Abstract
The importance of founder events in promoting evolutionary changes on islands has been a subject of long-running controversy. Resolution of this debate has been hindered by a lack of empirical evidence from naturally founded island populations. Here we undertake a genetic analysis of a series of historically documented, natural colonization events by the silvereye species-complex (Zosterops lateralis), a group used to illustrate the process of island colonization in the original founder effect model. Our results indicate that single founder events do not affect levels of heterozygosity or allelic diversity, nor do they result in immediate genetic differentiation between populations. Instead, four to five successive founder events are required before indices of diversity and divergence approach that seen in evolutionarily old forms. A Bayesian analysis based on computer simulation allows inferences to be made on the number of effective founders and indicates that founder effects are weak because island populations are established from relatively large flocks. Indeed, statistical support for a founder event model was not significantly higher than for a gradual-drift model for all recently colonized islands. Taken together, these results suggest that single colonization events in this species complex are rarely accompanied by severe founder effects, and multiple founder events and/or long-term genetic drift have been of greater consequence for neutral genetic diversity.
Keywords: islands‖silvereyes‖colonization‖microsatellites
The idea that establishing a population from a small number of founders can result in a cascade of genetic changes leading to evolutionary differentiation was first developed by Mayr (1) in his seminal “genetic revolution” model. In this model, the key role of the founder event is to reduce levels of heterozygosity that subsequently affect the nature of coadapted gene complexes (1). Other founder-effect models have since been developed (2, 3), leading to much controversy surrounding the role of founder events in population differentiation (4–10).
One way to estimate the likelihood that founder events play an important role in natural island systems is to determine whether neutral genetic changes occur abruptly (by means of initial founder events) or in a more gradual manner (by long-term drift and new mutations). The rate of genetic change can be estimated by comparing the level of neutral genetic variation in island populations that have been established over a range of time periods. Recently founded populations can be used to gauge the relative impact of drift associated with the founding event itself, whereas older populations will bear additional genetic consequences of persisting as relatively small populations over evolutionary time.
The major difficulty faced when attempting to compare island forms of different ages is lack of information on the timing of natural founding events. As a result, most studies of the genetic consequences of founder events in free-living animals have been on populations artificially introduced by humans (11–18, but see ref. 19). Such studies illustrate whether extreme founder events can influence genetic parameters, but tell us little about the genetic consequences of the sorts of colonizations that really do occur in nature. Hence, they cannot tell us whether founder events are important in explaining natural diversity.
Members of the silvereye species complex (Zosterops lateralis), which have repeatedly colonized islands in the southwest Pacific from the Australian mainland (20; Fig. 1a), provide an unusual opportunity to redress this lack of evidence. Because of the silvereye's propensity to colonize islands and subsequently differentiate with respect to morphology (Fig. 1b), Mayr used this species complex to support his original genetic revolution founder effect model of differentiation (1). The Tasmanian subspecies, Z. l. lateralis, is particularly useful in this context, because a detailed record exists of its colonization history for the past 200 years (20–22) (Fig. 1a). This detailed account of “recent” colonization events is complemented by many island silvereye forms representing evolutionarily old colonization events, including those on Norfolk Island (Z. tenuirostris), Lord Howe Island (Z. tephropleurus), and Heron Island (Z. l. chlorocephalus) (Fig. 1a; refs. 23 and 24). Based on a combination of molecular and paleobiological evidence, the ages of these three evolutionarily “old” forms are in the order of millions of years, hundreds of thousands of years, and between 3,000 and 4,000 years, respectively (25–27). By using this system, therefore, we could make the crucial contrast between genetic changes in recent vs. old island populations.
Figure 1.
The distribution of silvereye forms used in this study, with the timing and sequence of colonization events by Z. l. lateralis indicated by dates and arrows. (a) Geographic pattern of silvereye colonization. White and black circles show sampling locations for the recent colonizer, Z. l. lateralis, and evolutionarily old populations (Australian mainland; Norfolk Island; Lord Howe Island; and Heron Island), respectively. (b) Morphological divergence among silvereye forms. Abbreviations: ML, Australian mainland Z. l. familiaris; T, Tasmania; SI, South Island New Zealand; CI, Chatham Island; PN Palmerston North, south end of North Island New Zealand; NIlat, Norfolk Island (all Z. l. lateralis); HI, Heron Island Z. l. chlorocephalus; LHI, Lord Howe Island Z. tephropleurus; NIten, Norfolk Island Z. tenuirostris. Note that no morphological information is available for Auckland and that some SE bars are smaller than data points.
