Abstract
The classic wheat evolutionary history is one of adaptive radiation of the diploid Triticum/Aegilops species (A, S, D), genome convergence and divergence of the tetraploid (Triticum turgidum AABB, and Triticum timopheevii AAGG) and hexaploid (Triticum aestivum, AABBDD) species. We analyzed Acc-1 (plastid acetyl-CoA carboxylase) and Pgk-1 (plastid 3-phosphoglycerate kinase) genes to determine phylogenetic relationships among Triticum and Aegilops species of the wheat lineage and to establish the timeline of wheat evolution based on gene sequence comparisons. Triticum urartu was confirmed as the A genome donor of tetraploid and hexaploid wheat. The A genome of polyploid wheat diverged from T. urartu less than half a million years ago (MYA), indicating a relatively recent origin of polyploid wheat. The D genome sequences of T. aestivum and Aegilops tauschii are identical, confirming that T. aestivum arose from hybridization of T. turgidum and Ae. tauschii only 8,000 years ago. The diploid Triticum and Aegilops progenitors of the A, B, D, G, and S genomes all radiated 2.5–4.5 MYA. Our data suggest that the Acc-1 and Pgk-1 loci have different histories in different lineages, indicating genome mosaicity and significant intraspecific differentiation. Some loci of the S genome of Aegilops speltoides and the G genome of T. timophevii are closely related, suggesting the same origin of some parts of their genomes. None of the Aegilops genomes analyzed is a close relative of the B genome, so the diploid progenitor of the B genome remains unknown.
Keywords: evolution‖plant‖grass‖acetyl-CoA carboxylase‖3-phosphoglycerate kinase
Three members of the grass family (Poaceae)—wheat (genus Triticum), rice (genus Oryza) and maize (genus Zea)—are staple crops. Their domestication 8,000 to 12,000 years ago—wheat in South-West Asia, rice in China, and maize in the Americas—permitted the founding of great civilizations. The domestication of wheat involved multiple polyploidization events between several species of the Triticum and Aegilops genera (Triticeae tribe of the Pooideae subfamily of grasses). Hexaploid bread wheat (Triticum aestivum) is the most prominent member of the tribe, which also includes domesticated diploid and tetraploid wheats as well as rye (Secale cereale) and barley (Hordeum vulgare). The evolution of the diploid Triticum and Aegilops species, the origin of the homoeologous genomes, and the timeline of the polyploidization events that established tetraploid wheat in nature are the subject of this report.
The major grass subfamilies, including Pooideae, radiated 50–80 million years ago (MYA; ref. 1). It was estimated previously that the Triticeae and Poeae (Lolium rigidum) tribes diverged ≈35 MYA, and Hordeum and Secale diverged from the Triticum/Aegilops lineage ≈11 MYA and ≈7 MYA, respectively (1). Subsequent events increased wheat ploidy to four and later to six approximately 8,000 years ago. The general evolution of the Triticeae tribe, as revealed by meiotic pairing analysis, has been defined by divergence at the diploid level from a common diploid ancestor and convergence at the polyploid level involving the diverged diploid genomes (2). Various Aegilops species contributed significantly to the genetic makeup of the polyploid wheats. Extensive classical analyses and more recent molecular studies have provided information on the identity of donors and some of the patterns of genome evolution of the Triticum/Aegilops species (3). A review of Triticum and Aegilops taxonomy and related literature is available from the Wheat Genetic Resource Center at ksu.edu/wgrc.
The Triticum and Aegilops genera contain 13 diploid and 18 polyploid species. The diploid species contain eight distinct genomes that were given the following names: A (A and Ab/Am), D, S (S, Ss, Sb, Sl, Ssh), M, C, U, N, and T. Two genomes found in polyploid wheats were given new names, B and G, because their diploid progenitors are not known. The allopolyploids arose from interspecific hybridization events followed by spontaneous chromosome doubling. Species containing a common genome, either A, D, or U, resemble the diploid donor of the common genome in morphology and mode of seed dispersal.
The wild and cultivated wheats include diploid, tetraploid, and hexaploid species for which either Triticum urartu or Triticum monococcum was the A genome donor. These two diploid einkorn wheats produce sterile hybrids, indicating that they are valid biological species (4). T. urartu exists only in its wild form. T. monococcum includes the wild form, T. monococcum ssp. aegilopoides (syn. Triticum boeoticum) and the cultivated form T. monococcum ssp. monococcum (limited to mountainous regions of Yugoslavia and Turkey). There are two tetraploid wheat species: Triticum turgidum (AABB genome) and Triticum timopheevii (AAGG genome). T. turgidum includes the wild ssp. dicoccoides and several cultivated subspecies such as T. turgidum ssp. durum (durum or macaroni wheat) grown in semiarid areas such as the Mediterranean basin, India, and the Northern Great Plains of the United States and Canada. The tetraploid T. timopheevii includes the wild-form T. timopheevii ssp. armeniacum (syn. Triticum araraticum) and the cultivated form, T. timopheevii ssp. timopheevii (grown in the Transcaucasian region). Finally, there are two hexaploid wheats: T. aestivum (AABBDD genome), including several subspecies, and Triticum zhukovskyi (AmAmAAGG genome). Genetic studies have revealed that the polyploid wheat species constitute two evolutionary lineages. T. turgidum (AABB) and T. aestivum (AABBDD) comprise one lineage, and T. timopheevii (AAGG) and T. zhukovskyi (AmAmAAGG) comprise the other. Early cytogenetic studies suggested that the A genomes of the tetraploids in both lineages were contributed by T. monococcum (5–7). More recent studies, as well as our own reported below, showed that T. urartu contributed the A genome in both lineages (8–10). Also, it was suggested that the wild tetraploids T. turgidum ssp. dicoccoides and T. timopheevii ssp. armeniacum arose from hybridization between T. urartu and two different plasmon types of another wild diploid (8, 11). T. aestivum arose under cultivation 8,000 years ago from spontaneous hybridization between T. turgidum ssp. dicoccon and the diploid goatgrass Aegilops tauschii ssp. strangulata (12–16). T. zhukovskyi (AmAmAAGG) also originated under cultivation from hybridization of T. timopheevii with T. monococcum (17); one set of A genomes was contributed by T. urartu and the other by T. monococcum (8).
