Abstract
Mutation of the functionally redundant Hoxa 11/Hoxd 11 genes gives absent or rudimentary kidneys resulting from a dramatic reduction of the growth and branching of the ureteric bud. To understand better the molecular mechanisms of Hoxa 11/Hoxd 11 function in kidney development, it is necessary to identify the downstream target genes regulated by their encoded transcription factors. To this end, we conducted a screen for Hoxa 11-responsive genes in two kidney cell lines. HEK293 cells, which usually do not express Hoxa 11, were modified to allow inducible Hoxa 11 expression. The mK10 cells, derived specifically for this study from Hoxa 11/Hoxd 11 double-mutant mice, were also modified to give cell populations with and without Hoxa 11 expression. Differential display, Gene Discovery Arrays, and Affymetrix genechip probe arrays were used to screen for genes up- or down-regulated by Hoxa 11. Nine genes, PDGF A, Cathepsin L, annexin A1, Mm.112139, Est2 repressor factor, NrCAM, ZNF192, integrin-associated protein, and GCM1, showed reproducible 3-fold or smaller changes in gene expression in response to Hoxa 11. One gene, the Integrin α8, was up-regulated approximately 20-fold after Hoxa 11 expression. The Integrin α8 gene is expressed together with Hoxa 11 in metanephric mesenchyme cells, and mutation of Integrin α8 gives a bud-branching morphogenesis defect very similar to that observed in Hoxa 11/Hoxd 11 mutant mice. In situ hybridizations showed a dramatic regional reduction in Integrin α8 expression in the developing kidneys of Hoxa 11/Hoxd 11 mutant mice. This work suggests that the Integrin α8 gene may be a major effector of Hoxa 11/Hoxd 11 function in the developing kidney.
The Hoxa 11 and Hoxd 11 genes have an important function in kidney development. Hoxa 11 is expressed in the metanephric mesenchyme before ureteric bud invasion and continues in the condensing mesenchyme around the ureteric bud tips (1). As the condensing mesenchyme further differentiates and some cells undergo a mesenchymal-to-epithelial conversion, Hoxa 11 expression is lost, suggesting a role for Hoxa 11 early in the process of nephrogenesis. Hoxd 11 expression in the developing kidney is similar (1). Gene-targeting studies show that the paralogous Hoxa 11 and Hoxd 11 genes are functionally redundant (2). Although the single knockout of each gene has no detectable renal phenotype, the Hoxa 11/Hoxd 11 double knockout results in renal agenesis or the formation of a small kidney rudiment that contains very few nephrons (2). The ureteric bud of the double mutant shows a dramatic reduction in branching (1), consistent with the reduced number of nephrons formed, which suggests that the mutant mesenchyme, where Hoxa 11 and Hoxd 11 are expressed, does not properly signal the bud to branch.
To understand better the mechanisms of Hox gene function it is necessary to identify their downstream effectors. Despite considerable effort, relatively little progress has been made in this area. One strategy used with success has been immunoprecipitation of target genes that have been fixed in vivo to the transcription factor of interest (3–6). Sequence scanning of promoters for potential Hox protein-binding sites has identified the β-amyloid (7), cytotactin (8), and neural cell adhesion (9) genes as candidate downstream targets. Cell-line studies have often used a candidate target-gene approach, for example, showing that Hoxb 7 up-regulates bFGF expression in melanoma cells (10). By using a combination of strategies the p53 and the progesterone receptor genes were identified as targets of Hoxa 5 (11, 12).
In this report we describe the results of a screen for the downstream targets of Hoxa 11. A cell-line strategy was used (13–15). Differential display, Gene Discovery Arrays (GDA), and Affymetrix genechip probe arrays were used to identify genes that were up- or down-regulated in response to Hoxa 11 expression. In both cell systems used, the Integrin α8 gene showed the greatest fold change in expression, which is of particular note because the kidney phenotypes resulting from the Hoxa 11/Hoxd 11 and Integrin α8 mutations are strikingly similar (2, 16), suggesting that Integrin α8 may be a key effector of Hoxa 11 function in kidney development. The role of Hoxa 11 in regulating Integrin α8 expression in vivo was confirmed by in situ hybridizations. Hoxa 11/Hoxd 11 mutant kidneys showed dramatic regional reduction in Integrin α8 expression.
Methods
Isolation and Establishment of Kidney Cell Lines.
Hoxa 11-SV40 Large Tag transgenic mice were bred with Hoxa 11+/− Hoxd 11+/− double-heterozygous mice. Triple-heterozygous mice (Hoxa 11+/−, Hoxd 11+/−, Hoxa 11-SV40 Large Tag+) were bred to produce embryos of the desired genotype. Kidney tissue was carefully dissected from E18.5 embryos and dissociated with trypsin, and the cells were plated on 100-mm tissue-culture dishes. Genotype was determined by PCR. The cells were cultured in DMEM (GIBCO/BRL/Invitrogen) with 10% FBS (GIBCO/BRL/Invitrogen) at 37°C in 5% CO2. Media were changed every 2–3 days, and cells were split when confluent. After 10 or more passages, clones were established by dilution cloning. The mK10 cell line has subsequently been grown for more than 20 additional passages, suggesting these cells are immortalized. HEK293 Tet-Off cells were purchased from CLONTECH and maintained according to the manufacturer's protocol.
