Abstract
As the closest outgroup to the Bilateria, the Phylum Cnidaria is likely to be critical to understanding the origins and evolution of body axes. Proteins of the decapentaplegic (DPP)/bone morphogenetic protein (BMP) 2/4 subfamily are central to the specification of the dorsoventral (D/V) axis in bilateral animals, albeit with an axis inversion between arthropods and chordates. We show that a dpp/BMP2/4 ortholog (bmp2/4-Am) is present in the reef-building scleractinian coral, Acropora millepora (Class Anthozoa) and that it is capable of causing phenotypic effects in Drosophila that mimic those of the endogenous dpp gene. We also show that, during coral embryonic development, bmp2/4-Am expression is localized in an ectodermal region adjacent to the blastopore. Thus, a representative of the DPP/BMP2/4 subfamily of ligands was present in the common ancestor of diploblastic and triploblastic animals where it was probably expressed in a localized fashion during development. A localized source of DPP/BMP2/4 may have already been used in axis formation in this ancestor, or it may have provided a means by which an axis could evolve in triploblastic animals.
Most animals have two obvious body axes, anterior-posterior and dorsal-ventral. The molecular mechanisms by which these axes are specified are common to arthropods and mammals (1, 2), implying a single origin for the higher animals (the Bilateria). The closest outgroup to the Bilateria is the Phylum Cnidaria. Understanding cnidarian axis formation will, therefore, provide important clues to the origins and evolution of body axes (3, 4).
Little is known about the molecular control of axis formation in the Cnidaria. Even the number of axes present in cnidarians remains contentious. Dating from the theories of Haeckel (5), it has been widely assumed that the earliest metazoans had a single axis, i.e., were radially symmetrical (e.g., refs. 6 and 7) and that, therefore, the Bilateria arose from a radially symmetrical ancestor, perhaps exhibiting a cnidarian-grade organization. Although several recent textbooks of invertebrate zoology (e.g., refs. 7 and 8) describe members of the Cnidaria as possessing a primary radial symmetry, the topic is controversial. Many authors have suggested that some cnidarians either are bilateral, or probably were bilateral at one stage in their evolution (e.g., refs. 3, 6, 9, and 10), thus raising the possibility that the overt radially symmetrical character of the phylum is derived. Another, far from settled, question concerns the relationship between the major longitudinal polarized axis of cnidarians, the oral-aboral axis, and the anterior-posterior axis of the Bilateria. Although homology between these axes is often assumed, compelling evidence is lacking. Moreover, there is no strong case for assigning the cnidarian oral pole as homologous to the anterior pole of bilaterians, as is often done, because, in the swimming cnidarian planula, the oral opening is posterior, and in cnidarian polyps the same orifice serves as both a mouth and an anus.
One way of gaining insights into the relationship of the cnidarian and bilaterian body axes is to investigate the expression patterns of conserved genes or gene families. Recent molecular data indicate that conserved patterning systems exist for both the anterior-posterior axis and the dorsal-ventral axis in bilaterian animals. The anterior-posterior axis is characterized by the ordered regional distribution of Hox gene expression domains (1, 11). Such a deployment of Hox genes has been proposed to be a universal and definitive metazoan character, the zootype (12). Whether the zootype hypothesis holds for the diploblastic phyla is presently controversial (13, 14), but it is generally held to be true for the Bilateria. The dorsal-ventral axis is defined by the actions of genes of the dpp/BMP4 class and their antagonists (15). In Drosophila, decapentaplegic (DPP) is expressed dorsally and acts to specify dorsal fates whereas its antagonist, the product of the short gastrulation gene (SOG) is expressed ventrally. In Xenopus, the situation is inverted, with the DPP ortholog bone morphogenetic protein (BMP) 4 being expressed ventrally and the SOG ortholog, Chordin, acting dorsally. These observations indicate that there is a conserved system for defining the polarity of the dorsal-ventral axis, although an axis inversion with respect to the substratum has occurred in the course of evolution between arthropods and vertebrates (16–18).