Here, we examine nuclear DNA variation across six microsatellite loci in ten silvereye populations representing source mainland populations, the recent island populations, and evolutionarily old island populations, to examine the genetic consequences of island dwelling. First, we made comparisons between populations with respect to within-population indices of genetic diversity: expected heterozygosity and allelic diversity. Reduced variation in both measures may result from population bottlenecks (28). Second, we used the degree of genetic differentiation of a population from its source as an indicator of genetic drift. In the presence of a bottleneck, especially a narrow one, genetic distance can increase rapidly (29).
If single founder events have been important in shaping genetic diversity and differentiation of island silvereye forms, then recently colonized populations of Z. l. lateralis are expected to have reduced diversity compared with their immediate source populations (28, 30), and the degree of genetic differentiation, could be “instantaneous” and bear no relationship to the age of the population (29, 31). The level of genetic change for recent populations should be of similar magnitude to that in old populations. Alternatively, if long-term drift has been more important in shaping the level of genetic diversity and differentiation of island forms, then loss of diversity and increased levels of differentiation will occur gradually over time (29, 32). Under this scenario, the extent of divergence in recent populations should be less than that in old populations.
Finally, we use a Bayesian approach based on computer simulations to investigate the demographic factors that have led to the observed pattern of genetic differentiation. On the basis of the findings of our empirical microsatellite data and the historically known pattern of colonization, we used the simulations to estimate the effective size of flocks that founded the recent populations. If founder events are important in this system, we expect the estimated effective founder flocks to be small. We then test whether or not it is necessary for the model to incorporate founder events at all by comparing the explanatory power of two models. The first is the “founder-event” model that incorporates population bottlenecks in each population, and thereby allows for a strong pulse of drift at founding. The second model, the “gradual-drift” model does not incorporate population bottlenecks and assumes a long-term effective population size equal to or less than the original source. Again, if founder events are important in this system, we expect the founder-event model to be a better fit than the model assuming gradual drift alone.
Methods
Sampling and Laboratory Techniques.
Blood samples for DNA analysis were collected from Zosterops taxa from nine sites in Australia and southwest Pacific islands. Samples from the nine sites represented three species, three subspecies, and six populations (Fig. 1a). A population from the east coast of the Australian mainland was used as a representative of a large, outbreeding continental population, and it was assumed that this population was equivalent to the ancestral mainland population for island populations. Likewise, we assumed that contemporary samples from the initial source (Tasmania, Fig. 1a) of the recent colonization sequence are representative of the population at the time of colonization. Birds were caught by using mist nets or traps and 20–40 μl of blood from the subbrachial wing vein was sampled and stored in 1 ml of lysis buffer (33). All sampling was nondestructive and birds were released at point of capture. DNA was extracted by using either standard phenol-chloroform protocols (34) or a “salting-out” method (35).
DNA variation was assayed at five microsatellite loci developed from a Zosterops lateralis chlorocephalus library (36) and at one locus developed from a swallow (Hirundo rustica) library (37). PCR conditions for Zosterops primer sets were as described (36), whereas the conditions for the swallow primer set were according to ref. 37, except that the PCR annealing temperature was lowered from 63°C to 55°C. A size standard was produced by sequencing 2 μg of pBluescript KS (+) plasmid (Stratagene) with [γ-33P]ATP end-labeled universal primer with standard cycle sequence conditions (38). The size standard and microsatellite PCR products were electrophoresed on a 6% denaturing polyacrylamide gel. Gels were exposed to Hyperfilm-MP (Amersham Pharmacia) for 1 to 5 days. Allele sizes were scored manually by comparison against the size standard.
Statistical Analyses.
Tests for linkage disequilibrium and heterozygote deficit were conducted in GENEPOP V.3.1d (39). The significance of linkage disequilibrium was determined for each locus pair across all populations. (With Bonferroni correction, the critical value was corrected for 60 multiple comparisons; adjusted α = 0.0008.) Global tests of heterozygote deficit were conducted for each population (with Bonferroni correction, critical value corrected for 10 comparisons; adjusted α = 0.005), each locus (critical value corrected for six comparisons; adjusted α = 0.008), and then for each locus-population comparison (critical value corrected for 60 comparisons; α = 0.0008).