The origin of the B and G genomes remains controversial. Much evidence suggests that an ancestor Aegilops speltoides species (S genome) was the donor of what became the B genome of the bread and durum wheats (18–26). It is possible that Ae. speltoides is a significantly diverged form of the ancestral B genome donor (27). Plasmon (the sum of extrachromosomal hereditary determinants) analysis also pointed to Ae. speltoides as the B genome donor (28, 29), but it remains uncertain whether Ae. speltoides is the sole source of the B genome or whether the genome resulted from an introgression of several parental species (30). The B genome in T. turgidum and the G genome in T. timopheevii were proposed to be closely related to each other (31, 32), but also it was suggested that the G genome of T. timopheevii is more closely related to the S genome of Ae. speltoides than to the B genome of T. turgidum (33). Analysis of organelle DNA in Ae. speltoides indicates that it may be the maternal (cytoplasmic) donor of all polyploid wheats (34).
Here, we present the results of a molecular phylogenetic analysis of the Triticum and Aegilops species including A, D, and S diploids and A genome polyploids by using a system based on sequences of large fragments of nuclear genes encoding plastid ACCase and plastid PGK, as described (1).
Materials and Methods
Multiple sequence alignments of Acc-1 and Pgk-1gene fragments (GenBank accession numbers AF343496–AF343536 and AF343474–AF343495, respectively) were described (1). These fragments were 1.5–1.6 kb long, spanned several introns and exons, and encoded most of the biotin carboxylase domain of ACCase and most of the mature PGK. CLUSTALX V.1.81 (35) and MACCLADE (Sinauer, Sunderland, MA) were used to create and analyze the alignments. The alignments were unambiguous within exons and required no manual adjustments within introns. The distribution of variable characters in different segments of the alignments is shown in Table 1. PAUP* V.4.0 (Sinauer, Sunderland, MA) and MEGA (S. Kumar, K. Tamura, I. Jakobsen and M. Nei, www.megasoftware.net) were used to calculate phylogenetic trees and nucleotide substitution rates.
Table 1.
Number of characters in the Acc-1 and Pgk-1 sequence alignments
| Gene | Characters | Introns | Exons | Synonymous | Nonsynonymous |
|---|---|---|---|---|---|
| Acc-1 | All (1,471) | 775 | 696 | 164 | 532 |
| Variable (208) | 165 | 43 | 40 | 3 | |
| Informative (94) | 75 | 19 | 18 | 1 | |
| Pgk-1 | All (1,603) | 709 | 894 | 220 | 674 |
| Variable (188) | 117 | 71 | 63 | 8 | |
| Informative (82) | 50 | 32 | 27 | 5 |
Phylogenetic trees were first calculated on the basis of intron sequences of Triticeae Acc-1 and Pgk-1 by the neighbor-joining method, without correction for multiple substitutions and with gaps excluded only from pairwise comparisons. Second, neighbor-joining trees were calculated without correction for multiple substitutions and with gaps excluded only from pairwise comparisons but including both intron sites and synonymous sites in exons merged into one character set. This Acc-1 character set consisted of a total of 939 nucleotides including 205 variable sites. The Pgk-1 character set consisted of a total of 929 nucleotides including 180 variable sites. Bootstrap values for the neighbor-joining trees were calculated as a percentage of 1,000 trials. Third, phylogenetic trees were generated by the heuristic maximum parsimony search (equally weighted characters and nucleotide transformations, gaps treated as missing data, 1,000 random-addition replicates, tree bisection-reconnection branch swapping) based on gene sequences (exons plus introns). Many best trees (length 244) were found for the Acc-1 gene (Consistency Index = 0.898, Retention Index = 0.951). Four best trees (length 263) were found for the Pgk-1 gene (Consistency Index = 0.757, Retention Index = 0.724). Parsimony bootstrap analysis followed the same scheme with 1,000 replicates each with 10 random-addition replicates for Pgk-1 and 10,000 replicates in a “fast step-wise addition” search for Acc-1 as implemented by PAUP*. Hordeum vulgare was used as an outgroup for both genes.