Data from the expression patterns of cnidarian homeobox genes related to those having conserved roles in anterior-posterior axis patterning in arthropods and chordates are inconclusive. This result may be in part due to the fact that no unequivocal Hox gene has been isolated from a cnidarian. To shed more light on the origin and evolution of the second body axis, we have turned our attention to genes of the DPP/BMP4 signaling pathway in the reef building coral Acropora millepora. We have previously reported the presence of DPP/BMP2/4-type signal transduction components, a type II receptor, and two receptor-mediated Smads, in this organism (19). Here, we describe the isolation and characterization of an A. millepora dpp/BMP2/4 ortholog, and the early embryonic expression of this gene.
Materials and Methods
PCR and cDNA Isolation.
An Acropora bmp2/4 PCR fragment was amplified from late embryonic stage cDNA by using the following strategy. A first stage reaction was carried out with the following primers: forward, 5′-TTYWSIGAYGTNGGNTGG-3′; reverse, 5′-TSIGTIGGNACRCARCA-3′ (35 cycles, 40°C annealing temperature). A fraction of this reaction was then amplified with the forward primer 5′-GGITGGSANGAYTGGAT-3′ and the same reverse primer (35 cycles, 57°C annealing temperature). Construction of the cDNA library from planula larvae is described elsewhere (20). Probes were made from the purified inserts of plasmids containing the cloned PCR products. Library screening and clone isolation were carried out by using standard techniques (21). Clones were recovered from the library as inserts in pBluescript SK(−) (Stratagene). Four cDNAs were isolated from 1 × 106 plaques, and one of these, pcAmbmp2/4-1, which contains a complete ORF, was sequenced on both strands by using an ABI377 analyzer and Applied Biosystems Big Dye terminator chemistry.
Northern Analysis.
Total RNA was prepared from material ground in liquid nitrogen according to the method of Chomczynski and Sacchi (22). Poly(A)+ RNA was isolated by using the PolyATtract mRNA isolation system (Promega). Approximately 2 μg of poly(A)+ RNA from each developmental stage, denatured in the presence of 2.2% formaldehyde, was separated on a 1% agarose gel in 1× Mops buffer and blotted on nylon membrane (Schleicher & Schuell). DNA probes were 32P-labeled by standard procedures. Prehybridization and hybridization were carried out at 65°C in 5× SSPE (1× = 0.15 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA), 5× Denhardt's solution (1× = 0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA), 0.5% SDS, and 20 μg/ml denatured salmon sperm DNA. Washes were carried out at 65°C with 2× SSC containing 0.1% SDS.
Whole-Mount in Situ Hybridization.
The procedures for fixation, delipification, and hybridization of coral embryos were carried out as described (23). Freehand sections of stained whole-mount embryos stored in 70% glycerol were cut by using a mounted fragment of a thin double-edged razor blade (Feather brand, Osaka).
Drosophila Genetic Transformation and in Situ Hybridization.
To make the UAS-Acropora bmp2/4 construct, a 1.6-kb HindIII/EcoRI fragment containing the entire ORF was first cloned between the HindIII and EcoRI sites of pBluescript KS(+) (Stratagene). The insert was then excised with XhoI and XbaI and cloned between the XhoI and XbaI sites of pUAST (24). Transformants were generated via P-element-mediated germ-line transformation (25). In situ hybridization to whole-mount embryos by using digoxigenin-labeled RNA probes was performed according to the Boehringer Mannheim protocol.
Results and Discussion
Isolation of an Acropora bmp2/4 cDNA.