Genetic diversity within populations was measured as unbiased expected heterozygosity (HE; ref. 40) and average number of alleles per locus (allelic diversity) by using the software TFPGA V.1.3 (41). Allelic diversity was corrected for sample size by using a personal program according to ref. 42. Significance of pairwise differences in heterozygosity and allelic diversity between each island population and its immediate source were assessed by two-sample t tests. Standard errors were calculated across loci. For the recently colonized populations, Spearman's Rank Correlation was used to test for associations between genetic diversity and the number of preceding colonization events. For example, Tasmania has zero preceding colonization events, and Norfolk Island has four such events. For Spearman's tests, islands were the independent data points.
Genetic divergence between taxa was quantified by using a relative measure of differentiation, the FST statistic of Weir and Cockerham (43). Pairwise FST values, and 95% confidence limits around each pairwise comparison, were generated by using the software TFPGA, version 1.3 (41), with 1,000 bootstrap replicates across loci. The absolute measure of divergence, Nei's standard genetic distance, DS (44), was also calculated. As the data is in matrix form, Mantel tests were used to test for associations between the number of colonization steps separating any two populations and average pairwise FST values and then average pairwise Nei's DS values, respectively, by using 10,000 randomizations in the program ADE-4 (45).
Bayesian Analysis of Colonization.
In our founder-event model, inferences about the effective number of founding individuals (Nfd) for each recently colonized population were made by using a Bayesian approach (46, 47). The analysis based on computer simulation combines microsatellite data with historical and demographical information on island colonization. The simulations were conducted according to methods outlined in ref. 47. A prior probability distribution for Nfd was set, being a uniform distribution between 2 and 300. This distribution was based on information about the size of migrating silvereye flocks (48). Prior distributions for other demographic parameters formulating our founder-event model were chosen according to published (20) and unpublished demographic information for the study species: a generation time of 3 years, a short duration of the bottleneck associated with the founding of any new island (i.e., one, two, or three generation(s) with equal probability for each duration) and a lognormal (6, 2) distribution (median Ne = 874, mean Ne = 4247) for the stable effective population size (i.e., in the source population and the colonized population following recovery). Parameters pertaining to the microsatellite markers (mutation rate of microsatellite loci and variance of the geometric distribution modeling the change in the number of repeats of mutant alleles) were determined as described in ref. 47. For reasons of tractability, the populations were treated by pair, with Tasmania as source population in each pair. We thus had four pairs, Tasmania–South Island, Tasmania–Chatham Island, Tasmania–North Island, and Tasmania–Norfolk Island. For each population pair, our founder-event model included all previous colonization steps. Posterior distributions of Nfd (and other parameters of the model) for each island population were then estimated by using a rejection algorithm (47), taking into account seven statistics summarizing the mean number of alleles (across loci), expected heterozygosity and variance in repeat number in the source (Tasmania) and the island population considered, and the level of differentiation between the source and island population as estimated by FST. A posterior distribution for Nfd was constructed for each island population from 1,000 values accepted by the rejection algorithm and compared with the original prior distributions. Mean, 5%, 50%, and 95% quantile values of Nfd were summarized for each population. It is noteworthy that the posterior distributions of Nfd were robust to prior beliefs on the demographic and mutation parameters formulating our colonization model (A.E. and S.M.C., unpublished results).
Additionally, to investigate the overall importance of founder events in this system, we examined an alternative form of our model, which we refer to as the gradual-drift model. This model assumed that the postcolonization effective population size in each island population has remained constant and equal to or less than that in the original source population. In contrast to the founder-event model, the gradual-drift model contains only two demographic parameters: the stable effective population size in the source (Tasmanian) population, Ns, and the stable effective population size in the colonized island population, Ni, the prior distribution for Ni being a lognormal (6,2) with Ni ≤ Ns. We then opposed the two models in a Bayesian framework by giving each model a prior weight probability of 0.5. The relative fit of the two models was then assessed for each population by using the acceptance rate of the rejection algorithm as an estimate of the posterior weight probability (47). The significance of differences between the posterior weight probabilities for each model were assessed by using the statistic G = 2ln(pm2/pm1) which follows a χ2 distribution with degrees of freedom equal to the difference in number of parameters in the founder-event and gradual-drift models (i.e., df = 1). This is analogous to a likelihood ratio test for nested models (49).
