domain 1 and profilin also bind to actin subdomains 1 and 3 but do so on nearly opposite sides of actin such that there is only limited overlap between their respective binding interfaces. The analogy with DBP is even greater in the case of gelsolin. That is, comparison of the actin–gelsolin and actin–DBP complexes, based on a superimposition of the Cα coordinates of the actins from the two structures, reveals an equivalence of the α-helical segments Ser-194–Lys-207 of DBP and Gln-71–Leu-84 of gelsolin domain 1 (22). Although there is no significant sequence similarity between these two superimposable α-helical segments, the nature of their interactions with actin is, in both cases, hydrophobic (Fig. 3). This interaction is, on the other hand, the only contact between DBP and actin that bares any structural similarity with a previously studied actin complex (18, 22, 24, 25). Notice that DBP residues Ser-194–Lys-207 are contained within the C-terminal helix of domain I, which is bent at Leu-206 (Fig. 1c). Full-length gelsolin comprises six homologous domains. In a different crystal structure, that of actin complexed with gelsolin domains 4–6 (24), an equivalent α-helix of gelsolin domain 4 (residues Glu-473–Leu-486) makes similar interactions with actin. In all three structures, hydrophobic residues from the helices interact with a patch of hydrophobic residues on the actin surface (Tyr-143, Thr-148, Ile-345, Leu-346, Leu-349, Phe-352, and Met-355).
Figure 4.
All-atom surface representations of the actin–DBP complex. The color scheme is as described for Figs. 1 and 2. Two views of the complex are shown rotated by 90°. The four subdomains of actin are numbered 1–4. All three domains of DBP participate in the interaction with actin subdomains 1 and 3. Notice how the three domains of DBP engulf approximately one half of actin subdomain 3, participating in extensive contacts on both sides of this subdomain.
Figure 3.
Common interactions of gelsolin and DBP with actin. The α-helical segments Ser-194–Lys-207 of DBP and Gln-71–Leu-84 of gelsolin domain 1 (in red) (22) share a similar site on the actin surface. They interact with a patch of hydrophobic amino acids (Tyr-143, Thr-148, Ile-345, Leu-346, Leu-349, Phe-352, and Met-355, yellow) in a cleft between actin subdomains 1 and 3 (green). Gelsolin residues Ala-76, Ala-78, Ile-79, Phe-80, and Val-82 and DBP residues Val-197, Leu-200, and Leu-204, which face the hydrophobic patch on actin, are shown (gray) but not labeled.
DBP is not generally recognized as an actin filament-binding protein but rather as a monomer-trapping molecule (7). To investigate this point further, the actin–DBP complex was superimposed onto the actin-filament model (28, 29). In that model, two parallel strands of head-to-tail-bound actin monomers are shifted along the filament axis by half a monomer length. As a result, at the so-called barbed end of the filament subdomains 1 and 3 of the first actin monomer of one of the strands are more exposed than the other. Thus, although the structure of gelsolin fragment 1 can be fit onto the first monomer of either of the filament strands, those of DBP and profilin can be added only onto the first monomer of the downward-shifted strand, but steric hindrance would prevent them from binding to the first monomer of the other, upward-shifted, strand. Although the region of DBP that interacts with actin at the profilin site does not share structural similarity with profilin, their sharing of this site may underline their monomer-binding preferences (30). Therefore, our analysis seems to suggest that similar to profilin, DBP potentially could bind to the most exposed actin monomer at the barbed end of the filament.
The contacts between actin and DBP are a combination of hydrophobic and electrostatic interactions including direct hydrogen bonds (a total of 10), two salt bridges (connecting actin residues Asp-288 and Lys-328 and DBP residues Arg-218 and Glu-143, respectively), and a large number of contacts mediated by solvent molecules (Fig. 2c). Although there are five amino acid segments of the two proteins that participate in the contacting interface, it is actin residues Tyr-279–Asn-296 and DBP residues Thr-196–Arg-218 that account for the largest number of interactions. The total area of the binding interface between the two proteins is 3,454 Å2. According to a classification (31) of protein–protein recognition sites into small (≈1,150 Å2), standard (≈1,600 Å2), and large (≈3,300 Å2) interfaces, the actin–DBP binding interface clearly qualifies as a large interface. It is also the largest interface among the four complexes of actin with actin-binding proteins known thus far (the binding interfaces for the DNase I, gelsolin fragment 1, and profilin being 1,781, 2,218, and 2,138 Å2, respectively). The size of the binding interface correlates well with the high affinity of the actin–DBP complex (Kd = 10−9 M; refs. 6–8 and 32).
