Abstract
Runx factors control lineage commitment and are transcriptional effectors of Smad signaling. Genetic defects in these pathways interfere with normal development. The in situ localization of Runx and Smad proteins must impact the mechanisms by which these proteins function together in gene regulation. We show that the integration of Runx and Smad signals is mediated by in situ interactions at specific foci within the nucleus. Activated Smads are directed to these subnuclear foci only in the presence of Runx proteins. Smad–Runx complexes are associated in situ with the nuclear matrix, and this association requires the intranuclear targeting signal of Runx factors. The convergence of Smad and Runx proteins at these sites supports transcription as reflected by BrUTP labeling and functional cooperativity between the proteins. Thus, Runx-mediated intranuclear targeting of Smads is critical for the integration of two distinct pathways essential for fetal development.
Mammalian cells respond to a vast array of extracellular signals that play a critical role in determining cell fate. These regulatory signals are often transmitted to the nucleus by proteins capable of nucleocytoplasmic shuttling. After signal-induced activation, these proteins interact with transcription factors and act on target genes (1–4). A broad range of nuclear proteins including tissue-restricted transcription factors (e.g., Runx proteins), chromatin-remodeling complexes (e.g., SWI/SNF), components of basal transcription machinery (e.g., active form of RNA Pol II), and proteins involved in RNA processing (e.g., SC35) exhibit distinct subnuclear distributions (5–8). The subnuclear organization of these regulatory complexes may determine their optimal functions. Mechanisms underlying the activation of signaling proteins and their nuclear translocation have been extensively studied (1–4). It remains elusive whether the biological activities of these proteins require assembly of regulatory complexes within distinct subnuclear domains.
Runx factors exhibit a tissue-restricted pattern of expression and are required for definitive hematopoiesis and osteoblast maturation (9–12). Runx proteins have recently been shown to interact through their C-terminal segment with Smads, a family of signaling proteins that regulate a diverse array of developmental and biological processes in response to transforming growth factor (TGF)-β/bone morphogenetic protein (BMP) family of growth factors (1, 13–16). The C terminus of Runx proteins also mediates interactions with coregulators like Yes-associated protein (17) and groucho/TLE proteins (18). Moreover, subnuclear distribution of Runx proteins is mediated by the nuclear matrix-targeting signal, a protein motif present in the C terminus of Runx factors (19–22). Importantly, in vivo osteogenesis requires the C terminus of Runx2 containing the overlapping subnuclear targeting signal and the Smad interacting domain (23). The Runx and Smad proteins are jointly involved in the regulation of phenotypic gene expression and lineage commitment. Gene ablation studies have revealed that both Runx proteins and Smads are developmentally involved in hematopoiesis and osteogenesis (9–13, 24). Furthermore, Runx2 and the BMP-responsive Smads can induce osteogenesis in mesenchymal pluripotent cells (25–27). However, little is known about the involvement of higher-order nuclear structure in the assembly and organization of Runx–Smad complexes.
The interaction between Smad and Runx proteins is mediated through a region of the C terminus of Runx factors that overlaps with the subnuclear targeting signal (14, 15, 19, 20). We therefore postulated that the interaction of Runx proteins with Smads requires a distinct spatiotemporal organization of transcriptionally active complexes. Here we report that Smads are targeted to subnuclear sites only in cells that express endogenous Runx factors. Exogenous expression of Runx factors in cells that lack endogenous Runx proteins is sufficient for the subnuclear targeting of receptor Smads. We further show that, whereas nuclear import of receptor Smads is agonist-dependent, Runx proteins are required to target Smads to subnuclear sites. BrUTP labeling followed by double-label in situ immunofluorescence strikingly reveals that Runx proteins recruit Smads to subnuclear sites of active transcription. Taken together, our data suggest a mechanism for the in situ integration of signals by the assembly of regulatory complexes at transcriptionally active subnuclear sites.
Methods
Cell Culture and Transient Transfections.