Modification of Cell Lines.
The plasmid pcDNA 3.1/Hygro (Invitrogen) was used to drive the expression of full-length Hoxa 11 cDNA in mK10 cells. For the 293 Tet-Off cells, a FLAG epitope tagged Hoxa 11 cDNA was cloned into the pBI Tet expression vector (CLONTECH) and introduced by transfection. One day after calcium chloride transfection, cells were subjected to Hygromycin selection and colonies picked after sufficient growth. FLAG epitope tagging was introduced with PCR by using the following primers: 5′-CCCCAAGCTTCCACCATGGCTCCAAAGAAGAAGCGTAAGGTAGACTACAAGGACGACGATGACAAGGATTTTGATGAG-3′ and 5′-AGGAAGCTTAACCACGGAGATCTG-3′. The FLAG epitope was detected with the M5 monoclonal antibody (Sigma). Doxycycline (2 μg/ml) was added to the media for 2 days with media changes every 12 h to regulate Hoxa 11 expression in the 293 Tet-Off cells.
Gene Expression Profile Differences.
Differential display was performed by using GeneHunter (Nashville, TN) reagents and protocols. The first five sets of randomly designed hexamers were used on each sample and done in triplicate. For GDAs (Genome Systems/Incyte Pharmaceuticals, Palo Alto, CA), probe labeled from cell line mRNA with a poly(dT) oligonucleotide in a reverse transcription reaction with [33P]UTP was used to probe the arrays according to the manufacturer's protocol. Affymetrix genechip probe arrays were probed with biotinylated cRNA prepared according to the manufacturer (Affymetrix, Santa Clara, CA). The Human 6800 gene chip set was used in these studies.
Northern Blots and Reverse Transcription (RT)-PCR.
Total RNA was isolated from trypsinized cells by using RNAzol (Tel-Test, Friendswood, TX). Poly(A)+ was selected with the Oligotex mRNA isolation kit (Qiagen, Carlsbad, CA). Reverse transcription was performed with Superscript II (GIBCO/BRL/Invitrogen) and a random hexamer mix according to manufacturer's directions. After reverse transcription, PCR used Taq polymerase (Qiagen). Semiquantitative RT-PCR was done by adding 1 μCi of [32P]dATP to each 50-μl PCR. The linear amplification phase was determined for each primer pair by determining incorporation at 18, 20, 22, 24, and 26 cycles. Gels were exposed to phosphoimager screens, and plots of intensity vs. cycle number were generated. The lowest cycle number possible was selected for the comparison analysis. All gene expression was normalized to GAPDH. Probes used for Northern blots were made by RT-PCR. Primers used: Hoxa 11 RT 5′-AAAACCTCGCTTCCTCCGACTACC-3′ and 5′-CGCAATGTGGCTTGACCTTGTC-3′; GCMa 5′-GAACCTGACGACTCTGATTCTGAAG-3′ and 5′-TCACAGTTGGGACAGCGTTTCC-3′; Integrin α8 5′-AGCTACTTCGGCTACTCACTGGAC-3′ and 5′-TCCTCCCACTATAAGGTCTCCATTC-3′; PDGF-A 5′-GTGCTTTATTGCCAGTGTGCG-3′ and 5′-AAGACATTCCTGCTTCCTGCG-3′; GAPDH 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′; IAP 5′-TTGGAGCCATTCTTTTCGTCC-3′ and 5′-AAGGGTCTCATAGGTGACAACCAG-3′; ZFP192 5′-CTGTGCTTCTTCCCATCTCCT-3′ and 5′-AATAACGAGTGAGGTGCTGAGGC-3′; NrCAM 5′-CTCAAAATCTTGTGCTGTCCCC-3′ and 5′-GCAGTTCCCTGTTGTCCTTCAG-3′.
In Situ Hybridization.
For whole-mount in situ hybridizations the genitourinary block was isolated from E13.5 embryos. Whole mount and section in situ hybridizations were performed according to established protocols (17, 18).
Results
Strategy.
To better understand the developmental function of Hoxa 11 it is necessary to identify the downstream target genes it regulates. To this end we used a cell-line strategy similar to those described (13–15, 19). In essence, the gene expression patterns of otherwise identical cells with and without Hoxa 11 expression were compared. The Hoxa 11 responsive genes were altered in expression level.
Cell Lines.
The use of developmentally appropriate cell lines promotes the identification of legitimate downstream gene targets. Such cells have the correct cofactor combination required for proper target binding. We are particularly interested in the function of the Hoxa 11 gene in kidney development. Hoxa 11 is expressed in the metanephric mesenchyme of the developing kidney before induction by the ureteric bud and during the condensation of these cells to form the epithelia of the nephron (1). The Hoxa 11 gene is turned off during the later stages of epithelia differentiation.
Two cell lines were used. The Human Embryonic Kidney 293 (HEK293) cells represent a late stage of kidney epithelia development. No Hoxa 11 transcripts were detected in these cells by RT-PCR or Northern blot (data not shown). HEK293 cells are used extensively for study of channel and transporter proteins (20–22).