Nucleotide substitution rates at synonymous and intron positions were first calculated separately for all pairs of sequences without correction for multiple substitutions and with gaps excluded only from pairwise comparisons. Substitution rates then were calculated for the merged intron plus synonymous sites character sets described above without correction for multiple substitutions and with gaps excluded only from pairwise comparisons. Average substitution rates between major lineages were calculated with SDs for each character set. Substitution rate heterogeneity was assessed for major lineages by the simple relative rate test (36). Divergence times were calculated as described (37) by using the previously estimated divergence time between wheat and barley at 11.4 ± 0.6 MYA which was based on the divergence time between Pooideae and Panicoideae set at 60 MYA (1).
Results
Phylogenetic Trees.
Three different phylogenetic trees of the Triticeae tribe were calculated from the genomic DNA sequences of the Acc-1 and Pgk-1 genes. Neighbor-joining trees were based on either intron sites (Figs. 1 and 2) or on intron plus synonymous sites merged into one character set (not shown). The merged intron plus synonymous character sets were used to enhance tree resolution by including all available variable characters of the two types (see discussion of substitution rate calculations below). Finally, strict consensus maximum parsimony trees were calculated based on all informative characters found in the two genes. The topology of the two types of neighbor-joining trees was identical at all branch points that were well supported by bootstrap analysis (>70%) but differed at some branch points with low statistical support. The same conclusion was reached when the strict consensus maximum parsimony trees were compared with the neighbor-joining trees (Figs. 1 and 2). Several clades representing the major Triticum/Aegilops lineages, such as the A, B, and D genome clades, are well supported, but some of the earlier branch-points are not. Early events, such as the divergence of the diploid Triticum and Aegilops, most likely occurred within a narrow time window, so our analysis could not resolve their sequence.
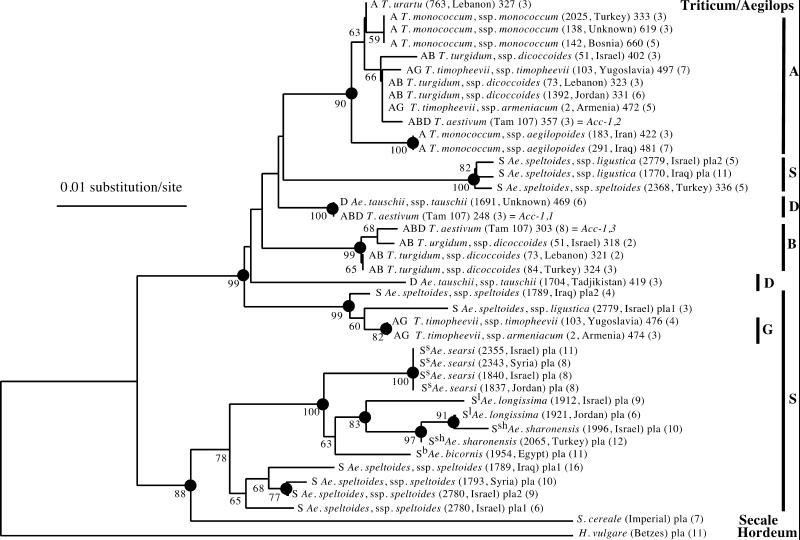
Figure 1.
Phylogenetic tree of the Triticeae/Aegilops complex based on intron sequences of the Acc-1 gene calculated by the neighbor-joining method. Taxon names include genome composition (Triticum/Aegilops species), species name, accession number/cultivar name, geographical origin (Triticum/Aegilops species), sequence name/number and number of clones analyzed. The species accession numbers are the same as in the Wheat Genetic Resource Center (Kansas State University) collection. This information allows identification of the corresponding DNA sequences in GenBank files (AF343496–AF343536). Bootstrap values >50% are shown. Dots indicate nodes supported by bootstrap values >70% and by strict consensus maximum parsimony tree.
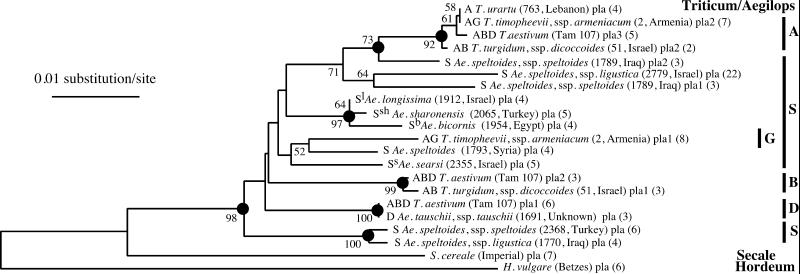
Figure 2.
Phylogenetic tree of the Triticeae/Aegilops complex based on intron sequence of the Pgk-1 gene calculated by the neighbor-joining method. Taxon names include genome composition (Triticum/Aegilops species), species name, accession number/cultivar name, geographical origin (Triticum/Aegilops species), sequence name/number, and number of clones analyzed. The species accession numbers are the same as in the Wheat Genetic Resource Center (Kansas State University) collection. This information allows identification of the corresponding DNA sequences in GenBank files (AF343474–AF343495). Bootstrap values >50% are shown. Dots indicate nodes supported by bootstrap values >70% and by strict consensus maximum parsimony tree.