Results
Heterozygote Deficit and Linkage Disequilibrium.
No global heterozygote deficit was observed in any population, indicating that samples were representative of each population. One locus, HrU6, showed a significant global heterozygote deficit (P < 0.001, adjusted α = 0.005) because of a heterozygote deficit in one population (Lord Howe Island, P < 0.0001, adjusted α = 0.0008). Because other populations had an expected number of heterozygotes at this locus, the locus itself is not faulty (i.e., it is not affected by null alleles), and the results for Lord Howe Island are likely to be a random sampling effect. We therefore kept this locus in the analysis. Significant linkage disequilibrium was observed between two pairs of loci of 60 pairwise comparisons, ZL12/ZL22 and ZL22/HrU6 (P < 0.0001, adjusted α = 0.0008), but the association between these loci is limited to one population (Auckland). All comparisons between these two sets of loci in the nine other populations were not significant; therefore, it is reasonable to consider these loci independent.
Genetic Diversity and Differentiation in Recently Colonized Populations.
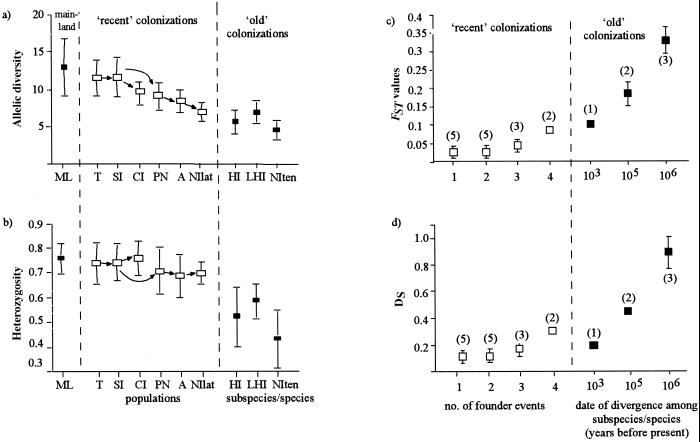
We found very little evidence that single founder events led to significant changes in genetic diversity or significant population differentiation. When each recently colonized population was compared with its immediate source population, no significant differences were found in terms of either allelic diversity (Fig. 2a; t < 1.95, P > 0.1 in all cases) or heterozygosity (Fig. 2b; t < 1.05, P > 0.2 in all cases). In any single founder event, allelic diversity decreased by between 0.1 and 21.5% and 0 and 3.1% for heterozygosity. The degree of divergence was generally low with little evidence of significant divergence between single-step source-founder population pairs in terms of FST (Fig. 2c; 95% confidence intervals for FST include zero in 4 of 5 source-founder comparisons). Estimates of divergence based on Nei's DS showed the same qualitative pattern (Fig. 2d).
Figure 2.
Genetic diversity and divergence of silvereye forms as measured by (a) heterozygosity; (b) allelic diversity; (c) pairwise FST, and (d) Nei's DS. Error bars indicate standard errors calculated across loci. Arrows indicate recent colonization sequence. Abbreviations for populations are the same as in Fig. 1b, with the addition of A, Auckland, north end of North Island of New Zealand. Number of founder events is the number of island colonizations separating any two populations. Numbers in parentheses indicate the number of pairwise comparisons among populations or among subspecies/species. Age of divergence among subspecies and species were based on molecular and paleobiological evidence (26, 27).
In contrast, we did find evidence that the level of genetic diversity and the extent of genetic differentiation are influenced by multiple colonization events. The number of preceding colonization events affected measures of diversity and differentiation in the recently founded populations. Spearman's Rank Correlation showed that an increased number of founder steps was associated with a significant decrease in allelic diversity (Rho = −0.99, P < 0.05), corresponding to a 40% reduction in average allelic diversity between the Tasmanian and Norfolk Island populations. Also, a significant positive association was found between both pairwise FST and pairwise Nei's DS and the number of colonization steps separating each population pair (Mantel test of matrix comparisons, r = 0.50, P < 0.05 and r = 0.52, P < 0.05 for FST and Nei's DS, respectively). The number of previous colonization events did not significantly affect heterozygosity levels (Spearman's test, Rho = −0.64, P > 0.10) with only a 5.5% difference in mean heterozygosity levels between the Tasmanian and Norfolk Island populations.