Rat osteosarcoma ROS 17/2.8 or human cervical carcinoma HeLa cells were maintained in F12 medium containing 5% FCS or in DMEM containing 10% FBS, respectively. Transfections were carried out with standard protocols. For BMP2 or TGF-β treatment, cells were incubated with medium containing 1% charcoal-stripped serum containing BMP2 (300 ng/ml) or TGF-β (2.5 ng/ml) for indicated time.
Plasmid Constructs.
Hemagglutinin (HA)-tagged Runx2 expression construct (pHA-Runx2) has been reported (19). The expression vector for the Y428A mutant of Runx2 was generated by PCR-based site-directed mutagenesis. Flag-tagged Smad constructs and the dominant negative mutant of BMPR type I [BMPRI (KR)] have been reported (28, 29).
In Situ Immunofluorescence and Digital Microscopy.
HeLa and ROS 17/2.8 cells were grown on gelatin-coated coverslips and transfected with 0.5 μg of cytomegalovirus (CMV)-driven Flag-tagged receptor Smads. Cells treated with TGF-β or BMP2 (Genetics Institute, Cambridge, MA, a kind gift from V. Rosen) or untreated cells were processed in situ for whole-cell (WC) or nuclear matrix-intermediate filament (NM-IF) preparations and digital microscopic analyses essentially as described (17). Runx2 was detected by a rabbit polyclonal Ab against the HA tag at a dilution of 1:3000 (Santa Cruz Biotechnology). Rabbit polyclonal Abs against Runx1, Runx2, and Runx3 were a generous gift from S. Hiebert (Vanderbilt University). Smads were detected with a mouse monoclonal M2 Ab against Flag tag (Sigma–Aldrich) at a dilution of 1:1000. Secondary Abs used were either Alexa 488 anti-rabbit or Alexa 568 anti-mouse (Molecular Probes) at a dilution of 1:800.
BrUTP Labeling.
HeLa cells grown on gelatin-coated glass coverslips were transfected with the indicated constructs and treated with BMP2 as described above. To label sites of active transcription, cells were washed twice with ice-cold PBS and incubated with glycerol buffer (20 mM Tris⋅HCl, pH 7.4/5 mM MgCl2/0.5 mM EGTA/25% glycerol) for 3 min on ice. Cells were washed with glycerol buffer containing 0.05% Triton X-100 and 2 mM proteinase inhibitor 4-(2-aminoethyl)benzenesulfinyl fluoride (AEBSF). Cells were then incubated with transcription buffer [50 mM Tris⋅HCl, pH 7.4/10 mM MgCl2/0.5 mM EGTA/100 mM KCl/25% glycerol/0.025 mM S-adenosylmethionine/0.75 mM BrUTP/4 mM AEBSF/1 μl RNase inhibitor (RNasin, Roche Molecular Biochemicals), and 0.5 mM each of ATP, CTP, and GTP] for 15 min at room temperature. After BrUTP incorporation, cells were washed with ice-cold PBS containing 2 mM AEBSF and processed for WC or NM-IF preparations.
Immunoprecipitation and Western Blotting.
ROS 17/2.8 cells were transfected with 10 μg of indicated expression constructs in 100-mm plates. Cells were treated with BMP2 for 24 h as described above and harvested 48 h after transfection. Cells were processed for immunoprecipitations and Western blot analyses as described (30).
Results
Smads Exhibit Differences in Subnuclear Distribution in Cells Expressing or Lacking Runx Factors.