The Acc-1 and Pgk-1 trees show significant similarities but also some striking differences (Figs. 1 and 2). First, rye is an outgroup for all of the Triticum/Aegilops Pgk-1 genes, but it forms a distinct clade with some Aegilops Acc-1 genes. Second, Aegilops searsi, Aegilops longissima, Aegilops sharonensis, and Aegilops bicornis Acc-1 genes form one well supported clade, whereas Ae. searsi is not in the same clade with the other three Aegilops species in the Pgk-1 tree. Third, the two Ae. speltoides subspecies, speltoides and ligustica, show different associations with other species when Acc-1 and Pgk-1 genes are compared. These issues are discussed below.
Phylogenetic Inferences Based on the Acc-1 and Pgk-1 Gene Sequences.
All Triticum species are present in the same well supported clade and their genes, placed on A, B, D, and G genome branches, each with good statistical support, are approximately equidistant. This observation is true for both Acc-1 and Pgk-1 genes (Figs. 1 and 2). This result suggests that the progenitors of these genomes in modern diploids and polyploids radiated at approximately the same time. The Acc-1 genes from both A genome species, T. urartu and T. monococcum, were analyzed. They all cluster in one clade, as expected, with T. monococcum ssp. aegilopoides being the most distant relative. The two subspecies of T. monococcum, ssp. monococcum and ssp aegilopoides (syn. T. boeoticum), are very similar in their morphology but can be clearly distinguished at the DNA sequence level (Fig. 1).
The A genome Acc-1 gene of T. turgidum (AABB genome), T. timopheevii (AAGG genome) and T. aestivum (AABBDD genome) are most closely related to T. urartu. The close relationship between T. urartu and the A genome in those polyploids also is evident from the Pgk-1 gene comparison (Fig. 2) as well as from a similar analysis of the Acc-2 gene (38). Our results suggest a relatively recent origin of the A genome-containing tetraploids.
Two accessions of Ae. tauschii ssp. tauschii, 1691 (var. meyeri) and 1704 (var. typica), were analyzed. The sequences of the Acc-1 and Pgk-1 genes from 1691 are identical to the sequences of the D genome orthologs in T. aestivum (Figs. 1 and 2). This is also true for the Acc-2 gene, the Pgk-2 gene (38), and the Ψ-Acc-2 pseudogene (1). This result is in agreement with previous studies that showed that var. meyeri is closely related to ssp. strangulata and the D genome of bread wheat (16). The sequence of the Acc-1 gene from 1704 is significantly different (Fig. 1), although the sequences of Acc-2 and Ψ-Acc-2 from 1704 are very similar to those of 1691 and T. aestivum (1, 38).
Ae. speltoides is a member of the sitopsis section which includes also Ae. searsi, Ae. bicornis, Ae. sharonensis, and Ae. longissima. Ae. speltoides was suggested to be the closest living relative of diploid species that contributed the B and G genome to polyploid wheats. Interpretation of the placement of the Aegilops Acc-1 and Pgk-1 genes on the phylogenetic trees is complicated. First, some of the analyzed Ae. speltoides accessions have two copies of the genes. Second, relationships among these genes are different depending on which gene (Acc-1 or Pgk-1) and which copy of the duplicated genes is analyzed (Figs. 1 and 2). One significant difference is that rye is an outgroup for Pgk-1 from all Triticum and Aegilops species, but for the Acc-1 gene, it is not. In the latter case, some Aegilops species (Ae. searsi, longissima, sharonensis, bicornis, and some Ae. speltoides ssp. speltoides) seem to be more closely related to rye than to Triticum and to some other Aegilops species (Ae. tauschii and Ae. speltoides ssp. ligustica). Barley, as expected, is an outgroup for all Acc-1 and Pgk-1 genes from Triticum and Aegilops species and for rye.
In agreement with the taxonomy, Acc-1 genes from Ae. speltoides ssp. speltoides (accession no. 2368; a single copy) and Ae. speltoides ssp. ligustica (accession 1770, a single copy, and accession 2779, one of two copies) are closely related (Fig. 1). The same is true for Pgk-1 genes from 2368 and 1770 (Fig. 2). Furthermore, their position on the phylogenetic tree relative to both the Triticum species as well as to rye is very similar for both genes. However, the similarities end there. The second copy of the Ae. speltoides ssp. ligustica 2779 Acc-1 gene is similar to a copy of the gene from Ae. speltoides ssp. speltoides 1789, and they are both similar to the G genome Acc-1 gene from AAGG tetraploids (Fig. 1). All these genes are found in the Triticum clade. The second copy of the Ae. speltoides ssp. speltoides 1789 Acc-1 gene is similar to the gene in Ae. speltoides ssp. speltoides 1793 (a single copy) and to two copies of the gene from Ae. speltoides ssp. speltoides 2780. All these genes are found in the rye clade together with genes from the other five Sitopsis species (Fig. 1).
One of the two Pgk-1 gene copies from Ae. speltoides ssp. speltoides 1789 is a chimera of unknown origin with some similarity to the A genome genes. Both copies of the gene from 1789 are significantly different from the Pgk-1 gene from Ae. speltoides ssp. speltoides 1793, and all three are significantly different from genes found in Ae. speltoides ssp. speltoides 2368 and Ae. speltoides ssp. ligustica 1770. The similarity of Pgk-1 in 2368 and 1770 agrees with the subspecies classification of these two accessions. Acc-1 genes of 2368 and 1770 are also very similar. The other Pgk-1 genes from speltoides place distantly on the tree, suggesting a different origin.