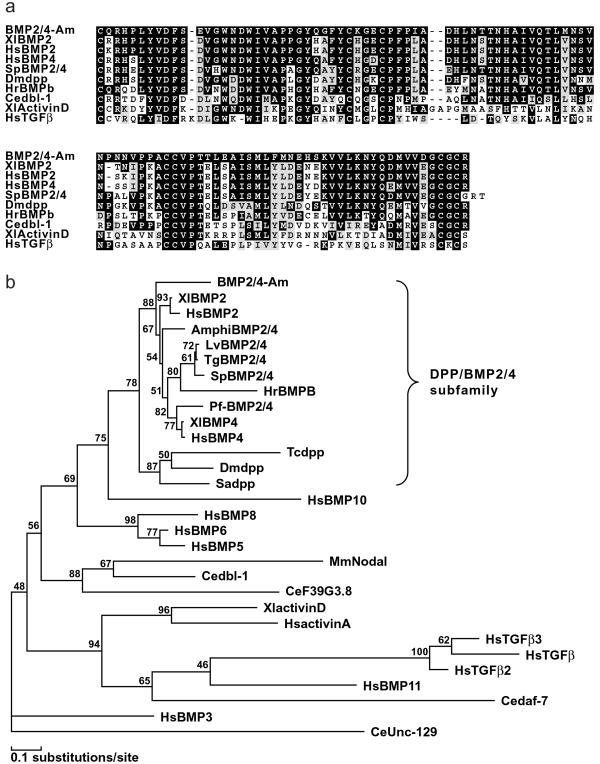
An Acropora bmp2/4 cDNA clone was isolated as described in Materials and Methods. Conceptual translation of the complete ORF of the cDNA results in a putative protein, BMP2/4-Am. Fig. 1a shows an alignment of the 102-aa C-terminal region of BMP2/4-Am with a range of related transforming growth factor β (TGF-β) family proteins, starting with the first conserved cysteine residue. BMP2/4-Am, is most closely related to the vertebrate BMP2/4 class. Matches with orthologous invertebrate protein sequences (including Drosophila DPP) score significantly lower. In the C-terminal region, identity with (for example) Xenopus laevis BMP2 is 80% (82/102), and with Drosophila melanogaster DPP is 67% (69/102). The levels of identity with vertebrate sequences are surprisingly high. Other proteins that are likely to mediate DPP signaling in Acropora also show higher identity with deuterostome sequences than with their Drosophila or Caenorhabditis orthologs (19). A preliminary Acropora expressed sequence tag (EST) project indicates that this pattern applies more broadly, an observation that will be discussed elsewhere (G.S., D. Kortschak, D.J.M., and R.S., unpublished observations). It is also striking that BMP2/4-Am more closely matches vertebrate BMP2/4s than do the arthropod DPPs. Phylogenetic analysis, shown in Fig. 1b, confirms that BMP2/4-Am belongs to the DPP/BMP2/4 subclass of the TGF-β family. Note that the putative Caenorhabditis DPP ortholog, DBL-1 (27), is relatively distantly related to the orthologous group to which BMP2/4-Am belongs.
Figure 1.
Sequence comparisons. (a) Alignment of the C-terminal domain of BMP2/4-Am with a range of related proteins. (b) Phylogenetic analysis of the C-terminal region of the BMP2/4-Am sequence in relation to those of other TGF-β family proteins. The conserved C-terminal regions of a representative range of TGF-β family proteins were aligned and subjected to maximum likelihood analyses in molphy Version 2.3 (26) using the Dayhoff matrix and local rearrangement search mode. The numbers on the branches are percentages of 1,000 bootstrap replicates supporting the topology shown. The Caenorhabditis elegans UNC-129 sequence (GenBank accession no. AF029887) was defined as outgroup. Note that the DPP subfamily, which includes BMP2/4-Am, is clearly resolved.
Acropora BMP2/4 Has Dorsalizing Activity in Drosophila.