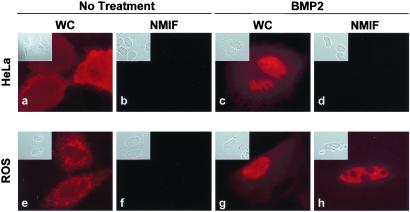
The interaction between Runx and Smad proteins provides a paradigm to understand mechanisms underlying the in situ assembly of multiprotein complexes. Here, we examine subnuclear distribution of Smads (BMP-regulated Smad1 and Smad5 and TGF-β-regulated Smad2 and Smad3) by using cell lines expressing (ROS 17/2.8) or lacking (HeLa) endogenous Runx factors (31, 32). Fig. 1 shows that Smad5 is distributed in the cytoplasm in both HeLa and ROS17/2.8 cells in the absence of agonist (Fig. 1 a and e). After activation with BMP2, Smad5 is translocated into the nucleus in both cell lines (Fig. 1 c and g). Extraction of cells to remove soluble proteins and chromatin revealed that Smad5 is not associated with the nuclear matrix of HeLa cells (Fig. 1d). In contrast, Smad5 was retained in the nuclear matrix of ROS 17/2.8 cells (Fig. 1h). Similar results were obtained with the BMP-responsive Smad1 as well as with the TGF-β-responsive Smad2 and Smad3 (data not shown). These findings indicate distinct subnuclear compartmentalization of Smads in cells expressing (ROS 17/2.8) or lacking (HeLa) endogenous Runx factors and suggest that this differential distribution may involve Runx proteins present in ROS 17/2.8 cells.
Figure 1.
Differential subnuclear organization of Smad5 in nonosseous and osseous cells. Human cervical carcinoma HeLa cells (a–d) or rat osteosarcoma ROS 17/2.8 cells (e–h) were grown on gelatin-coated coverslips and transfected with 0.5 μg of expression construct coding for Flag Smad5. After 24 h of transfection, cells were processed in situ for the WC and the NM-IF preparation and immunofluorescence microscopy. A mouse mAb against Flag tag (dilution: 1:1000) was used to detect Smad5.
Runx Subnuclear Foci Are a Target of Smad Signaling.
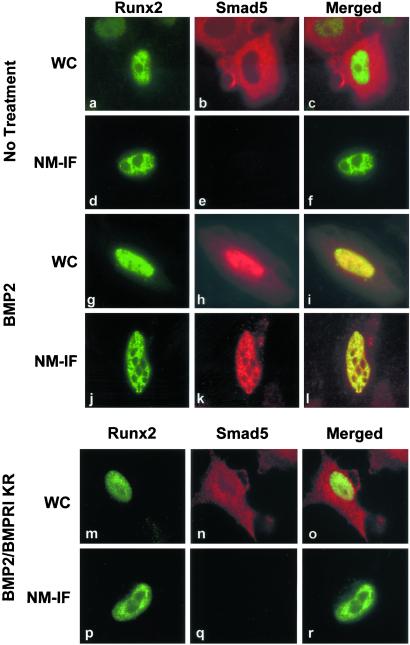
To address the involvement of Runx factors in differential subnuclear distribution of Smads, we exogenously expressed Runx factors together with Smads in cells that do not express any of the Runx proteins. In the absence of BMP2 or TGF-β, expression of Runx1, Runx2, or Runx3 has no effect on subcellular distribution of Smads (Fig. 2 a–c and data not shown). We treated cells with BMP2 (which activates Smad1 and Smad5) or TGF-β (which activates Smad2 and Smad3). In response to BMP2 (Fig. 2 g–i) or TGF-β (data not shown), Smads are translocated into the nucleus and colocalize with Runx2 in the WC preparation. Importantly, Smads show punctate subnuclear distribution and colocalize with Runx2 in the NM-IF preparation (Fig. 2 j–l). In the absence of agonist, Smads remain in the cytoplasm whereas Runx2 exhibits distinct subnuclear distribution, indicating that interaction must occur in the nucleus (Fig. 2 a–c). To confirm that the agonist-dependent activation of the pathway is an upstream event in subnuclear targeting of Smads, we used a dominant negative inhibitor [BMPRI (KR)] of the BMP signaling pathway (28). In the presence of BMPRI (KR), the distribution of Smad5 remains cytoplasmic even when cells are treated with BMP2 (Fig. 2n). Consequently, Smad5 is not associated with the nuclear matrix in BMP2-treated cells expressing Runx2 (Fig. 2q). Expression of BMPRI (KR) does not affect nuclear localization (Fig. 2m) or nuclear matrix association (Fig. 2p) of Runx2. These data demonstrate that the exogenous expression of Runx2 as well as activation of the BMP/TGF-β pathway are necessary for targeting Smads to subnuclear sites.