The Acc-1 and Pgk-1 genes in Ae. bicornis, Ae. Sharonensis, and Ae. longissima are found in one clade (Figs. 1 and 2), although the four Acc-1 sequences from sharonensis and longissima are intertwined. Acc-1 sequences from four Ae. searsi accessions are identical to each other and similar to sequences from bicornis, sharonensis, and longissima (Fig. 1). The sequence of the Pgk-1 gene from only one Ae. searsi accession was analyzed, but it is also the least closely related gene of the four Aegilops species (Fig. 2). This result is in agreement with earlier suggestions based on other methods. Sharonensis and longissima are very similar in morphology, which is a possible explanation for the structure of their clade, but they are clearly distinguishable at the DNA sequence level. Finally, the Acc-1 and Pgk-1 genes from the Aegilops species analyzed in this study show no significant similarity to the B genome.
Nucleotide Substitution Rates and Divergence Times.
Nucleotide substitution rates were calculated by using the merged intron and synonymous character sets. As a result, these rates are average rates for intron and synonymous sites where the synonymous rates have a lesser weight because of the lower number of such sites in the merged character set. This approach was justified by very similar nucleotide substitution rates observed for introns and synonymous sites in the Acc and Pgk genes in Triticeae (1). The same rate similarity, with only a few exceptions, was observed for the major Triticum/Aegilops lineages. The average ratio of the nucleotide substitution rates in introns and at synonymous sites, calculated from average rates between the major lineages, was 1.08 ± 0.55 and 0.76 ± 0.26 for the Acc-1 and Pgk-1 gene, respectively. The variation in the ratio can be explained, in part, by a high variation of the synonymous rates because of too few nucleotide changes at synonymous sites being counted. This explanation is especially true for the Acc-1 gene, where some of the calculations were based on fewer than five changes. These pairwise rates were used to calculate average substitution rates between the major Triticum and Aegilops lineages (see the supporting information, which is published on the PNAS web site, www.pnas.org). Standard deviations were less than 10%, except for four pairs of sequences including sequences from the Ae. sharonensis/Ae. longissima/Ae. bicornis clade. The corresponding rates between all these lineages also were calculated separately for intron and synonymous sites and then averaged (supporting information). In this average, both intron and synonymous rates have the same weight. In most cases the two types of averages gave very similar results.
The divergence times between major Triticum and Aegilops lineages (supporting information) were estimated from the substitution rates calculated as described above by using a molecular clock model calibrated with the 11.4 ± 0.6 MYA divergence time between barley and the Triticum/Aegilops lineage (1). The error for such estimates is ≈30%, primarily because of the error of the fossil-based estimate of the divergence time of the Pooideae and Panicoideae subfamilies of grasses (50–80 MYA) used to set the clock.
Our system has inherent limitations for species which are too distantly or too closely related. For more divergent species, at and above the tribe level, reliable alignment of intron sequences becomes a problem (1, 38). Exon sequences could then be used to address some of the questions by the analysis of nucleotide substitution rates at synonymous positions, providing a time window between 10 and over 100 MYA (1). The method also fails for very closely related species or for different accessions of the same species or populations. There are not enough substitutions to count, even in introns.
The merged intron plus synonymous character sets provided better resolution and consistency for the more closely related species. On average, 4.8 and 4.5 nucleotide changes per million years were counted in Acc-1 and Pgk-1 introns (Table 1), respectively. Synonymous sites (Table 1) added approximately one change per million years to that count for Acc-1 and three changes for Pgk-1. This gain for the Pgk-1 gene is significant. Calculation of the divergence times based on all sites in introns and synonymous sites in exons combined allows analysis within a 0.5–20 MYA window. Estimates of the divergence time between rye and Triticeae illustrates well the outcome of the two averaging methods described above. The divergence time based on the merged intron plus synonymous character set was 7.4 ± 0.9 MYA. This result is the average of two estimates, one for Acc-1 and one for Pgk-1. The corresponding estimate based on intron and exon sites, calculated separately, was 7.6 ± 1.8 MYA. This result is the average of four estimates: two for Acc-1 and two for Pgk-1. Both of these estimates are similar to the 7.2 ± 1.6 MYA calculated previously (1). As we already noted (1), the rye case is rather extreme in its high nucleotide substitution rate variability between introns and synonymous sites and between these rates calculated for Acc-1 and Pgk-1 genes.
Substitution rate heterogeneity was assessed for the major lineages by the simple relative rate test (36). In this test, the relative rate equals 1 for two lineages evolving at the same rate. The relative rate calculated for Pgk-1 and Acc-1 varied within the range of 0.6 to 2.0. This finding is illustrated in Table 2 for the B genome. With the exception of the Acc-1 relative rates for Ae. speltoides ssp. ligustica, these differences are two-fold or less, indicating a rather low-rate heterogeneity in the Acc-1 genes and an even lower rate in the Pgk-1 genes. Some of these apparent rate heterogeneities may be the results of horizontal transfer of genome fragments between species.
Table 2.