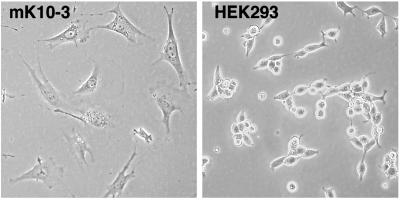
The other cell line used, mK10, was derived specifically for these studies. The Hoxa 11 promoter was used to drive restricted expression of SV40 large T antigen (Tag) in transgenic mice, to immortalize and developmentally arrest early kidney cells. The transgene DNA construct included 5.1 kb upstream of the Hoxa 11 transcription start site, 3.8 kb downstream, and the intact Hoxa 11 intron. Ten Hoxa 11-SV40 transgenic founder mice were made, and one was mated with Hoxa 11, Hoxd 11 double-heterozygous mutants to make Hoxa 11+/− Hoxd 11+/−, Hoxa 11-SV40 transgenics. These mice were then mated, and fetuses collected at E18.5. The mK10 kidney cell line was made from a single Hoxa 11−/− Hoxd 11−/− Hoxa11-SV40 Tag fetus. After ten passages dilution cloning was used to derive the mK10 cells, which have now been passaged more than 20 times, suggesting they represent an immortalized cell line. The morphologies of the HEK293 and mK10 cells are shown in Fig. 1.
Figure 1.
Morphology of mK10 and HEK293 cells. The mK10 cells, double-homozygous mutant for Hoxa 11 and Hoxd 11, have a flattened, often spindle-shaped appearance, typical of fibroblasts. The HEK293 cells are more rounded, often appearing polygonal at confluence, which suggests an epithelial character.
Making Cell Lines With and Without Hoxa 11 Expression.
HEK293 cells with inducible Hoxa 11 expression were made by using the Tet-Off system developed by Bujard and colleagues (23). An amino-terminal FLAG-tagged Hoxa 11 cDNA construct was cloned into the Tet-responsive vector (CLONTECH) and transfected into modified HEK293 cells that express the tet-Transactivator. Hygromycin-resistant clones were picked and expression levels of FLAG-Hoxa 11 in the presence and absence of doxycycline determined by Western blot (Fig. 2). A single clone showing greater than 10-fold induction in the absence of doxycycline was selected for subsequent studies.
Figure 2.
Induction of Hoxa 11 expression in HEK293 cells. Western blot detection of Hoxa11-FLAG fusion protein in cells not induced (+doxycycline) and induced (−doxycycline). Actin served as a loading control.
The mK10 cells were transfected with pCMV-Hoxa 11, which drives expression of Hoxa 11 from the CMV promoter. Among the resulting hygromycin-resistant clones, the mK10/pCMV-Hoxa 11 clone with the highest level of Hoxa 11 expression, was selected for comparison studies.
Screens for Hoxa 11 Responsive Genes.
Differential display, GDA, and Affymetrix genechip probe arrays were used to compare gene expression profiles of cells, with and without Hoxa 11 expression, to search for Hoxa 11 downstream targets.
RNAs from mK10 and mK10/pCMV-Hoxa11 cells were examined by differential display. Each PCR was done with three independent RNA isolations. More than 100 differences were identified and tested by reverse Northern (RNA labeled, PCR products blotted), which identified 33 clones apparently differently expressed. Repeated Northern analysis, however, confirmed only two genes as reproducibly differently expressed. GAPDH was used as a loading control, with phosphoimager quantitation. Both genes were down-regulated approximately 2-fold by Hoxa 11 expression. Cathepsin L is a widely expressed cysteine protease found in lysosomes that is sometimes secreted by expressing cells (24). Cathepsin L has been implicated in tumor invasion (25), trophoblast invasion during implantation (26, 27), and endochondral bone formation (28). Annexin AI (Lipocortin I) is a member of the calcium–phospholipid-binding family of proteins thought to be involved in signal transduction (29). Annexin AI is expressed in restricted epithelial kidney structures in the adult including Bowman's capsule, macula densa, and collecting ducts (30).
The double-mutant cell line was also used in a screen with GDAs, with more than 18,000 cDNAs blotted onto a nylon filter. Duplicate arrays were probed with 33P-labeled cDNA made by reverse transcription of each mRNA sample. Analysis of the phosphoimage files by Genome Systems (Incyte Pharmaceuticals, Palo Alto, CA) listed 705 cDNAs altered at least 2-fold between the mK10 and mK10/pCMV-Hoxa 11 RNA samples (420 up, 285 down). Each blotted cDNA was examined manually to eliminate artifacts caused by blotting errors, and forty of the most promising cDNAs were selected for confirmation by Northern blot analysis. Expression levels were again normalized to GAPDH expression by using a phosphoimager. Two clones showed a reliable, although small, expression difference (Table 1). One clone represented an expressed sequence tag cluster designated Mm.112139 with weak homology to EHD1, an EH domain-containing protein expressed in the developing limb buds, testis, and kidney (31). EH domain proteins are implicated in signaling and ligand-induced endocytosis (32). The Est2 repressor factor (ERF) gene is an ets-domain transcription factor expressed widely in the adult (33) that acts as an antagonist to other ets-domain transcriptional activators (34).
Table 1.