Genetic Diversity and Differentiation in Evolutionary Old Island Populations.
The evolutionarily old island forms almost always contained significantly fewer alleles (Fig. 2a; t tests between each island population and the mainland: in 5 of 6 cases, t > 2.5, P < 0.05, for Lord Howe Island, t = 2.29, P = 0.07) and significantly fewer heterozygotes (Fig. 2b; t > 2.5, P < 0.05 in all cases) than the source Australian mainland population. Significant genetic divergence also occurred between evolutionarily old populations in terms of FST (Fig. 2c, FST > 0 in all cases, and is highest for comparisons involving the oldest form, Z. tenuirostris). Nei's DS showed an association with age of the island form, with the oldest form being the most differentiated (Fig. 2d).
Bayesian Analysis of Colonization.
Our founder-event model did not support the expectation of small effective founder sizes. Posterior distributions for founder sizes were shifted toward large values for all but one of the islands, giving low statistical support for small founding flocks. Point estimates (50% quantiles) for the effective size of the founding flocks ranged from 132 to 216 effective founders and 5% quantiles were greater than 34 (Table 1). The one exception was Norfolk Island, where the posterior distribution gave strong statistical support to small values of effective founders, with a 50% quantile of 24 and a 5% quantile as small as 8 (Table 1).
Table 1.
Prior and posterior values for effective number of founding individuals (Nfd) of Z. l. lateralis on each island (Is.) based on 20,000 and 1,000 values, respectively
| Population | Quantiles
|
|||
|---|---|---|---|---|
| Mean | 5% | 50% | 95% | |
| Prior | 152 | 17 | 152 | 286 |
| Posterior | ||||
| South Is. | 198 | 72 | 207 | 292 |
| North Is. | 143 | 34 | 132 | 278 |
| Chatham | 207 | 90 | 216 | 294 |
| Norfolk | 24 | 8 | 24 | 43 |
When we opposed the founder-event model and the gradual-drift model in a Bayesian framework, the posterior weight probability was higher for the gradual-drift model for three of the four populations (South Island NZ, North Island NZ, and Chatham Island), and higher for the founder-event model in only one population (Norfolk Island) (Table 2). However none of these differences were statistically significant (South Island, NZ, pG = 0.12; North Island NZ, pG = 0.20; Chatham Island, pG = 0.23; Norfolk Island, pG = 0.23).
Table 2.
Comparison of the fit of the founder-event model vs. the gradual-drift model by using posterior weight probabilities of islands colonized by Z. l. lateralis
| Opposing model | Posterior probabilities
|
|||
|---|---|---|---|---|
| SI | PN | CI | NI | |
| Founder event | 0.84 | 0.80 | 0.67 | 0.41 |
| vs. | vs. | vs. | vs. | vs. |
| Gradual drift | 0.16 | 0.20 | 0.33 | 0.59 |
| Significance test (pG) | 0.12 | 0.20 | 0.23 | 0.23 |
Abbreviations: SI, South Island New Zealand; PN, Palmerston North, south end of North Island New Zealand; CI, Chatham Island; NI, Norfolk Island.
Discussion
Our results show that, for most of the recently established island populations studied here, single founding events have had a limited effect on levels of neutral genetic diversity and divergence. Neither allelic diversity nor heterozygosity within populations show significant reductions after any single founder event. Heterozygosity is an effective indicator of population bottlenecks only if the event was severe in strength or duration (28, 29, 50); therefore, this result may be partly because of the low sensitivity of this particular measure. However, allelic diversity is a more sensitive indicator of changes in population size than heterozygosity, because rare alleles can be lost easily (28). Our results therefore support laboratory-based experimental work that has suggested that single colonization events are unlikely to have a marked effect on genetic diversity (8, 51, 52).
Likewise, the degree of genetic differentiation associated with any single colonization event suggests that colonization itself does not result in a strong pulse of genetic drift. Pairwise estimates of relative and absolute divergence measures between each recently colonized population and its source indicates that the prevalent pattern is one of little genetic differentiation. The one possible exception is the most recently colonized population on Norfolk Island, which is discussed below. Overall however, these results indicate that single-step founder events are unlikely to provide sufficient genetic perturbation to lead to the reductions in genetic variation and pronounced genetic differentiation assumed by the original founder effect model (1).