Figure 2.
Runx2 transcription factor targets Smads to subnuclear sites. HeLa cells, which lack endogenous Runx proteins, were transfected with 0.5 μg of expression construct for Flag Smad5 together with HA-Runx2 expression vector. BMP2-treated or untreated cells were processed for WC (a–c, untreated cells; g–i, BMP2-treated cells) or NM-IF (d–f, untreated cells; j–l, BMP2-treated cells) preparation and in situ immunofluorescence 24 h after transfection. Smad5 was detected with mouse mAb against Flag tag whereas Runx2 was detected by a rabbit polyclonal Ab against HA tag. To confirm that the activation of Smad5 by BMP2 is required for nuclear accumulation and consequent subnuclear targeting of Smad5, a dominant negative inhibitor of BMP signaling, BMPRI (KR), was expressed along with expression constructs for HA-Runx2 and Flag-Smad5. Cells were treated with BMP2 and were processed for WC (m–o) or NM-IF (p–r) preparation and in situ immunofluorescence as described.
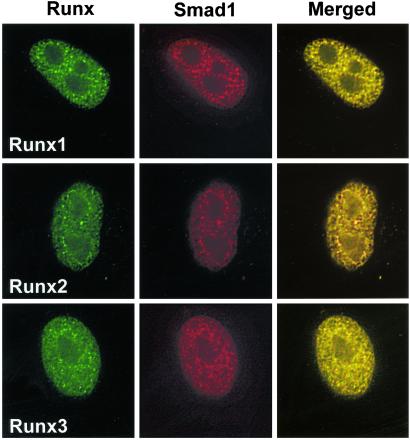
Runx1 and Runx3, the other family members, also possess subnuclear targeting signal and Smad interaction domain homologous to that of Runx2. To assess whether other Runx factors can also target Smads to intranuclear sites, we used Runx1 and Runx3 in coexpression experiments. Fig. 3 shows that Runx1 and Runx3 also colocalize with Smads and mediate their association with subnuclear foci (Fig. 3). Thus, these studies demonstrate that the ability of Runx factors to target Smad coregulatory proteins to specific subnuclear sites is a general property of this class of transcription factors.
Figure 3.
Runx factors specify subnuclear localization of Smads. HeLa cells were transfected with 0.5 μg of expression construct for Flag Smad1 along with Runx1, Runx2, or Runx3 expression vectors. Cells were treated with BMP2, and Smad1 was examined for association with Runx factors in the nuclear matrix (NM-IF preparation) by in situ immunofluorescence 24 h after transfection. Smad1 was detected with mouse mAb against Flag tag. Runx proteins were detected with rabbit polyclonal Abs at a dilution of 1:200. Images were taken by a Zeiss Axioplan microscope coupled with a charge-coupled device (CCD) camera and were processed for deconvulation microscopy with METAMORPH bioimaging software.
Runx2 Is Required for Subnuclear Targeting of Smads.
Smads interact with Runx factors through a region of the C terminus that overlaps with the subnuclear targeting signal (14, 15, 19, 20). Hence, Smads may rely on the subnuclear targeting signal of Runx factors to organize into transcriptional complexes. To confirm whether subnuclear targeting of Smads requires the nuclear matrix-targeting signal (NMTS) of Runx proteins, we introduced a point mutation (Y428A) in the NMTS of Runx2 (Fig. 4a). To validate that the point mutation (Y428A) does not affect Smad–Runx interaction, we carried out coimmunoprecipitation studies. Fig. 4b shows that Runx2 Y428A mutant protein remains capable of interaction with Smads. Moreover, this point mutation has no effect on nuclear import of Runx2 (Fig. 4c); however, it abolishes association of Runx2 with the nuclear matrix (Fig. 4f). When Runx2 Y428A mutant and Smad5 are coexpressed, Runx2 Y428A mutant colocalizes with Smad5 in WC preparations (Fig. 4 c–e). In contrast, the mutant Runx2 protein or Smad5 are not detected in NM-IF preparations (Fig. 4 f–h). Similar results were obtained with the BMP-responsive Smad1 as well as the TGF-β-responsive Smad2 and Smad3 (data not shown). Taken together, our findings provide direct evidence that Runx proteins are required for targeting Smads to the nuclear matrix-associated subnuclear foci. Furthermore, the cotargeting of a developmental transcription factor and a transducer of morphogenic cues to specific subnuclear sites is a mechanism for the in situ integration of two signaling pathways to regulate transcription.