Confirmed gene expression differences
| Screen/cells used | Unigene cluster | Description | Fold change |
|---|---|---|---|
| Differential display: mK10 & Pcmv-Hoxa 11 | Mm.930 | Cathepsin L | −2.0 |
| Mm.14860 | Annexin A1 | −2.0 | |
| GDA: mK10 & Pcmv-Hoxa 11 | Mm.112139 | Expressed sequence tags | −1.8 |
| Mm.8068 | Est2 repressor factor | −1.4 | |
| Affymetrix GeneChip: Tet inducible Hoxa | Hs.91296 | Integrin α8 | +23.0 |
| 11/HEK 293 | Hs.37040 | PDGF-A | −2.0 |
| Hs.28346 | GCMa | +1.6 | |
| Hs.7912 | hBRAVO/NrCAM | +2.0 | |
| GB Z25521 | IAP/CD47 | +3.0 | |
| Hs.57679 | Zinc finger protein 192 | +2.2 |
+, Increased expression with Hoxa 11 expression.
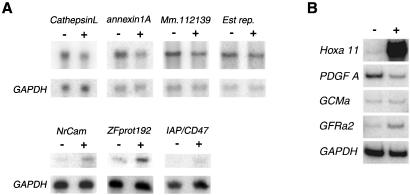
Affymetrix genechip probe arrays were used to compare gene expression profiles of Hoxa 11 induced and uninduced HEK293 cells. Of the approximately 6,800 genes screened, 309 were reported as 1.5-fold or greater differently expressed. Further analysis by Northern blot or quantitative RT-PCR confirmed expression differences for six genes (Fig. 3 and Table 1). Integrin α8 is a cell-membrane protein that interacts with the extracellular matrix. Integrins regulate cell growth and differentiation, creating a link between the extracellular matrix environment and cell behavior (35). Platelet-Derived Growth Factor A chain (PDGF-A) is a well-characterized growth factor expressed in epithelial cells in many tissues including the kidney. There, PDGF-A expression is initiated in newly formed epithelial cells as they are formed from condensed mesenchymal cells around the ureteric bud tips, whereas the receptors for PDGF are expressed in mesenchymal cells (36). Hoxa 11 is expressed in the condensing mesenchymal cells that are beginning the epithelialization process and turns off as the cells differentiate further (1). This observed pattern, with Hoxa 11 expression decreasing as PDGF expression increases, is consistent with an in vivo role for Hoxa 11 in repressing the PDGF gene. GCM1 is a transcription factor gene cloned by homology to the Drosophila Glial Cells Missing α gene. Although responsible for neuronal cell-fate decisions in the fly, both known mammalian homologs are expressed in E16.5 kidneys and in the placenta, suggesting other roles in mammalian development (37). Gene knockout of GCM1 disrupted branching morphogenesis of trophoblast populations in the placenta, resulting in lethality at E10, before kidney formation (38). Neuronal cell adhesion molecule (NrCAM) plays an important role in the growth cone during neural development (39). Although no role for NrCAM in kidney development has been ascribed, other neural cell adhesion molecules have been described in the ureteric epithelium and ductal epithelial portions of the nephron (40). Integrin-associated protein (IAP/CD47) is a membrane protein important in the inflammatory response to infection (41). Zinc finger protein 192 is a Kruppel family member expressed in the adult kidney (42).
Figure 3.
Candidate Hoxa 11 downstream targets. (A) Northern blots showing expression differences in mutant mK10 cells (Upper) and HEK293 cells (Lower) with and without Hoxa 11 expression. + indicates with Hoxa 11 expression. Blots were stripped and then reprobed with GAPDH as a loading control. (B) Semiquantitative RT-PCR confirmation of candidate target gene expression differences in HEK293 cells. Observed differences for these genes were small but reproducible.
The Integrin α8 gene stood out as the most interesting of the ten candidate targets identified by the three screens for several reasons. First, this gene showed an approximately 20-fold up-regulation in response to Hoxa 11 expression, whereas the other genes showed more modest changes. Second, the expression of Integrin α8 closely matches that of Hoxa 11 in the metanephric mesenchyme surrounding the branching ureteric bud, consistent with Hoxa 11 being an important in vivo regulator of Integrin α8 expression. Third, the kidney phenotype of the Integrin α8 mutant mouse mirrors that of the Hoxa 11 mutant, suggesting that Integrin α8 may be a key mediator of Hoxa 11 function in the developing kidney. Further studies therefore focused on the Integrin α8 gene.
Integrin α8 Is Also Up-Regulated by Hoxa 11 in mK10 Cells.
The up-regulation of Integrin α8 after Hoxa 11 expression was seen in the HEK293 cells by using the Affymetrix genechip, but was not detected by Differential Display or GDA in the mK10 cells. This lack of detection could have resulted from the incomplete nature of the Differential Display and GDA screens, or could have been the result of Integrin α8 not responding to Hoxa 11 in mK10 cells. To distinguish these possibilities Integrin α8 expression was first measured by standard RT-PCR in mK10 cells, double mutant for Hoxa 11 and Hoxd 11, and in mK10/pCMV-Hoxa 11cells, with Hoxa 11 expression. Higher Integrin α8 expression was seen in the mK10/pCMV-Hoxa 11cells (Fig. 4A), which was then confirmed by multiple repetitions of a semiquantitative RT-PCR assay monitoring label incorporation in the linear amplification phase (Fig. 4B). Integrin α8 expression was up-regulated approximately 20-fold in the Hoxa 11-expressing cells.