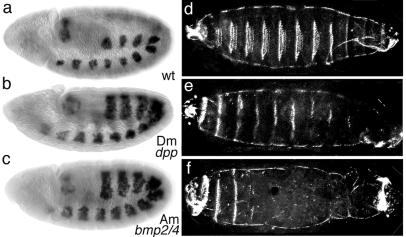
The high similarity between BMP2/4-Am and vertebrate and Drosophila orthologs suggested that the coral protein might have biological activity in bilateral animals. To investigate the morphogenetic properties of BMP2/4-Am, two independent Drosophila lines carrying an Acropora bmp2/4 cDNA transgene under the control of the yeast GAL4 UAS (upstream activation sequence) were generated. The UAS sequence upstream of the cDNA results in its transcription when yeast GAL4 is expressed in Drosophila cells (24). A UAS-Drosophila dpp cDNA line was used as a positive control for the effects of induced dpp expression. In Drosophila, dpp is expressed in dorsal ectoderm and induces dorsal fates. In addition, it signals to the underlying cells to specify dorsal mesoderm. Expression of the Drosophila dpp cDNA construct in the ventral mesodermal tissues by using a twist-GAL4 line results in dorsalization of the ventral tissues (28). To examine whether the Acropora bmp2/4 cDNA had any dorsalizing effect in this assay, the UAS-Acropora bmp2/4 cDNA lines were crossed to the twist-GAL4 line. The resulting phenotypes were dramatic and indistinguishable from those of the twist-GAL4;UAS-Drosophila dpp cDNA embryos. Dorsalization of the ventral tissues and 100% larval lethality occurred in both cases. For example, the expression of bagpipe (bap) extended ventrally relative to its normal dorsal mesodermal expression (Fig. 2 a–c). This is the expected outcome of the induction of tinman (tin) by the ectopic ventral dpp expression and the consequent induction of bap.
Figure 2.
Ectopic expression of dpp or bmp2/4-Am in the mesoderm leads to the mis-expression of bagpipe and affects the patterning of the ventral epidermis. (a and d) Wild type embryo. (b and e) Embryo carrying twist-GAL4;UAS-Drosophila dpp. (c and f) Embryo carrying twist-GAL4;UAS-Acropora bmp2/4. (a, b, and c) Distribution of bagpipe mRNA in stage 10 embryos (lateral view). In wild-type embryos, bagpipe expression is confined to the dorsal mesoderm (a). bagpipe expression extends ventrally in embryos carrying either twist-GAL4;UAS-Drosophila dpp or twist-GAL4;UAS-Acropora bmp2/4 (b and c). (d, e, and f) Stage 17 embryos, ventral view of cuticle. (d) Wild-type embryo. (e) Embryo carrying twist-GAL4;UAS-Drosophila dpp. (f) Embryo carrying twist-GAL4;UAS-Acropora bmp2/4. Embryos expressing either cDNA exhibit fewer of the prominent ventral denticle belts.
Cuticles from the twist-GAL4;UAS-Acropora bmp2/4 cDNA embryos were analyzed to determine whether the twist enhancer-activated expression of bmp2/4-Am was also able to dorsalize the ventral epidermis. Although no dorsal hairs were detected on the ventral surface of these embryos, there was a marked reduction in both number and size of the ventral-specific denticle belts (Fig. 2 d–f). This result was most prominent at the posterior of the embryos where, in extreme cases, no belts could be seen. A similar apparent dorsalization of the ventral epidermis is seen in the case of the twist-GAL4;UAS-Drosophila dpp embryos (Fig. 2e).
Expression of bmp2/4-Am.