Figure 4.
Runx2 is required for subnuclear targeting of Smad5. (a) A schematic of Runx2 protein. The region where Smads interact is shown. The Y428A mutation, which disrupts association of Runx2 with the nuclear matrix, was introduced by PCR-based site-directed mutagenesis. QA, poly glutamine-poly alanine stretch; RHD, runt homology domain; NMTS, nuclear matrix-targeting signal; VWRPY, a highly conserved motif that mediate interactions of Runx2 with groucho/TLE proteins. (b) ROS17/2.8 cells grown in 100-mm plates were transfected with 10 μg each of expression constructs for Flag-Smad1 and wild-type HA-Runx2 or HA-Runx2 Y428A mutant. Cells were treated with 300 ng/ml of BMP2 for 24 h and processed for immunoprecipitation. One microgram Ab against Flag tag was used for immunoprecipitation. The immunoprecipitated complex was resolved by 8% SDS/PAGE. Wild-type or mutant Runx2 proteins were detected with mouse mAb against HA tag (dilution 1:2000). HeLa cells were transfected with 0.5 μg of expression constructs for Flag-Smad1 along with HA-tagged wild-type or Runx2 Y428A mutant proteins. Cells were treated with BMP2 and subjected to WC (c–e) or NM-IF (f–h) preparation and double-labeled in situ immunofluorescence with mouse mAb against Flag tag and rabbit polyclonal Ab against HA tag.
Runx2 Recruits Smads to Subnuclear Sites of Active Transcription.
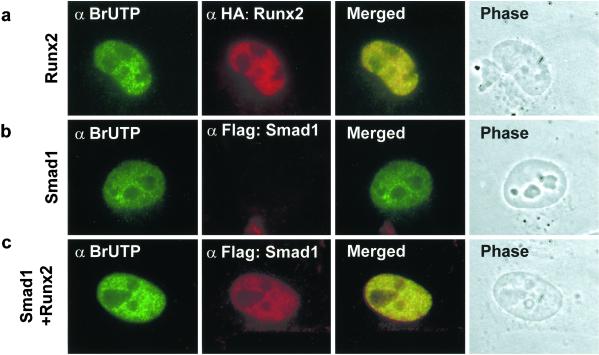
Runx factors cooperate with Smads to regulate gene expression and lineage commitment (14–16, 27). This cooperation necessitates subnuclear interactions in situ between Runx factors and Smads at sites that support transcription. Therefore, we examined the extent to which Smad–Runx complexes are associated with transcriptionally active intranuclear foci that were labeled with BrUTP. Cells expressing Runx2 and/or Smads were incubated with Abs against BrUTP and HA or Flag tags to visualize BrUTP incorporation at sites containing Runx2–Smad complexes. The localization of Smads with sites of transcription was assessed in the NM-IF preparation in the presence or absence of Runx2. The majority of Runx2 subnuclear foci is transcriptionally active (Fig. 5a). As expected, Smads are not directed to the nuclear matrix in the absence of Runx2 (Fig. 5b). However, in the presence of Runx2, Smads (i.e., Smad1 and Smad2) colocalize with BrUTP-labeled subnuclear sites (Fig. 5c and data not shown). These data indicate that Runx proteins can recruit Smads to subnuclear sites of active transcription. In addition, these data provide evidence that the structural compartments of the nucleus mediate in situ functional cooperation between Runx and Smad proteins in transcriptional control.
Figure 5.