The architecture of DBP is such that after formation of its complex with actin, the C-terminal α-helix of domain I and the loop connecting domains II and III are brought into direct contact with actin. As discussed above, these two regions of DBP differ dramatically from their HSA counterparts (Fig. 1 c and d), suggesting an evolutionary relationship between the domain organization of DBP and its unique actin-binding properties.
In summary, the structures of uncomplexed DBP and that of its complex with actin described here demonstrate how a divergent three-dimensional organization of the three domains of DBP, relative to its homologue HSA, give rise to an extremely efficient actin-binding fold. The fact that both DBP and actin undergo only minor changes after complex formation indicates that DBP in the bloodstream is “primed” for actin binding. It has been proposed, however, that the rapid clearance of the actin–DBP complex from circulation is caused by a conformational change in DBP induced by actin binding (33). Our results do not support this view but instead would indicate that it is the complex itself that is recognized for clearance. The structure of DBP complexed with a vitamin D metabolite has been reported very recently (34). In agreement with a study that used limited proteolysis to identify the sterol-binding site (35), Verboven et al. localize this site to the N terminus of DBP (34). Such a location is distal to the actin-binding interface identified in this work, which provides a structural understanding of previous reports that the sterol-binding and actin-binding properties of DBP are independent (35–38). It also seems reasonable to propose that the large structural differences between DBP and HSA are directed toward its actin-binding function, which involves specific interactions with all three domains of DBP and not toward its sterol-binding property, which is confined to the first four α-helices of DBP. As revealed in this work, the actin-binding interface of DBP can be viewed approximately as the sum of the areas covered by profilin (25) and gelsolin fragment 1 (22). Moreover, some specific hydrophobic interactions of DBP mimic interactions seen in the actin-gelsolin complex. This feature may explain the ability of DBP to compete effectively with gelsolin for the binding of monomeric actin (39), which may help free gelsolin for further filament severing while allowing DBP to eliminate actin monomers from circulation.
Acknowledgments
We thank the staff members at BioCARS (Advanced Photon Source, Argonne, IL) for assistance during data collection and John Gergely for a critical reading of the manuscript. This work was supported by National Institutes of Health Grants R01 AR46524 (to R.D.) and R01 HL66219 (to P.G.). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38. Use of the BioCARS facilities was supported by National Institutes of Health Grant RR07707.
Abbreviations
- DBP
vitamin D-binding protein
- HSA
human serum albumin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1KXP for the actin–DBP complex and 1KW2 for uncomplexed DBP).
References
- 1.Sheterline P, Clayton J, Sparrow J. Protein Profile. 1995;2:1–103. [PubMed] [Google Scholar]
- 2.Pollard T D, Blanchoin L, Mullins R D. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 3.White P, Cooke N. Trends Endocrinol Metab. 2000;11:320–327. doi: 10.1016/s1043-2760(00)00317-9. [DOI] [PubMed] [Google Scholar]
- 4.Lee W M, Galbraith R M. N Engl J Med. 1992;326:1335–1341. doi: 10.1056/NEJM199205143262006. [DOI] [PubMed] [Google Scholar]
- 5.Haddad J G, Harper K D, Guoth M, Pietra G G, Sanger J W. Proc Natl Acad Sci USA. 1990;87:1381–1385. doi: 10.1073/pnas.87.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mc Leod J F, Kowalski M A, Haddad J G., Jr J Biol Chem. 1989;264:1260–1267. [PubMed] [Google Scholar]
- 7.Van Baelen H, Bouillon R, De Moor P. J Biol Chem. 1980;255:2270–2272. [PubMed] [Google Scholar]