Figure 4.
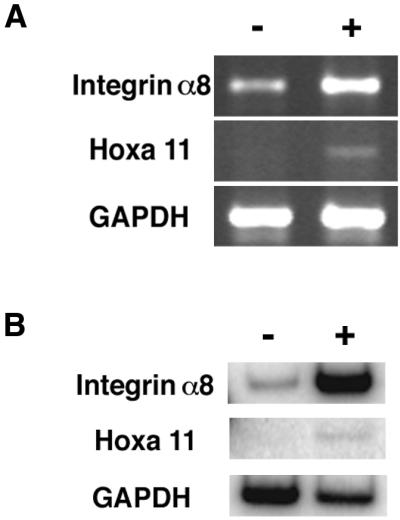
Up-regulation of Integrin α8 in mK10 cells in response to Hoxa 11 expression. (A) Nonradiolabel RT-PCR showed Integrin α8 expression increased with expression of Hoxa 11. (B) Semiquantitative radiolabel RT-PCR further confirmed that Hoxa 11 increased Integrin α8 expression. Intensities were normalized to GAPDH. + marks lane with Hoxa 11 expression.
In Vivo Confirmation.
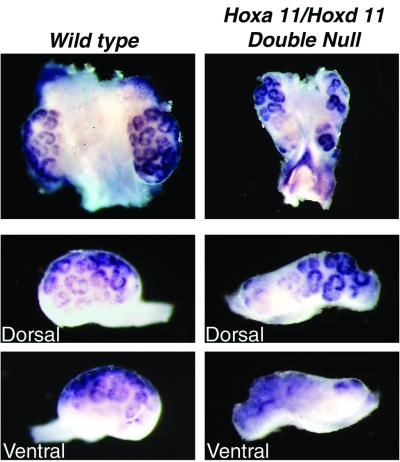
If Hoxa 11 plays a critical role in the expression of Integrin α8, then we would predict the expression pattern to be altered in the Hoxa 11/Hoxd 11 double mutants. To test this prediction, E13.5 kidneys were isolated from Hoxa 11/Hoxd 11 double-mutant embryos and wild-type littermates and examined for Integrin α8 expression by in situ hybridization. A dramatic, but localized reduction of Integrin α8 expression was seen in the mutant kidneys (Fig. 5). Some regions showed near-normal Integrin α8 expression, but other regions, and in particular the ventral side of the kidney, showed an almost complete loss of Integrin α8 expression.
Figure 5.
Regional reduction of Integrin α8 expression in Hoxa 11/Hoxd 11 double-mutant kidneys. Whole-mount in situ hybridizations with E13.5 mutant and wild-type littermate kidneys. (Top) The more medial positioning of the mutant kidneys, with the posterior poles nearly touching. Part of the dorsal region of the mutant kidney, and the entire ventral region, showed reduced Integrin α8 expression.
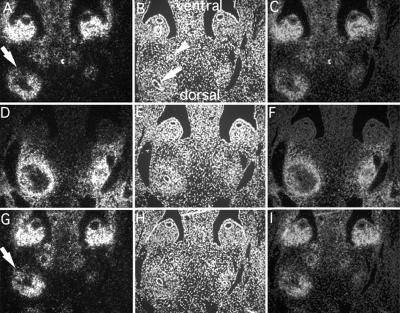
The altered expression of Integrin α8 in Hoxa 11/Hoxd 11 double-mutant kidneys was further confirmed by section in situ hybridization. The ventral renal mesenchyme adjacent to the ureteric bud showed a consistent reduction of Integrin α8 expression (Fig. 6 A and G). This altered expression pattern was coincident with the observed severe reduction of branching morphogenesis in the ventral domain of the mutant kidney. The normal domain of Hoxa 11 expression is outlined by Foxd1 expression, a marker of stromal cells (Fig. 6D). In the poles of the kidney, where branching of the bud was more normal, we observed symmetric Integrin α8 expression surrounding the bud (data not shown).
Figure 6.
Section in situ hybridizations further define the altered Integrin α8 expression in Hoxa 11/Hoxd 11 mutant kidneys. Serial transverse sections through E13.5 Hoxa 11/Hoxd 11 double-mutant embryos were hybridized with antisense riboprobes to Integrin α8 (A and C, and G and I) or Foxd1 (a marker of stromal cells) (D and F). A, D, and G, darkfield images; B, E, and H, 4′,6-diamidino-2-phenylindole fluorescent-stained images; C, F, and I, merged darkfield and fluorescent images. B shows the dorsoventral orientation of the embryo and the structures present. Arrow, ureteric bud; arrowhead, mesonephric duct; g, gonad. The arrows in A and G indicate the ventral renal mesenchyme, where Integrin α8 expression is severely reduced compared with dorsal expression. For this double mutant the kidney shown on the right-hand side was extremely malformed, with an absence of detectable ureteric bud branching.
Discussion
To understand the genetic networks controlling kidney organogenesis it is necessary to identify the downstream targets of the transcription factors, such as Hoxa 11, which regulate this process. The cell-line-probe array strategy used in this study provides a relatively rapid screen that does not require target guessing or laborious testing of genes one at a time. As probe arrays provide more complete coverage of the genome these screens become more universal in nature. This strategy has been surprisingly successful, even when developmentally inappropriate cell lines have been used. For example, osteosarcoma cells with inducible WT1 expression were used to identify amphiregulin as a downstream target of the Wilms tumor gene (15) in kidney development.