Although single founder events had a negligible effect on genetic diversity and differentiation, we did find evidence for the effects of sequential founder events. Allelic diversity significantly decreased and genetic differentiation significantly increased as the number of founder events separating populations increased. Heterozygosity levels were not affected, despite the increased potential for multiple vs. single population bottlenecks to affect this measure (30, 53). To test the potential evolutionary impact of sequential founder effects we compared the level of change among the recently founded populations with that seen among evolutionarily old populations, corresponding to colonizations that took place between several thousand and millions of years ago. Three or four sequential founder events are required for recently colonized populations to reach levels of (allelic) diversity and divergence similar to the levels of the youngest of the old island forms. This result is interesting given that the former populations have only been separated from one another for about 50 generations, while the latter (Heron Island, Z. l. chlorocephalus) has been genetically isolated for more than a thousand generations (20, 25) and has likely persisted with relatively small population sizes (current Ne for Heron Island is approximately 173 individuals, based on demographic data (the mean age of breeding birds and the survival of young to that age) J. Kikkawa, unpublished data). This finding again supports models and laboratory-based experimental work that has confirmed the potential power of sequential founder events (53, 54).
The general lack of strong founder effects accompanying island colonization by silvereyes is particularly striking given that members of the species complex have been repeatedly invoked by evolutionary biologists in discussions of island colonization and founder effects (1, 55). Why then do founder effects seem to be so weak in this species complex? Our simulations of the colonization process suggest that new island populations are probably not founded by small flocks. Instead, our estimates of effective founding flock size were relatively large with mean values of more than 100 individuals. Large founding-flock sizes would increase the likelihood of the new population being genetically representative of the source population. When the founder-event model was opposed to the gradual-drift model, we could not clearly reject one model in favor of the other for any recently colonized population. Taken together, these results indicate that the focus on founder effects in this species complex has been overemphasized and that large flock sizes and/or gradual drift are more parsimonious explanations for the observed genetic changes.
The most recent colonization event of Z. l. lateralis to Norfolk Island provides the exception to most of the general trends described above. This population is the most differentiated at neutral loci of all of the recently colonized populations. Two explanations are possible for this. First, it is possible that introgressive hybridization between the new arrival and Z. tenuirostris may have occurred. Initial hybridization between the two species immediately after colonization has been suggested but is not thought to have continued (20, 56). No evidence was found of introgressive hybridization between Z. tenuirostris and Z. l. lateralis from the mtDNA analysis (S.M.D., L. Kelleman, and C.M., unpublished results) or from the distribution of microsatellite alleles characteristic of Z. tenuirostris. A more likely explanation, therefore, is that a smaller flock founded this geographically remote population, or greater variance occurred in reproductive success than in other cases examined. The estimates of the effective founder size of this population are consistent with this explanation, being approximately an order of magnitude less than other recently colonized populations. This population was also the only one for which better statistical support existed for the founder-event model, although this difference was not statistically significant.
In sum, our results demonstrate that the natural colonization behavior of the silvereye is not consistent with the assumptions of traditional, single-step founder-effect models of divergence. Instead, the evidence suggests that the low genetic variation and pronounced genetic differentiation of old island forms is a result of multiple founder events possibly by large numbers of individuals, gradual drift in relatively small isolated populations (57), or a combination of these two mechanisms. Future work will assess whether the same mechanisms can be applied to the phenotypic divergence observed in the recently colonized populations.
Acknowledgments
We thank B. Degnan, A. Hugall, A. Fletcher, P. Park, P. Gray, B. Robertson, D. Lambert, P. Schweigman, D. Onley, M. Bell, P. Smith, and W. Smith for help in catching birds; M. Blows, F. Frentiu, N. Johnson, I. Lovette, S. Masta, D. Paetkau, T. Price, S. Scott, R. Slade, D. Reznick, and S. Robinson for discussion and/or comments on this or earlier versions of the manuscript. This work was approved by the University of Queensland Ethics Committee and was funded by the Australian Research Council and Stuart Leslie Fund (Birds Australia).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 7818.