Runx2 recruits Smad1 to subnuclear sites. HeLa cells were transfected with 0.5 μg of indicated expression constructs. BMP2-treated cells were labeled with BrUTP and subjected to NM-IF preparation and in situ immunofluorescence. A rat mAb (dilution 1:20) was used to detect BrUTP. Flag-Smad was detected with a mouse mAb against Flag tag.
Recruitment of Smads to Sites of Active Transcription Is Coupled with the Regulation of Gene Expression.
We have shown that Runx transcription factors recruit Smads to sites of active transcription. To address whether presence of Smad–Runx complex at transcriptionally active sites is directly coupled with the control of a gene responsive to both Runx and Smads regulatory proteins, we used a luciferase reporter under the control of multiple copies of TGF-β-responsive element (TβRE). As shown in Fig. 6, wild-type Runx2 protein activates the reporter to 3–4-fold. In contrast, expression of TGF-β-responsive Smad3 or BMP-responsive Smad5 does not influence reporter activity. When cells expressing Runx2 and Smads are treated with either TGF-β (for Smad3) or BMP2 (for Smad5), Runx2 and Smads cooperate to activate the reporter to an extent of 100-fold. Notably, this increase is functionally linked to Runx2-mediated recruitment of Smads to sites of active transcription. Importantly, this functional cooperation between Runx2 and Smads to transactivate a reporter gene is not observed when the Runx2 Y428A mutant, which does not associate with the nuclear matrix, is coexpressed with Smads (Fig. 6). Taken together, these data suggest a coupling of transcriptional regulation with Runx-mediated recruitment of Smads to sites of active gene expression.
Figure 6.
Recruitment of Smads to sites of active transcription is coupled with the regulation of gene expression. HeLa cells were transfected with 0.45 95 g of pTβRE-Luc (Upper) along with 250 ng of pcDNA 3.1 (EV), Runx2 (wild-type or Y428A mutant), and/or Smad3 or 5. A promoterless luciferase gene was used as internal control for the transfection efficiency. Cells were treated with TGF-β (10 ng/ml) or BMP2 (300 ng/ml) for 6 h and harvested in passive lysis buffer 24 h after transfection. Ten microliters of the cell lysate was used for dual luciferase assay. The activity of the firefly luciferase was normalized with that of Renilla luciferase. The graphs represent three independent experiments with n = 6. TβRE, TGF-β-responsive element.
Discussion
We have addressed the fundamental question of how distinct developmental signals involving proteins capable of nucleocytoplasmic shuttling converge within the nucleus. With biochemical and in situ immunofluorescence approaches, we have shown that tissue-restricted Runx factors can mediate subnuclear targeting of receptor-regulated Smads to transcriptionally active sites. Our data support a novel mechanism by which signaling pathways functionally integrate at intranuclear sites to exert biological control.
Components of the transcriptional regulatory machinery are organized in discrete subnuclear foci. This subnuclear concentration of gene regulatory factors may govern biological functions by facilitating the spatiotemporal assembly and activity of complexes that contain gene-specific transcription factors, their corresponding coregulators, as well as histone-modifying enzymes, chromatin-remodeling factors, and RNA polymerase II (5–8, 33). We and others have previously identified intranuclear targeting motifs that direct regulatory proteins to specific foci that may accommodate the assembly of transcriptional complexes. The intranuclear recruitment of Smads by Runx factors and their intrinsic subnuclear targeting signal to subnuclear sites demonstrates that Runx factors can function as subnuclear acceptor proteins for signal transduction (19–21, 34–36). These key findings support the concept that targeting signals provide the requisite specificity for intranuclear trafficking and functional compartmentalization within the nucleus.