To optimize the screen for Hoxa 11 targets in kidney development we used kidney cell lines. The HEK293 cells seem to represent a late stage in kidney epithelia development, after Hoxa 11 expression is turned off. The other cell line used, mK10, was designed to represent metanephric mesenchyme at an early stage of development, when Hoxa 11 would usually be expressed. To remove Hoxa 11 expression the cells were made from mice mutant for both Hoxa 11 and the functionally redundant Hoxd 11. To make the mK10 cell line we used Hoxa 11 promoter-driven expression of SV40 Tag in transgenic mice, an approach with a long history of success. Surprisingly, SV40 Tag expression is able to not only immortalize cells, but to arrest them developmentally. For example, a 1.8-kb promoter for the glycoprotein hormone α-subunit was used to immortalize and developmentally arrest cells that expressed the α-subunit and responded to gonadotrophin-releasing hormone (43). A longer version of this promoter (5.5 kb), which expressed earlier, was used subsequently to establish more primitive α-subunit expressing cells, and the β-subunit leutinizing hormone promoter was used to make cell lines from later stages in the gonadotroph lineage (44). These and other cell lines generated in similar fashion have made significant contributions to our understanding of the genetic pathways of pituitary lineage-specific development (45–47). The mK10 cell might also be developmentally arrested. The same Hoxa 11-SV40 Tag transgene was used to make the mK3 kidney cell line from mice with wild-type Hoxa 11 and Hoxd 11 genes. The mK3 cells are able to drive branching morphogenesis of the ureteric bud in organ coculture, showing they maintain early metanephric mesenchyme function (48).
Most of the candidate targets identified showed only a modest response to Hoxa 11 expression, consistent with the results of similar studies (13–15). The biological significance of such 1.5- to 3-fold changes in expression level is uncertain. Nevertheless, at least two of these genes are of potential interest in kidney development. The growth factor PDGF A is expressed in newly formed epithelial cells derived from the condensing mesenchyme (35). Hoxa 11 is expressed in the condensing mesenchyme until the cells begin to convert to epithelial cells. These complementary patterns are consistent with the observed repression of PDGF A by Hoxa 11 in HEK293 cells. Mutation of PDGF A results in defects in lung (49) and central nervous system (50) development, but no apparent kidney malformations. This could be the result of continued expression of functionally redundant PDGF B and PDGF C in the metanephric mesenchyme (51). The GCM1 gene is of interest because its mutation gives a branching morphogenesis defect in the chorioallantoic placenta, resulting in embryo death at E10, before kidney development. Expression of GCM1 has been detected in the E16.5 kidney by RT-PCR (37).
The Integrin α8 gene showed a striking up-regulation in response to Hoxa 11 expression in both the HEK293 and mK10 cells. The Integrin α8 gene is usually expressed in the metanephric mesenchyme cells abutting the ureteric bud, where Hoxa 11 is also expressed (1,16). Mutation of the Integrin α8 gene gave a severe defect in the growth and branching of the ureteric bud (16), similar to that seen in the Hoxa11/Hoxd 11 mutants (1). The Integrin α8 gene therefore represented an excellent candidate effector of Hoxa 11 in kidney development.
In vivo confirmation of the requirement of Hoxa 11/Hoxd 11 function for normal Integrin α8 expression was provided by in situ hybridizations showing reduction of Integrin α8 expression in Hoxa 11/Hoxd 11mutant kidneys. Of interest, this reduction was regional, with the ventral domain of the kidney most severely effected. The relatively normal expression of Integrin α8 in restricted areas may explain the frequent formation of a rudimentary kidney, rather than no kidney at all. At least seven Hox genes are expressed in the metanephric mesenchyme (48), some of which may provide localized functional redundancy with Hoxa 11/Hoxd 11, thereby explaining the limited continued expression of Integrin α8.
The integrins, as extracellular matrix receptors, are generally associated with receiving, not sending signals. Yet mutation of Hoxa 11/Hoxd 11 and Integrin α8, which are expressed in the mesenchyme, give defects in the branching of the ureteric bud. These defects likely reflect the considerable crosstalk between the mesenchyme and bud during kidney development. Failure of the mesenchyme to receive a signal from the bud properly could then result in a deficiency of signaling from the mesenchyme to the bud.
Acknowledgments
We thank the members of the Affymetrix Academic User Center for their assistance. This work was supported by National Institutes of Health Grants HD24517 (to S.S.P.) and DK02702 (to L.T.P.).