References
- 1.Mayr E. In: Evolution as a Process. Huxley J, Hardy A C, Ford E B, editors. London: Allen & Unwin; 1954. pp. 157–180. [Google Scholar]
- 2.Carson H L. In: Population Biology and Evolution. Lewontin R C, editor. NY: Syracuse Univ. Press; 1968. pp. 123–137. [Google Scholar]
- 3.Templeton A R. Genetics. 1980;94:1011–1038. doi: 10.1093/genetics/94.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lande R. Am Nat. 1980;116:463–479. [Google Scholar]
- 5.Carson H L, Templeton A R. Annu Rev Ecol Syst. 1984;15:97–131. [Google Scholar]
- 6.Barton N H, Charlesworth B. Annu Rev Ecol Syst. 1984;15:133–164. [Google Scholar]
- 7.Barton N. In: Speciation and Its Consequences. Otte D, Endler J A, editors. Sunderland, MA: Sinauer; 1989. pp. 229–256. [Google Scholar]
- 8.Rice W, Hostert E. Evolution (Lawrence, Kans) 1993;47:1637–1653. doi: 10.1111/j.1558-5646.1993.tb01257.x. [DOI] [PubMed] [Google Scholar]
- 9.Grant P R. Evolution on Islands. Oxford: Oxford Univ. Press; 1998. [Google Scholar]
- 10.Grant P R. Oikos. 2001;92:385–403. [Google Scholar]
- 11.Baker A J, Moeed A. Evolution (Lawrence, Kans) 1987;41:525–538. doi: 10.1111/j.1558-5646.1987.tb05823.x. [DOI] [PubMed] [Google Scholar]
- 12.Baker A J, Dennison M D, Lynch A, LeGrand G. Evolution (Lawrence, Kans) 1990;44:981–999. doi: 10.1111/j.1558-5646.1990.tb03819.x. [DOI] [PubMed] [Google Scholar]
- 13.Baker A J, Peck M K, Goldsmith M A. Condor. 1990;92:76–88. [Google Scholar]
- 14.Berry R J. Philos Trans R Soc London B. 1996;351:753–764. doi: 10.1098/rstb.1996.0070. [DOI] [PubMed] [Google Scholar]
- 15.Ardern S L, Lambert D L, Rodrigo A G, McLean I G. J Hered. 1997;88:179–186. [Google Scholar]
- 16.Cabe P R. Heredity. 1998;80:519–525. [Google Scholar]
- 17.Tarr C L, Conant S, Fleischer R C. Mol Ecol. 1998;7:719–731. [PubMed] [Google Scholar]
- 18.Broders H G, Mahoney S P, Montevecchi W A, Davidson W S. Mol Ecol. 1999;8:1309–1315. doi: 10.1046/j.1365-294x.1999.00695.x. [DOI] [PubMed] [Google Scholar]
- 19.Grant P R, Grant B R. Evolution (Lawrence, Kans) 1995;49:229–240. doi: 10.1111/j.1558-5646.1995.tb02235.x. [DOI] [PubMed] [Google Scholar]
- 20.Mees G F. Zool Verh (Leiden) 1969;102:1–390. [Google Scholar]
- 21.North A J. Rec Aust Mus. 1904;5:337–338. [Google Scholar]
- 22.Falla R A, Sibson R B, Turbott E G. A Field Guide to the Birds of New Zealand and Outlying Islands. London: Collins; 1966. [Google Scholar]
- 23.Hutton I. Birds of Lord Howe Island, Past and Present. Coffs Harbour: I. Hutton; 1991. [Google Scholar]
- 24.Collar N J, Crosby M J, Stattersfield A J. Birds to Watch. 2. The World List of Threatened Birds. Cambridge: Birdlife International; 1994. [Google Scholar]
- 25.Degnan S M, Owens I P F, Clegg S M, Moritz C C, Kikkawa J. In: Proc. 22nd Int. Ornith. Cong., Durban. Adams N J, Slotow R H, editors. Johannesburg: Birdlife South Africa; 1999. pp. 1881–1898. [Google Scholar]
- 26.Degnan S M, Moritz C. Auk. 1993;109:800–811. [Google Scholar]
- 27.Hopley D. The Geomorphology of the Great Barrier Reef: Quaternary Development of Coral Reefs. New York: Wiley; 1982. [Google Scholar]