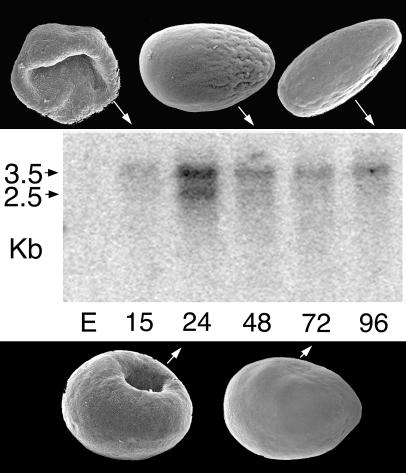
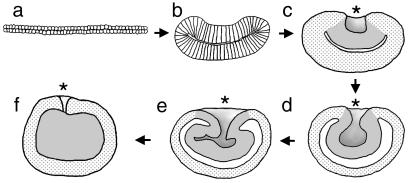
Expression of bmp2/4-Am was examined by using Northern blot analysis with mRNA from a series of developmental stages of A. millepora, as shown in Fig. 3. This analysis revealed two transcripts and a peak in expression at the stage when the blastopore (Fig. 3, 24 h) is closing. The detailed embryology of Acropora, especially as it relates to the process by which endoderm is created, has yet to be described. However, an overview (29) and a diagrammatic summary (30) of embryonic development have been published. A schematic representation of the morphological changes that occur during gastrulation is shown in Fig. 4. Immediately before the start of expression of bmp2/4-Am, the embryo consists of a relatively flat bilayer of cells (Fig. 4a) containing a large amount of evenly dispersed lipid. This bilayer then increases in thickness while decreasing in circumference (Fig. 4b). The edges of the thickened disk begin to move upward and inward, forming a depression in one side (Fig. 4b). As the inward movement continues, cells of one of the layers (the presumptive endoderm) become internalized (Fig. 4 c–f). During this process, the two cell layers separate on their internal faces, resulting in the formation of a blastocoel that appears to have a high lipid content. This lipid will eventually be taken up by the endoderm. As the blastopore closes, the internalized layer loses its epithelial character and apparently redifferentiates as endoderm; a clear boundary between the outer layer (ectoderm) and the internal tissue (endoderm) can be seen at this stage. The larva produced by this process next develops an elongated shape, and an oral pit appears at the narrower posterior end, as defined by the direction of swimming. Because the blastopore closes before the formation of the oral pit, the relationship between them is unknown, although in hydrozoans it is known that the site at which gastrulation is initiated corresponds to the future posterior pole of the larva (31).
Figure 3.
The morphology of the corresponding developmental stages is shown in association with a Northern blot hybridized with a bmp2/4-Am probe. Two transcripts, of ≈3.5 and 2.5 kb, are apparent, with expression peaking as the blastopore is closing. E, eggs; numbers indicate hours postfertilization.
Figure 4.
Acropora gastrulation shown in diagrammatic sectioned view. (a) The early embryo is an extended disk two cells thick. (b) The disk shrinks in circumference and thickens while the edges start to fold inward. (c) The cavity thus formed continues to deepen and the two cell layers separate on their inner faces. (d and e) The process continues, and the blastopore begins to close. As this closure occurs, the internalized tissue differentiates as endoderm. (f) By the time the blastopore has almost closed, the endoderm has differentiated to a relatively uniform appearance. Tissue layers, rather than individual cells, are portrayed in c–f. Ectoderm, stippled; endoderm and presumptive endoderm, gray; blastocoel, white. The blastopore is marked with an asterisk.
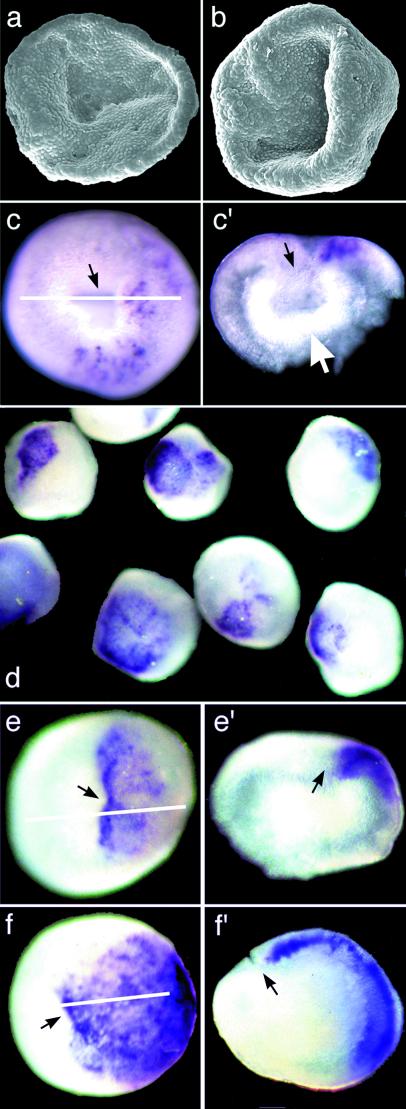
Expression of bmp2/4-Am during this process is revealed by in situ hybridization (Fig. 5 c–f). bmp2/4-Am mRNA is initially detected at scattered positions in the ectoderm near the blastopore (Fig. 5 c and c′). This expression then becomes more intense and solid in one quadrant of the surface ectoderm, adjacent to the blastopore (Fig. 5 d, e, e′, f, and f′).