Subnuclear targeting of gene regulatory proteins to specific foci has been implicated in fidelity of tissue-specific transcriptional regulation. The composition of these subnuclear foci determines whether genes are activated or repressed (33, 34, 37) and that abrogation of subnuclear targeting results in pathological conditions (37). The findings we present here indicate mutations that prevent subnuclear localization of Runx proteins but retain Runx–Smad interactions disrupt the normal targeting of Runx–Smad complex to transcriptionally active sites. Indeed, interfering with subnuclear targeting of Runx factors and their interaction with Smads results in human disorders and lethal mouse phenotypes. For example, chromosomal translocations in human leukemia involving Runx1 result in fusion proteins that lack the subnuclear targeting signal as well as the Smad interacting domain (38). Recently, a nonsense mutation in Runx2 has been described in patients with the skeletal disease cleidocranial dysplasia (CCD) (39). This mutant protein lacks both subnuclear targeting and Smad interaction. Moreover, eliminating the Smad interaction domain and subnuclear targeting signal, which overlap in the C terminus of either Runx1 or Runx2, abrogates hematopoiesis and bone formation in the mouse (23, 40), respectively. The pathological consequences of mutations that compromise targeting of the protein to subnuclear sites suggest direct coupling of subnuclear organization, gene expression, and biological functions.
We propose that Runx proteins represent key organizers that integrate signals in situ at subnuclear foci by recruiting coregulators with repressive or activating potential. Our data show that the recruitment of Smads to intranuclear sites is Runx-dependent and spatially linked to active transcription. Thus, Runx factors may accommodate the dynamic targeting of signal transducers to sites of active transcription and functionally alter the activity of specific subnuclear foci.
Acknowledgments
This work was supported by National Institutes of Health Grants AR39588, AR45688, and AR48818.
Abbreviations
- TGF
transforming growth factor
- HA
hemagglutinin
- WC
whole cell
- NM-IF
nuclear matrix-intermediate filament
- BMP
bone morphogenetic protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Massague J. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 2.Finidori J. Vitam Horm. 2000;59:71–97. doi: 10.1016/s0083-6729(00)59004-9. [DOI] [PubMed] [Google Scholar]
- 3.Ihle J N. Curr Opin Cell Biol. 2001;13:211–217. doi: 10.1016/s0955-0674(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 4.Akiyama T. Cytokine Growth Factor Rev. 2000;11:273–282. doi: 10.1016/s1359-6101(00)00011-3. [DOI] [PubMed] [Google Scholar]
- 5.Merriman H L, van Wijnen A J, Hiebert S, Bidwell J P, Fey E, Lian J, Stein J, Stein G S. Biochemistry. 1995;34:13125–13132. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 6.Reyes J-C, Muchardt C, Yaniv M. J Cell Biol. 1997;137:263–274. doi: 10.1083/jcb.137.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei X, Samarabandu J, Devdhar R S, Siegel A J, Acharya R, Berezney R. Science. 1998;281:1502–1505. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- 8.Shopland L S, Lawrence J B. J Cell Biol. 2000;150:F1–F4. doi: 10.1083/jcb.150.1.f1. [DOI] [PubMed] [Google Scholar]
- 9.Ito Y. Genes Cells. 1999;4:685–696. doi: 10.1046/j.1365-2443.1999.00298.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto F, Thornell A P, Crompton T, Denzel A, Gilmour K C, Rosewell I R, Stamp G W H, Beddington R S P, Mundlos S, Olsen B R, et al. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 12.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson R T, Gao Y-H, Inada M, et al. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 13.Derynck R, Zhang Y, Feng X H. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 14.Hanai J, Chen L F, Kanno T, Ohtani-Fujita N, Kim W Y, Guo W H, Imamura T, Ishidou Y, Fukuchi M, Shi M J, et al. J Biol Chem. 1999;274:31577–31582. doi: 10.1074/jbc.274.44.31577. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y W, Yasui N, Ito K, Huang G, Fujii M, Hanai J, Nogami H, Ochi T, Miyazono K, Ito Y. Proc Natl Acad Sci USA. 2000;97:10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. EMBO J. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yagi R, Chen L F, Shigesada K, Murakami Y, Ito Y. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Javed A, Guo B, Hiebert S, Choi J-Y, Green J, Zhao S-C, Osborne M A, Stifani S, Stein J L, Lian J B, et al. J Cell Sci. 2000;113:2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- 19.Zaidi S K, Javed A, Choi J-Y, van Wijnen A J, Stein J L, Lian J B, Stein G S. J Cell Sci. 2001;114:3093–3102. doi: 10.1242/jcs.114.17.3093. [DOI] [PubMed] [Google Scholar]
- 20.Zeng C, van Wijnen A J, Stein J L, Meyers S, Sun W, Shopland L, Lawrence J B, Penman S, Lian J B, Stein G S, et al. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang L, Guo B, Javed A, Choi J-Y, Hiebert S, Lian J B, van Wijnen A J, Stein J L, Stein G S, Zhou G W. J Biol Chem. 1999;274:33580–33586. doi: 10.1074/jbc.274.47.33580. [DOI] [PubMed] [Google Scholar]
- 22.Zeng C, McNeil S, Pockwinse S, Nickerson J A, Shopland L, Lawrence J B, Penman S, Hiebert S W, Lian J B, van Wijnen A J, et al. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi J-Y, Pratap J, Javed A, Zaidi S K, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen A J, Lian J B, et al. Proc Natl Acad Sci USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goumans M J, Mummery C. Int J Dev Biol. 2000;44:253–265. [PubMed] [Google Scholar]
- 25.Lee M H, Javed A, Kim H J, Shin H I, Gutierrez S, Choi J Y, Rosen V, Stein J L, van Wijnen A J, Stein G S, et al. J Cell Biochem. 1999;73:114–125. [PubMed] [Google Scholar]
- 26.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney J M, Fujisawa-Sehara A, Suda T. J Cell Biol. 1994;127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K S, Kim H J, Li Q L, Chi X Z, Ueta C, Komori T, Wozney J M, Kim E G, Choi J Y, Ryoo H M, et al. Mol Cell Biol. 2000;20:8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hata A, Lo R S, Wotton D, Lagna G, Massague J. Nature (London) 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- 29.Liu F, Hata A, Baker J C, Doody J, Carcamo J, Harland R M, Massague J. Nature (London) 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 30.Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. EMBO J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Javed A, Gutierrez S, Montecino M, van Wijnen A J, Stein J L, Stein G S, Lian J B. Mol Cell Biol. 1999;19:7491–7500. doi: 10.1128/mcb.19.11.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armesilla A L, Calvo D, Vega M A. J Biol Chem. 1996;271:7781–7787. doi: 10.1074/jbc.271.13.7781. [DOI] [PubMed] [Google Scholar]
- 33.Stein G S, van Wijnen A J, Stein J L, Lian J B, Montecino M, Choi J-Y, Zaidi K, Javed A. J Cell Sci. 2000;113:2527–2533. doi: 10.1242/jcs.113.14.2527. [DOI] [PubMed] [Google Scholar]
- 34.DeFranco D B, Guerrero J. Crit Rev Eukaryotic Gene Expression. 2000;10:39–44. [PubMed] [Google Scholar]
- 35.Mancini M G, Liu B, Sharp Z D, Mancini M A. J Cell Biochem. 1999;72:322–338. [PubMed] [Google Scholar]
- 36.McNeil S, Guo B, Stein J L, Lian J B, Bushmeyer S, Seto E, Atchison M L, Penman S, van Wijnen A J, Stein G S. J Cell Biochem. 1998;68:500–510. [PubMed] [Google Scholar]
- 37.Stein G S, van Wijnen A J, Stein J L, Lian J B, Montecino M, Zaidi S K, Javed A. J Cell Biochem. 2000;35:84–92. [PubMed] [Google Scholar]
- 38.McNeil S, Zeng C, Harrington K S, Hiebert S, Lian J B, Stein J L, van Wijnen A J, Stein G S. Proc Natl Acad Sci USA. 1999;96:14882–14887. doi: 10.1073/pnas.96.26.14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Yasui N, Kakazu N, Abe T, Takada K, Imai S, Sato M, Nomura S, Ochi T, Okuzumi S, et al. Gene. 2000;244:21–28. doi: 10.1016/s0378-1119(99)00558-2. [DOI] [PubMed] [Google Scholar]
- 40.North T, Gu T L, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck N A. Development (Cambridge, UK) 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]