- 28.Nei M, Maruyama T, Chakraborty R. Evolution (Lawrence, Kans) 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 29.Chakraborty R, Nei M. Evolution (Lawrence, Kans) 1977;31:347–356. doi: 10.1111/j.1558-5646.1977.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 30.Motro U, Thomson G. Evolution (Lawrence, Kans) 1982;36:1059–1066. doi: 10.1111/j.1558-5646.1982.tb05474.x. [DOI] [PubMed] [Google Scholar]
- 31.Hedrick P W. Evolution (Lawrence, Kans) 1999;53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. [DOI] [PubMed] [Google Scholar]
- 32.Wright S. Variability Within and Among Natural Populations. Chicago: Chicago Univ. Press; 1978. [Google Scholar]
- 33.Seutin G, White B N, Boag P T. Can J Zool. 1991;69:82–90. [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 35.FitzSimmons N N, Moritz C, Moore S S. Mol Biol Evol. 1995;12:432–440. doi: 10.1093/oxfordjournals.molbev.a040218. [DOI] [PubMed] [Google Scholar]
- 36.Degnan S M, Robertson B C, Clegg S M, Moritz C C. Mol Ecol. 1999;8:157–158. doi: 10.1046/j.1365-294x.1999.00799.x. [DOI] [PubMed] [Google Scholar]
- 37.Primmer C R, Møller A P, Ellegren H. Mol Ecol. 1995;4:493–498. doi: 10.1111/j.1365-294x.1995.tb00243.x. [DOI] [PubMed] [Google Scholar]
- 38.Hillis D M, Mable B K, Larson A, Davis S K, Zimmer E A. In: Molecular Systematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. pp. 321–381. [Google Scholar]
- 39.Raymond M, Rousset F. J Hered. 1995;8:248–249. [Google Scholar]
- 40.Nei M. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller M P. TOOLS FOR POPULATION GENETICS: TFPGA, V.1.3. Flagstaff: Northern Arizona Univ.; 1997. [Google Scholar]
- 42.Ewens W J. Theor Pop Biol. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- 43.Weir B S, Cockerham C C. Evolution (Lawrence, Kans) 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 44.Nei M. Am Nat. 1972;106:283–292. [Google Scholar]
- 45.Thioulouse J, Chessel D, Dolédec S, Olivier J M. Stat Comput. 1997;7:75–83. [Google Scholar]
- 46.Shoemaker J S, Painter I S, Weir B S. Trends Genet. 1999;15:354–358. doi: 10.1016/s0168-9525(99)01751-5. [DOI] [PubMed] [Google Scholar]
- 47.Estoup A, Wilson I J, Sullivan C, Cornuet J-M, Moritz C. Genetics. 2001;159:1671–1687. doi: 10.1093/genetics/159.4.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan K, Sutton P. Corella. 1993;17:41–42. [Google Scholar]
- 49.Hilborn R, Mangel M. The Ecological Detective: Confronting Models with Data. Princeton: Princeton Univ. Press; 1997. [Google Scholar]
- 50.Leberg P L. Evolution (Lawrence, Kans) 1992;46:477–494. doi: 10.1111/j.1558-5646.1992.tb02053.x. [DOI] [PubMed] [Google Scholar]
- 51.Charlesworth D, Charlesworth B. Annu Rev Ecol Syst. 1987;18:237–268. [Google Scholar]
- 52.Moya A, Galiana A, Ayala F J. Proc Natl Acad Sci USA. 1995;92:3983–3986. doi: 10.1073/pnas.92.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le Corre V, Kremer A. J Evol Biol. 1998;11:495–512. [Google Scholar]
- 54.Meffert L M, Bryant E H. Evolution (Lawrence, Kans) 1991;45:293–306. doi: 10.1111/j.1558-5646.1991.tb04404.x. [DOI] [PubMed] [Google Scholar]
- 55.Lack D. Ecological Isolation in Birds. Oxford: Blackwell Scientific; 1971. [Google Scholar]
- 56.Gill F B. Condor. 1970;72:481–482. [Google Scholar]
- 57.Mundy N I, Winchell C S, Burr T, Woodruff D S. Proc R Soc London Ser B. 1997;264:869–875. [Google Scholar]