Figure 5.
bmp2/4-Am expression in various developmental stages of Acropora. In parts of the figure where the same letter is used twice (e.g., c and c′), the part labeled with a prime is a section through the embryo whose whole mount staining is shown labeled with the corresponding letter. The white line on the intact embryo indicates the plane of section. (a and b) Stages before (a) and just after (b) the detectable onset of bmp2/4-Am expression. (c) bmp2/4-Am expression is initially patchy within a domain associated with the blastopore. (c′) a section reveals that expression is limited to the ectoderm. At this stage, the engulfed cell layer is still intact, forming the inner layer of a double-walled bowl (white arrow). (d) A field of stained embryos all fixed at a single point in time. There is some variability in the extent of staining, probably because of the nonsynchronous development of the embryos, but in all cases the staining is associated with the blastopore. (e and f) Higher magnification view of two embryos from the batch shown in d. In both cases, the staining is restricted to the ectoderm, as evident in the sections shown in e′ and f′). Black arrows indicate the position of the blastopore.
Thus, in common with arthropods and chordates, Acropora exhibits a localized distribution of dpp/BMP2/4 mRNA during gastrulation. One major difference between the pattern seen in Acropora and that in bilateral animals concerns the relationship of bmp2/4 expression with the embryonic blastopore. In embryos of bilateral species, cells producing the BMP2/4 signals are initially distal to the earliest morphogenetic movements of gastrulation (see discussion in ref. 32). This generalization is true for the protostome Drosophila, as well as for representatives of two deuterostome phyla, the Echinodermata and Chordata. In apparent contrast, bmp2/4-Am expression is associated with the blastopore, rather than a region distal to it. However, the relationship between the process of gastrulation in Acropora and that in higher organisms remains unclear. In particular, the first morphogenetic movements in Acropora involve changes in the shape of a flattened cellular bilayer that may be the topological equivalent of a blastocyst. The “blastopore,” rather than being the site at which gastrulation is initiated, appears to be formed near the endpoint of the process.
The distribution of bmp2/4-Am mRNA during embryonic development is consistent with a role in axial patterning. If such a role is confirmed, it would imply the existence of a cnidarian body axis having an evolutionary correspondence to the dorsal/ventral axis of higher animals. If cnidarians are considered to be truly radially symmetrical, then such an axis would correspond to the oral-aboral axis. However, at 24 h post fertilization [Figs. 3 (24 h) and 5 d–f], the embryo is morphologically radially symmetrical about an axis defined by the blastopore, and the asymmetrical distribution of bmp2/4-Am transcripts relative to the blastopore at this stage imparts a second polarized axis to the embryo. The result is a single plane of symmetry (a definition of bilaterality) running through the closing blastopore and bisecting the bmp2/4-Am expression domain. The symmetry-breaking nature of the bmp2/4-Am expression domain is thus consistent with a DPP/BMP2/4 signaling pathway serving to define a second body axis.