- 8.Bogaerts I, Verboven C, Rabijns A, Waelkens E, Van Baelen H, De Ranter C. Acta Crystallogr D. 2001;57:740–742. doi: 10.1107/s090744490100350x. [DOI] [PubMed] [Google Scholar]
- 9.Lee W M, Galbraith R M, Watt G H, Hughes R D, McIntire D D, Hoffman B J, Williams R. Hepatology. 1995;21:101–105. [PubMed] [Google Scholar]
- 10.Dahl B, Schiodt F V, Kiaer T, Ott P, Bondesen S, Tygstrup N. Crit Care Med. 1998;26:285–289. doi: 10.1097/00003246-199802000-00027. [DOI] [PubMed] [Google Scholar]
- 11.Daiger S P, Schanfield M S, Cavalli-Sforza L L. Proc Natl Acad Sci USA. 1975;72:2076–2080. doi: 10.1073/pnas.72.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto N, Homma S. Proc Natl Acad Sci USA. 1991;88:8539–8543. doi: 10.1073/pnas.88.19.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He X M, Carter D C. Nature (London) 1992;358:209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- 14.Spudich J A, Watt S. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 15.Vogelaar N J, Lindberg U, Schutt C E. J Mol Biol. 1991;220:545–547. doi: 10.1016/0022-2836(91)90097-p. [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 17.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 18.Kabsch W, Mannherz H G, Suck D, Pai E F, Holmes K C. Nature (London) 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 19.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.Cowtan K, Main P. Acta Crystallogr D. 1998;54:487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 22.McLaughlin P J, Gooch J T, Mannherz H G, Weeds A G. Nature (London) 1993;364:685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- 23.Otterbein L R, Graceffa P, Dominguez R. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 24.Robinson R C, Mejillano M, Le V P, Burtnick L D, Yin H L, Choe S. Science. 1999;286:1939–1942. doi: 10.1126/science.286.5446.1939. [DOI] [PubMed] [Google Scholar]
- 25.Schutt C E, Myslik J C, Rozycki M D, Goonesekere N C, Lindberg U. Nature (London) 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 26.Estes J E, Selden L A, Kinosian H J, Gershman L C. J Muscle Res Cell Motil. 1992;13:272–284. doi: 10.1007/BF01766455. [DOI] [PubMed] [Google Scholar]
- 27.Orlova A, Egelman E H. J Mol Biol. 1995;245:582–597. doi: 10.1006/jmbi.1994.0048. [DOI] [PubMed] [Google Scholar]
- 28.Holmes K C, Popp D, Gebhard W, Kabsch W. Nature (London) 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 29.Lorenz M, Popp D, Holmes K C. J Mol Biol. 1993;234:826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- 30.Coue M, Constans J, Viau M, Olomucki A. Biochim Biophys Acta. 1983;759:137–145. doi: 10.1016/0304-4165(83)90305-7. [DOI] [PubMed] [Google Scholar]
- 31.Lo Conte L, Chothia C, Janin J. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 32.Lees A, Haddad J G, Lin S. Biochemistry. 1984;23:3038–3047. doi: 10.1021/bi00308a030. [DOI] [PubMed] [Google Scholar]
- 33.Goldschmidt-Clermont P J, Williams M H, Galbraith R M. Biochem Biophys Res Commun. 1987;146:611–617. doi: 10.1016/0006-291x(87)90572-9. [DOI] [PubMed] [Google Scholar]
- 34.Verboven C, Rabijns A, De Maeyer M, Van Baelen H, Bouillon R, De Ranter C. Nat Struct Biol. 2002;9:131–136. doi: 10.1038/nsb754. [DOI] [PubMed] [Google Scholar]
- 35.Haddad J G, Hu Y Z, Kowalski M A, Laramore C, Ray K, Robzyk P, Cooke N E. Biochemistry. 1992;31:7174–7181. doi: 10.1021/bi00146a021. [DOI] [PubMed] [Google Scholar]
- 36.Guoth M, Murgia A, Smith R M, Prystowsky M B, Cooke N E, Haddad J G. Endocrinology. 1990;127:2313–2321. doi: 10.1210/endo-127-5-2313. [DOI] [PubMed] [Google Scholar]
- 37.Petrini M, Galbraith R M, Emerson D L, Nel A E, Arnaud P. J Biol Chem. 1985;260:1804–1810. [PubMed] [Google Scholar]
- 38.Yamamoto N, Homma S, Millman I. J Immunol. 1991;147:273–280. [PubMed] [Google Scholar]
- 39.Janmey P A, Stossel T P, Lind S E. Biochem Biophys Res Commun. 1986;136:72–79. doi: 10.1016/0006-291x(86)90878-8. [DOI] [PubMed] [Google Scholar]