Relative nucleotide substitution rates comparing the B genome and other major Triticum/Aegilops genomes
| Genome | A | D | G | S (ssp. speltoides) | S (ssp. ligustica) | Sl/Ssh/Sb | Ss |
|---|---|---|---|---|---|---|---|
| Acc-1 | 1.23 (1.04) | 0.59 (0.55) | 1.03 (0.97) | 0.76 (0.80) | 2.01 (2.02) | 1.01 (1.09) | 1.06 (1.14) |
| Pgk-1 | 1.18 (0.97) | 0.93 (0.90) | 1.04 (0.92) | 0.57 (0.69) | 1.03 (0.98) | 0.65 (0.68) | 0.66 (0.65) |
Our analysis is based on the assumption that different parts of the gene have the same evolutionary history. However, it is important to identify all those genes in which a single intron shows a significantly higher number of mutations compared with other introns in the same gene as well as those genes in which one part is much more similar to another gene than another part. The first case could represent a local accumulation of changes caused by events other than neutral drift; for example, changes that are a result of a recent transposition. The second case could indicate recombination events. An increased substitution rate in single introns was observed for some of the Acc-1 and Pgk-1 genes (data not shown). However, because of the low number of nucleotide changes found in individual introns, it was impossible to draw firm conclusions based on such single intron comparisons. Our analysis also indicated possible recombination. For example, the 5′-half of one of the Ae. speltoides ssp. speltoides Pgk-1 genes (pla2, Fig. 2) is very similar to genes in the A clade, whereas the 3′-half is divergent. As a result, this gene shows a greater affinity to the A clade than to the other Aegilops Pgk-1 genes. The origin of this possibly chimeric gene is not known. This was a clear-cut example. Detecting such chimeric molecules from closely related species is hampered by the small number of nucleotide changes. These observations underscore the importance of multigene and multitaxa analysis. Furthermore, multiple introns were analyzed as one set of characters to minimize the effect of such phenomena on the outcome of the analysis.
The information content of our data set is robust, allowing comparisons and error estimates at several different levels. First, individual pairwise distances of each type were averaged and, in most cases, evaluated by calculating SDs (supporting information), taking into account information provided by the analysis of phylogenetic trees derived from genomic sequences of the two genes (Figs. 1 and 2). Second, heterogeneity of nucleotide substitution rates in different lineages was probed (Table 2). Third, divergence times estimated for different major lineages were compared with the deduced time-line of some evolutionary events. Two different methods were used to calculate divergence times. Finally, divergence times estimated for different genes were compared, to provide a time window for each event or to indicate the possibility of a different gene history in some species (supporting information).
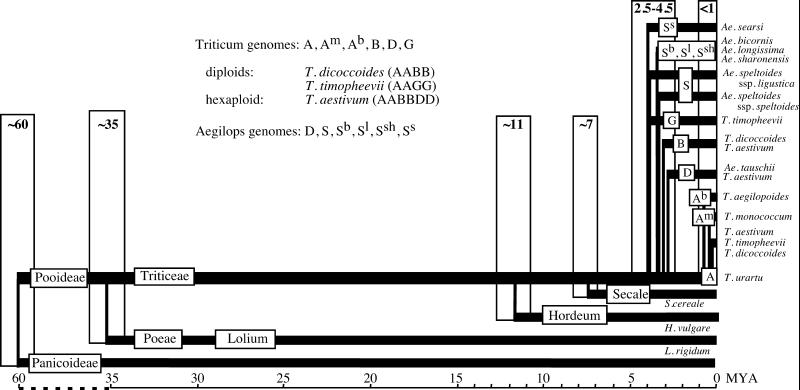
The diploid Triticum/Aegilops progenitors of the A, B, D, and G genomes in diploid, tetraploid, and hexaploid wheats all radiated at approximately the same time, 2.5–4.5 MYA (supporting information). These estimates are based on average values for the Acc-1 and Pgk-1 genes, which, in most cases, were very similar to each other. However, the divergence times between Triticum species (A, B, D, and G genomes) and the Ae. searsi and Ae. sharonensis/Ae. longissima/Ae. bicornis lineages is consistently lower for the Pgk-1 gene than for the Acc-1 gene. This ≈2-fold difference suggests a different evolutionary history for the two loci in different lineages. The A genome species radiated more recently, 0.5–1 MYA, and wheat tetraploids appeared less than 0.5 MYA.
Discussion
Although a significant amount of information is already available, many aspects of wheat evolution remain unknown or require independent verification. A gene sequence comparison-based approach seems suitable for the task, based on the assumption that analysis of a sufficiently large fragment of DNA will yield enough variable characters for the phylogenetic analysis. The success of this approach depends on proper selection and understanding of the origin, evolution, structure, and function of the genes being analyzed. Gene-specific and chromosome locus-specific effects on the nucleotide substitution rates, gene duplications and introgression, assignment of individual genes to genomes in polyploid species, and establishing orthologous relationships between them are some of the key issues. Corroborating evidence from two independent data sets for genes with different characteristics is essential.