The suggestion that bilaterality may have evolved before the separation of the Cnidaria from the rest of the Bilateria is not as revolutionary as it would have been even ten years ago. The idea that the ancestral cnidarian had a fundamental radial symmetry stems from the belief that the medusoid, rather than the polypoid, generation represents the ancestral cnidarian life form. The Class Hydrozoa, which includes many medusoid forms, and also contains truly radial representatives, was widely assumed to be the basal cnidarian Class (6). However, this argument has been greatly weakened by the finding that the mitochondrial genomes of two Classes with medusoid stages, the Hydrozoa and Scyphozoa, are linear, whereas those of the Anthozoa, which are exclusively polypoid, are circular, in common with those of other animals (33). More recently, phylogenetic studies using complete sequences of the small and large subunit ribosomal RNAs have also strongly rejected the hypothesis of a monophyletic Cnidaria with Hydrozoa basal in favor of a scheme with Anthozoa basal (34). The possibility that the Anthozoa may have been a bilateral group originally has been suggested by several authors (e.g., ref. 6).
The localized expression of bmp2/4-Am need not necessarily be associated with axis formation. In other animals, for example, DPP/BMP2/4 is involved in the partitioning of cells to adopt different fates. In sea urchin embryos, BMP2/4 determines the position of the boundary between ectoderm and endoderm, and in vertebrates, as well as in invertebrates, DPP/BMP activity distinguishes nonneurogenic from neurogenic ectoderm (32, 35–37). BMP2/4-Am may perform such roles in Acropora, and not be involved in axis formation.
In summary, it is possible that dpp/BMP ligands were already playing a role in axis formation in a putative common ancestor of Acropora and the Bilateria, consistent with the proposal that cnidarians either are, or were at one stage in their evolution, biradial or bilateral (3, 6, 9, 10). Alternatively, this family of ligands may have evolved to play one of the non-axis-forming roles of DPP/BMP2/4 before the divergence of the Cnidaria from the rest of the Metazoa. If the latter is true, the localized expression of BMP2/4-Am in the region of the blastopore provides a basis from which a DPP/BMP-dependent second axis could have evolved in an ancestral radial animal.
Acknowledgments
We thank M. Frasch for the bap probe.
Abbreviations
- DPP or dpp
decapentaplegic
- BMP
bone morphogenetic protein
- TGF-β
transforming growth factor β
- UAS
upstream activation sequence
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF285166).
References
- 1.McGinnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 2.Holley S A, Jackson P D, Sasai Y, Lu B, De Robertis E M, Hoffmann F M, Ferguson E L. Nature (London) 1995;376:249–253. doi: 10.1038/376249a0. [DOI] [PubMed] [Google Scholar]
- 3.Martindale M, Henry J Q. Am Zool. 1998;38:672–684. [Google Scholar]
- 4.Holland L Z. Curr Opin Genet Dev. 2000;10:434–442. doi: 10.1016/s0959-437x(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 5.Haeckel E. Q J Microsc Sci. 1874;14:142–165. [Google Scholar]
- 6.Hyman L H. The Invertebrates: Protozoa Through Ctenophora. New York: McGraw–Hill; 1940. [Google Scholar]
- 7.Brusca R C, Brusca G J. The Invertebrates. Sunderland, MA: Sinauer; 1990. [Google Scholar]
- 8.Ruppert E E, Barnes R D. Invertebrate Zoology. Philadelphia: Saunders; 1994. [Google Scholar]
- 9.Willmer P. Invertebrate Relationships: Patterns in Animal Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1990. [Google Scholar]