Abbreviations
- GDA
Gene Discovery Array
- RT
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Patterson L T, Pembauer M, Potter S S. Development (Cambridge, UK) 2001;128:2153–2161. doi: 10.1242/dev.128.11.2153. [DOI] [PubMed] [Google Scholar]
- 2.Davis A P, Witte D P, Hsieh-Li H M, Potter S S, Capecchi M R. Nature (London) 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- 3.Gould A P, Brookman J J, Strutt D I, White R A. Nature (London) 1990;348:308–312. doi: 10.1038/348308a0. [DOI] [PubMed] [Google Scholar]
- 4.Serrano N, Brock H W, Demeret C, Dura J M, Randsholt N B, Kornberg T B, Maschat F. Development (Cambridge, UK) 1995;121:1691–1703. doi: 10.1242/dev.121.6.1691. [DOI] [PubMed] [Google Scholar]
- 5.Tomotsune D, Shoji H, Wakamatsu Y, Kondoh H, Takahashi N. Nature (London) 1993;365:69–72. doi: 10.1038/365069a0. [DOI] [PubMed] [Google Scholar]
- 6.Chauvet S, Maurel-Zaffran C, Miassod R, Jullien N, Pradel J, Aragnol D. Dev Dyn. 2000;218:401–413. doi: 10.1002/1097-0177(200007)218:3<401::AID-DVDY1009>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Violette S M, Shashikant C S, Salbaum J M, Belting H G, Wang J C, Ruddle F H. Proc Natl Acad Sci USA. 1992;89:3805–3809. doi: 10.1073/pnas.89.9.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones F S, Chalepakis G, Gruss P, Edelman G M. Proc Natl Acad Sci USA. 1992;89:2091–2095. doi: 10.1073/pnas.89.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones F S, Prediger E A, Bittner D A, De Robertis E M, Edelman G M. Proc Natl Acad Sci USA. 1992;89:2086–2090. doi: 10.1073/pnas.89.6.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Care A, Silvani A, Meccia E, Mattia G, Stoppacciaro A, Parmiani G, Peschle C, Colombo M P. Mol Cell Biol. 1996;16:4842–4851. doi: 10.1128/mcb.16.9.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raman V, Tamori A, Vali M, Zeller K, Korz D, Sukumar S. J Biol Chem. 2000;275:26551–26555. doi: 10.1074/jbc.C000324200. [DOI] [PubMed] [Google Scholar]
- 12.Raman V, Martensen S A, Reisman D, Evron E, Odenwald W F, Jaffee E, Marks J, Sukumar S. Nature (London) 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Schrick J J, Fewell G D, MacFarland K L, Witte D P, Bodenmiller D M, Hsieh-Li H M, Su C Y, Potter S S. Dev Biol. 1999;211:64–76. doi: 10.1006/dbio.1999.9304. [DOI] [PubMed] [Google Scholar]
- 14.Harkin D P, Bean J M, Miklos D, Song Y H, Truong V B, Englert C, Christians F C, Ellisen L W, Maheswaran S, Oliner J D, Haber D A. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 15.Lee S B, Huang K, Palmer R, Truong V B, Herzlinger D, Kolquist K A, Wong J, Paulding C, Yoon S K, Gerald W, et al. Cell. 1999;98:663–673. doi: 10.1016/s0092-8674(00)80053-7. [DOI] [PubMed] [Google Scholar]
- 16.Muller U, Wang D, Denda S, Meneses J J, Pedersen R A, Reichardt L F. Cell. 1997;88:603–613. doi: 10.1016/s0092-8674(00)81903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 18.Cho A C, Patterson L T, Brookhiser W T, Mah S, Kintner C, Dressler G R. Development (Cambridge, UK) 1998;125:803–812. doi: 10.1242/dev.125.5.803. [DOI] [PubMed] [Google Scholar]
- 19.Torban E, Goodyer P R. Biochim Biophys Acta. 1998;1401:53–62. doi: 10.1016/s0167-4889(97)00119-5. [DOI] [PubMed] [Google Scholar]
- 20.Kupper J. Eur J Neurosci. 1998;10:3908–3912. doi: 10.1046/j.1460-9568.1998.00441.x. [DOI] [PubMed] [Google Scholar]
- 21.Pedersen K A, Schroder R L, Skaaning-Jensen B, Strobaek D, Olesen S P, Christophersen P. Biochim Biophys Acta. 1999;1420:231–240. doi: 10.1016/s0005-2736(99)00110-8. [DOI] [PubMed] [Google Scholar]
- 22.Lee M G, Wigley W C, Zeng W, Noel L E, Marino C R, Thomas P J, Muallem S. J Biol Chem. 1999;274:3414–3421. doi: 10.1074/jbc.274.6.3414. [DOI] [PubMed] [Google Scholar]
- 23.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishidoh K, Kominami E. Biol Chem. 1998;379:131–135. doi: 10.1515/bchm.1998.379.2.131. [DOI] [PubMed] [Google Scholar]
- 25.Dohchin A, Suzuki J I, Seki H, Masutani M, Shiroto H, Kawakami Y. Cancer. 2000;89:482–487. [PubMed] [Google Scholar]
- 26.Afonso S, Romagnano L, Babiarz B. Development (Cambridge, UK) 1997;124:3415–3425. doi: 10.1242/dev.124.17.3415. [DOI] [PubMed] [Google Scholar]