We recently established a two-gene system to study grass evolution based on nuclear genes encoding the plastid multidomain (eukaryotic-type) ACCase and the plastid (prokaryotic-type) PGK. In previous papers, we reported the structure of wheat ACCases and their genes, gene copy number, and chromosome localization (38–41). The origin and evolution of the multidomain ACCase gene family in grasses is well understood (1). Contrary to the multidomain plastid ACCase, which was created by duplication of an ancestral cytosolic gene, the cytosolic PGK arose by duplication of the PGK gene of endosymbiont origin (42). Phylogenetic analysis of the PGK gene family in plants was presented earlier (1). Acc-1 and Pgk-1 are predominantly single-copy genes in grasses, allowing establishment of orthologous relationships among them. Exon sequences reveal relationships above the tribe level, as reported earlier, whereas intron sequences allow phylogenetic analysis below the tribe level, as demonstrated in this paper. Nucleotide substitution rates in intron sites and at synonymous positions also were analyzed on a relative basis to allow comparisons of different genes and, by using the molecular clock concept, to express the sequence of evolutionary events in historical time.
The information on Acc-1 and Pgk-1 gene families allowed us to revisit the question of phylogenetic relationships among wheat and its relatives. Our results agree with some well established facts: the close relatedness of T. monococcum (AmAm genome) and T. urartu (AA genome), T. urartu being the donor of the A genome to wheat tetraploids; T. turgidum (AABB genome) and T. timopheevii (AAGG genome) as well as Ae. taushii being the donor of the D genome to the T. aestivum (AABBDD genome). The latter event occurred only 8,000 years ago, making the D genome copies of the Acc-1 and Pgk-1 genes in the diploid and hexaploid species indistinguishable. We showed that the tetraploidization events occurred very recently (probably less then 0.5 MYA) relative to the radiation of the major diploid species, which shaped the structure of the Triticum and Aegilops complex of species 2.5–4.5 MYA. These results, together with our earlier estimates of the divergence time of Poeae, Hordeum and Secale, provide the timeline of wheat evolution illustrated in Fig. 3 for the T. urartu lineage.
Figure 3.
Timeline of the evolution of Triticum and Aegilops lineages. The estimates of the divergence times (in MYA) are based on the data presented in the supporting information.
Three points need to be stressed with respect to the timeline estimates. First, the resolution of the DNA sequence analysis is not sufficient to determine the order of events that occurred less than one million years apart. Second, the molecular clock in our calculations is based on the divergence time of the grass subfamilies of 60 MYA, a value in the middle of the rather broad range of 50–80 MYA available from estimates based on fossil records. This range is a significant source of error. Furthermore, fossil records tend to underestimate the divergence times. Nevertheless, our timeline of wheat evolution puts the process in historical perspective and allows direct comparisons with the conclusions of other such studies. Finally, the divergence time estimates presented in this paper are based on a clock model in which the nucleotide substitution rate is the same for all branches. Our analysis suggests that this may not always be the case. To take into account one of the possible sources of error—rate variation among different lineages—different methods need to be used (43).
It has been suggested that Ae. speltoides (SS genome) contributed to the genetic makeup of tetraploid wheats. Our evidence suggests that some loci of the S genome of Ae. speltoides found in Iraq and Syria and the G genome of T. timopheevii are closely related, leading to the conclusion that parts of their genomes may have the same origin and were donated relatively recently by an Ae. speltoides progenitor. This evidence agrees with results of other studies implicating Ae. speltoides as the donor of the G genome. On the other hand, none of the loci of the Aegilops genome analyzed in our study seems to be a close relative of the B genome, so the diploid progenitor (or progenitors) of the B genome remains unknown. Our data also suggest that Acc-1 and Pgk-1 loci have different histories in different lineages, indicating genome mosaicity caused by an exchange of genetic material during the evolution of Triticeae. The analysis of Ae. speltoides was further complicated by an apparent duplication/heterogeneity of the Acc-1 and Pgk-1 genes in some accessions. The latter event also could be the result of a recent hybridization involving distinct lineages of speltoides because it is an outcrossing species. Such genome mosaicity could affect taxonomic assignments at both the species and subspecies level.
Supplementary Material
Acknowledgments
We thank Michael Clegg and Manyuan Long for helpful comments on the manuscript. This work was supported in part by gifts from Monsanto Corporation and the Harris and Frances Block Fund at the University of Chicago. DNA sequencing was performed by the University of Chicago Cancer Center Sequencing Facility.