- 10.Groger H, Schmid V. Genesis. 2001;29:110–114. doi: 10.1002/gene.1013. [DOI] [PubMed] [Google Scholar]
- 11.Hirth F, Reichert H. BioEssays. 1999;21:677–684. doi: 10.1002/(SICI)1521-1878(199908)21:8<677::AID-BIES7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 12.Slack J M, Holland P W, Graham C F. Nature (London) 1993;361:490–492. doi: 10.1038/361490a0. [DOI] [PubMed] [Google Scholar]
- 13.Ferrier D E, Holland P W. Nat Rev Genet. 2001;2:33–38. doi: 10.1038/35047605. [DOI] [PubMed] [Google Scholar]
- 14.Schierwater B, Desalle R. J Exp Zool. 2001;291:169–174. doi: 10.1002/jez.1066. [DOI] [PubMed] [Google Scholar]
- 15.Holley S A, Ferguson E L. BioEssays. 1997;19:281–284. doi: 10.1002/bies.950190404. [DOI] [PubMed] [Google Scholar]
- 16.Arendt D, Nubler-Jung K. Nature (London) 1994;371:26. doi: 10.1038/371026a0. [DOI] [PubMed] [Google Scholar]
- 17.Arendt D, Nubler-Jung K. Development (Cambridge, UK) 1999;126:2309–2325. doi: 10.1242/dev.126.11.2309. [DOI] [PubMed] [Google Scholar]
- 18.DeRobertis E M, Sasai Y. Nature (London) 1996;380:37–40. [Google Scholar]
- 19.Samuel G, Miller D, Saint R. Evol Dev. 2001;3:241–250. doi: 10.1046/j.1525-142x.2001.003004241.x. [DOI] [PubMed] [Google Scholar]
- 20.Brower D L, Brower S M, Hayward D C, Ball E E. Proc Natl Acad Sci USA. 1997;94:9182–9187. doi: 10.1073/pnas.94.17.9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 22.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 23.Hayward D C, Catmull J, Reece-Hoyes J S, Berghammer H, Dodd H, Hann S J, Miller D J, Ball E E. Dev Genes Evol. 2001;211:10–19. doi: 10.1007/s004270000112. [DOI] [PubMed] [Google Scholar]
- 24.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 25.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 26.Adachi J, Hasegawa M. MOLPHY, Programs for Molecular Phylogenetics Based on Maximum Likelihood. Tokyo: Institute of Statistical Mathematics; 1996. , Version 2.3. [Google Scholar]
- 27.Suzuki Y, Yandell M D, Roy P J, Krishna S, Savage-Dunn C, Ross R M, Padgett R W, Wood W B. Development (Cambridge, UK) 1999;126:241–250. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- 28.Staehling-Hampton K, Hoffmann F M, Baylies M K, Rushton E, Bate M. Nature (London) 1994;372:783–786. doi: 10.1038/372783a0. [DOI] [PubMed] [Google Scholar]
- 29.Hayashibara T, Ohike S, Kakinuma Y. Proc 8th Int Coral Reef Symp. 1997;2:1231–1236. [Google Scholar]
- 30.Miller D J, Ball E E. BioEssays. 2000;22:291–296. doi: 10.1002/(SICI)1521-1878(200003)22:3<291::AID-BIES11>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Goldstein B, Freeman G. BioEssays. 1997;19:105–116. doi: 10.1002/bies.950190205. [DOI] [PubMed] [Google Scholar]
- 32.Angerer L M, Oleksyn D W, Logan C Y, McClay D R, Dale L, Angerer R C. Development (Cambridge, UK) 2000;127:1105–1114. doi: 10.1242/dev.127.5.1105. [DOI] [PubMed] [Google Scholar]
- 33.Bridge D, Cunningham C W, Schierwater B, DeSalle R, Buss L W. Proc Natl Acad Sci USA. 1992;89:8750–8753. doi: 10.1073/pnas.89.18.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medina M, Collins A G, Silberman J D, Sogin M L. Proc Natl Acad Sci USA. 2001;98:9707–9712. doi: 10.1073/pnas.171316998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferguson E L, Anderson K V. Cell. 1992;71:451–461. doi: 10.1016/0092-8674(92)90514-d. [DOI] [PubMed] [Google Scholar]
- 36.Sasai Y, Lu B, Steinbeisser H, De Robertis E M. Nature (London) 1995;376:333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- 37.Wilson P A, Hemmati-Brivanlou A. Nature (London) 1995;376:331–333. doi: 10.1038/376331a0. [DOI] [PubMed] [Google Scholar]