- 27.Hemberger M, Himmelbauer H, Ruschmann J, Zeitz C, Fundele R. Dev Biol. 2000;222:158–169. doi: 10.1006/dbio.2000.9705. [DOI] [PubMed] [Google Scholar]
- 28.Nakase T, Kaneko M, Tomita T, Myoui A, Ariga K, Sugamoto K, Uchiyama Y, Ochi T, Yoshikawa H. Histochem Cell Biol. 2000;114:21–27. doi: 10.1007/s004180000162. [DOI] [PubMed] [Google Scholar]
- 29.Alldridge L C, Harris H J, Plevin R, Hannon R, Bryant C E. J Biol Chem. 1999;274:37620–37628. doi: 10.1074/jbc.274.53.37620. [DOI] [PubMed] [Google Scholar]
- 30.McKanna J A, Chuncharunee A, Munger K A, Breyer J A, Cohen S, Harris R C. J Cell Physiol. 1992;153:467–476. doi: 10.1002/jcp.1041530305. [DOI] [PubMed] [Google Scholar]
- 31.Mintz L, Galperin E, Pasmanik-Chor M, Tulzinsky S, Bromberg Y, Kozak C A, Joyner A, Fein A, Horowitz M. Genomics. 1999;59:66–76. doi: 10.1006/geno.1999.5800. [DOI] [PubMed] [Google Scholar]
- 32.Tong X K, Hussain N K, Adams A G, O'Bryan J P, McPherson P S. J Biol Chem. 2000;275:29894–29899. doi: 10.1074/jbc.M004096200. [DOI] [PubMed] [Google Scholar]
- 33.Liu D, Pavlopoulos E, Modi W, Moschonas N, Mavrothalassitis G. Oncogene. 1997;14:1445–1451. doi: 10.1038/sj.onc.1200965. [DOI] [PubMed] [Google Scholar]
- 34.Sgouras D N, Athanasiou M A, Beal G J, Jr, Fisher R J, Blair D G, Mavrothalassitis G J. EMBO J. 1995;14:4781–4793. doi: 10.1002/j.1460-2075.1995.tb00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boudreau N J, Jones P L. Biochem J. 1999;339:481–488. [PMC free article] [PubMed] [Google Scholar]
- 36.Seifert R A, Alpers C E, Bowen-Pope D F. Kidney Int. 1998;54:731–746. doi: 10.1046/j.1523-1755.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- 37.Kim J, Jones B W, Zock C, Chen Z, Wang H, Goodman C S, Anderson D J. Proc Natl Acad Sci USA. 1998;95:12364–12369. doi: 10.1073/pnas.95.21.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anson-Cartwright L, Dawson K, Holmyard D, Fisher S J, Lazzarini R A, Cross J C. Nat Genet. 2000;25:311–314. doi: 10.1038/77076. [DOI] [PubMed] [Google Scholar]
- 39.Davis J Q, Bennett V. J Biol Chem. 1994;269:27163–27166. [PubMed] [Google Scholar]
- 40.Debiec H, Christensen E I, Ronco P M. J Cell Biol. 1998;143:2067–2079. doi: 10.1083/jcb.143.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindberg F P, Bullard D C, Caver T E, Gresham H D, Beaudet A L, Brown E J. Science. 1996;274:795–798. doi: 10.1126/science.274.5288.795. [DOI] [PubMed] [Google Scholar]
- 42.Lee P L, Gelbart T, West C, Adams M, Blackstone R, Beutler E. Genomics. 1997;43:191–201. doi: 10.1006/geno.1997.4806. [DOI] [PubMed] [Google Scholar]
- 43.Windle J J, Weiner R I, Mellon P L. Mol Endocrinol. 1990;4:597–603. doi: 10.1210/mend-4-4-597. [DOI] [PubMed] [Google Scholar]
- 44.Alarid E T, Windle J J, Whyte D B, Mellon P L. Development (Cambridge, UK) 1996;122:3319–3329. doi: 10.1242/dev.122.10.3319. [DOI] [PubMed] [Google Scholar]
- 45.Bodner M, Castrillo J L, Theill L E, Deerinck T, Ellisman M, Karin M. Cell. 1988;55:505–518. doi: 10.1016/0092-8674(88)90037-2. [DOI] [PubMed] [Google Scholar]
- 46.Ingraham H A, Chen R P, Mangalam H J, Elsholtz H P, Flynn S E, Lin C R, Simmons D M, Swanson L, Rosenfeld M G. Cell. 1988;55:519–529. doi: 10.1016/0092-8674(88)90038-4. [DOI] [PubMed] [Google Scholar]
- 47.Therrien M, Drouin J. Mol Cell Biol. 1993;13:2342–2353. doi: 10.1128/mcb.13.4.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valerius M T, Patterson L T, Witte D P, Potter S S. Mech Dev. 2002;110:151–164. doi: 10.1016/s0925-4773(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 49.Bostrom H, Willetts K, Pekny M, Leveen P, Lindahl P, Hedstrand H, Pekna M, Hellstrom M, Gebre-Medhin S, Schalling M, et al. Cell. 1996;85:863–873. doi: 10.1016/s0092-8674(00)81270-2. [DOI] [PubMed] [Google Scholar]
- 50.Fruttiger M, Karlsson L, Hall A C, Abramsson A, Calver A R, Bostrom H, Willetts K, Bertold C H, Heath J K, Betsholtz C, Richardson W D. Development (Cambridge, UK) 1999;126:457–467. doi: 10.1242/dev.126.3.457. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Ponten A, Aase K, Karlsson L, Abramsson A, Uutela M, Backstrom G, Hellstrom M, Bostrom H, Li H, et al. Nat Cell Biol. 2000;2:302–309. doi: 10.1038/35010579. [DOI] [PubMed] [Google Scholar]