Abbreviations
- ACCase
acetyl-CoA carboxylase
- PGK
3-phosphoglycerate kinase
- MYA
million years ago
References
- 1.Huang S, Sirikhachornkit A, Su X, Faris J, Gill B, Haselkorn R, Gornicki P. Plant Mol Biol. 2002;48:805–820. doi: 10.1023/a:1014868320552. [DOI] [PubMed] [Google Scholar]
- 2.West J G, McIntyre C L, Appels R. Plant Syst Evol. 1988;160:1–28. [Google Scholar]
- 3.Cox T S. J Crop Prod. 1998;1:1–25. [Google Scholar]
- 4.Johnson B L, Dhaliwal H S. Am J Bot. 1976;63:1088–1094. [Google Scholar]
- 5.Sax K. Genetics. 1922;7:513–522. doi: 10.1093/genetics/7.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kihara H. Mem Coll Sci Univ Kyoto Ser B. 1924;1:1–200. [Google Scholar]
- 7.Lilienfeld F A, Kihara H. Cytologia. 1934;6:87–122. [Google Scholar]
- 8.Dvorak J, DiTerlizzi P, Zhang H-B, Resta P. Genome. 1993;36:21–31. doi: 10.1139/g93-004. [DOI] [PubMed] [Google Scholar]
- 9.Dvorak J, McGuire P E, Cassidy B. Genome. 1988;30:680–689. [Google Scholar]
- 10.Nishikawa K. In: Proceedings of the 6th International Wheat Genetics Symposium. Sakamoto S, editor. Kyoto, Japan: Kyoto University; 1983. pp. 59–63. [Google Scholar]
- 11.Takumi S, Nasuda S, Liu Y G, Tsunewaki K. Jpn J Genet. 1993;68:73–79. [Google Scholar]
- 12.Kihara H. Ag Hort (Tokyo) 1944;19:889–890. [Google Scholar]
- 13.McFadden E S, Sears E R. J Hered. 1946;37:81–89. doi: 10.1093/oxfordjournals.jhered.a105590. [DOI] [PubMed] [Google Scholar]
- 14.Dvorak J, Luo M-C, Yang Z-L, Zhang H-B. Theor Appl Genet. 1998;97:657–670. [Google Scholar]
- 15.Jaaska V. Plant Syst Evol. 1980;137:259–273. [Google Scholar]
- 16.Lubbers E L, Gill K S, Cox T S, Gill B S. Genome. 1991;34:354–361. [Google Scholar]
- 17.Upadhya M D, Swaminathan M S. Chromosoma. 1963;14:589–600. [Google Scholar]
- 18.Riley R, Unrau J, Chapman V. J Hered. 1958;49:91–98. [Google Scholar]
- 19.Friebe B, Badaeva E D, Gill B S. Plant Syst Evol. 1996;202:199–210. [Google Scholar]
- 20.Kerby K, Kuspira J. Genome. 1988;30:36–43. doi: 10.1139/g88-097. [DOI] [PubMed] [Google Scholar]
- 21.Johnson B L. Proc Natl Acad Sci USA. 1972;69:1398–1402. doi: 10.1073/pnas.69.6.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talbert L E, Blake N K, Storlie E W, Lavin M. Genome. 1995;38:951–957. doi: 10.1139/g95-125. [DOI] [PubMed] [Google Scholar]
- 23.Sasanuma T, Miyashita K T. Theor Appl Genet. 1996;92:928–934. doi: 10.1007/BF00224032. [DOI] [PubMed] [Google Scholar]
- 24.Pestsova E G, Goncharov N P, Salina E A. Theor Appl Genet. 1998;97:1380–1386. [Google Scholar]
- 25.Daud H M, Gustafson J P. Genome. 1996;39:543–548. doi: 10.1139/g96-069. [DOI] [PubMed] [Google Scholar]
- 26.Dvorak J, Zhang H B. Proc Natl Acad Sci USA. 1990;87:9640–9644. doi: 10.1073/pnas.87.24.9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blake N K, Lehfeldt B R, Laven M, Talbert L E. Genome. 1999;42:351–360. [PubMed] [Google Scholar]
- 28.Tsunewaki K, Ogihara Y. Genetics. 1983;104:155–171. doi: 10.1093/genetics/104.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsunewaki K. In: Nuclear and Organelle Genomes of Wheat Species. Sasakuma T, Kinoshita T, editors. Yokohama, Japan: Kihara Memorial Foundation; 1991. pp. 16–28. [Google Scholar]
- 30.Zohary D, Feldman M. Evolution (Lawrence, Kans) 1962;16:44–61. [Google Scholar]
- 31.Dvorak J, Appels R. Theor Appl Genet. 1982;63:349–360. doi: 10.1007/BF00303906. [DOI] [PubMed] [Google Scholar]
- 32.Feldman M. Can J Genet Cytol. 1966;8:144–151. [Google Scholar]
- 33.Dvorak J, Zhang H-B, Kota R S, Lassner M. Genome. 1989;32:1003–1016. [Google Scholar]
- 34.Wang G-Z, Miyashita N T, Tsunewaki K. Proc Natl Acad Sci USA. 1997;94:14570–14577. doi: 10.1073/pnas.94.26.14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li P, Bousquet J. Mol Biol Evol. 1992;9:1185–1189. doi: 10.1093/oxfordjournals.molbev.a040779. [DOI] [PubMed] [Google Scholar]
- 37.Gaut B S. Evol Biol. 1998;30:93–120. [Google Scholar]
- 38.Faris J, Sirikhachornkit A, Haselkorn R, Gill B, Gornicki P. Mol Biol Evol. 2001;18:1720–1733. doi: 10.1093/oxfordjournals.molbev.a003960. [DOI] [PubMed] [Google Scholar]
- 39.Gornicki P, Podkowinski J, Scappino L A, DiMaio J, Ward E, Haselkorn R. Proc Natl Acad Sci USA. 1994;91:6860–6864. doi: 10.1073/pnas.91.15.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gornicki P, Faris J, King I, Podkowinski J, Gill B, Haselkorn R. Proc Natl Acad Sci USA. 1997;94:14179–14185. doi: 10.1073/pnas.94.25.14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Podkowinski J, Sroga G E, Haselkorn R, Gornicki P. Proc Natl Acad Sci USA. 1996;93:1870–1874. doi: 10.1073/pnas.93.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin W, Schnarrenberger C. Curr Genet. 1997;32:1–18. doi: 10.1007/s002940050241. [DOI] [PubMed] [Google Scholar]
- 43.Sanderson M J. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.