Abstract
The strict orthology of mitochondrial (mt) coding sequences has promoted their use in phylogenetic analyses at different levels. Here we present the results of a mitogenomic study (i.e., analysis based on the set of protein-coding genes from complete mt genomes) of 60 mammalian species. This number includes 11 new mt genomes. The sampling comprises all but one of the traditional eutherian orders. The previously unrepresented order Dermoptera (flying lemurs) fell within Primates as the sister group of Anthropoidea, making Primates paraphyletic. This relationship was strongly supported. Lipotyphla (“insectivores”) split into three distinct lineages: Erinaceomorpha, Tenrecomorpha, and Soricomorpha. Erinaceomorpha was the basal eutherian lineage. Sirenia (dugong) and Macroscelidea (elephant shrew) fell within the African clade. Pholidota (pangolin) joined the Cetferungulata as the sister group of Carnivora. The analyses identified monophyletic Pinnipedia with Otariidae (sea lions, fur seals) and Odobenidae (walruses) as sister groups to the exclusion of Phocidae (true seals).
Keywords: Dermosimii‖Eutheria‖Mammalia‖phylogeny‖primate paraphyly
Mitogenomic (mtg) phylogenetics has contributed considerably to resolving evolutionary relationships among mammals. However, relatively few genomes have been sequenced for some orders and others are still unrepresented. The first eutherian mtg study (1) included five orders: Rodentia, Primates, Artiodactyla, Cetacea, and Carnivora. This study identified a sister group relationship between Artiodactyla and Cetacea and close affinities between these two orders and Carnivora. Because of the absence of an unequivocal outgroup (OG), the relationships relative to Primates and Rodentia could not be resolved, however. The first mtg rooting of the eutherian tree (2), using a marsupial as OG, reconstructed the relationship OG(Rodentia,(Primates,(Carnivora,(Artiodactyla,Cetacea)))), a topology that has been generally identified in subsequent mtg analyses. These two studies also showed that individual mitochondrial (mt) genes did not obligatorily reconstruct the same topology, underlining the necessity of using the concatenated sequences of different genes for maximizing the reliability of the analyses.
Most taxonomic schemes recognize 18 orders of extant eutherians (Table 1). It is likely, however, that this number is an underestimate because most molecular studies, both mtg (3, 4) and mt/nuclear (5–7), split Lipotyphla into separate lineages. Similarly, if Rodentia is nonmonophyletic (8–11), the number of eutherian orders may be still greater than suggested by only lipotyphlan polyphyly.
Table 1.
Mammalian taxa analyzed
| Monotremata, monotremes |
| Ornithorhynchus anatinus (platypus, X83427) |
| Tachyglossus aculeatus (echidna, AJ303116) |
| Marsupialia, marsupials |
| Didelphis virginiana (opossum, Z29573) |
| Macropus robustus (wallaroo, Y10524) |
| Vombatus ursinus (wombat, AJ304828) |
| Trichosurus vulpecula (brushtailed possum, AF357238) |
| Isoodon macrourus (bandicoot, AF358864) |
| Eutheria |
| Erinaceomorpha, hedgehogs, moon rats |
| Erinaceus europaeus (hedgehog, X88898) |
| Echinosorex gymnurus (moon rat, AF348079) |
| Rodentia, rodents |
| Rattus norvegicus (brown rat, AJ428514) |
| Mus musculus (mouse, J01420) |
| Cavia porcellus (guinea pig, AJ222767) |
| Thryonomys swinderianus (cane rat, AJ301644) |
| Glis glis (fat dormouse, Y11137) |
| Sciurus vulgaris (squirrel, AJ238588) |
| Volemys kikuchii (Taiwan vole, AF348082) |
| Primates, lemurs, lorises, tarsiers, monkeys, apes |
| Nycticebus coucang (slow loris, AJ309867) |
| Lemur catta (ring-tailed lemur, AJ421451) |
| Tarsius bancanus (tarsier, AF348159) |
| Cebus albifrons (pale-fronted capuchin, AJ309866) |
| Papio hamadryas (hamadryas baboon, Y18001) |
| Hylobates lar (gibbon, X99256) |
| Pongo abelii (Sumatran orangutan, X97707) |
| Gorilla gorilla (gorilla, X93347) |
| Homo sapiens (human, X93334) |
| Dermoptera, flying lemurs |
| Cynocephalus variegatus (flying lemur, AJ428849) |
| Lagomorpha, pikas, hares, rabbits |
| Ochotona collaris (pika, AF348080) |
| Oryctolagus cuniculus (rabbit, AJ001588) |
| Lepus europaeus (brown hare, AJ421471) |
| Scandentia, tree shrews |
| Tupaia belangeri (tree shrew, AJ421453, AF217811) |
| Chiroptera, bats |
| Artibeus jamaicensis (Jamaican fruit bat, AF061340) |
| Chalinolobus tuberculatus (NZ long-tailed bat, AF321051) |
| Pteropus scapulatus (little red flying fox, AF321050) |
| Soricomorpha, moles, shrews |
| Talpa europaea (European mole, Y19192) |
| Soriculus fumidus (Asiatic shrew, AF348081) |
| Pholidota, pangolins |
| Manis tetradactyla (long-tailed pangolin, AJ421454) |
| Carnivora, carnivores |
| Felis catus (cat, U20753) |
| Canis familiaris (dog, U96639) |
| Ursus maritimus (polar bear, AJ428577) |
| Odobenus rosmarus (walrus, AJ428576) |
| Eumetopias jubatus (northern sea lion, AJ428578) |
| Halichoerus grypus (grey seal, X72004) |
| Perissodactyla, horses, tapirs, rhinoceroses |
| Equus caballus (horse, X79547) |
| Equus asinus (donkey, X97337) |
| Rhinoceros unicornis (Indian rhinoceros, X97336) |
| Ceratotherium simum (white rhinoceros, Y07726) |
| Artiodactyla, pigs, camels, ruminants, hippopotamuses |
| Bos taurus (cow, J01394) |
| Ovis aries (sheep, AF010406) |
| Sus scrofa (pig, AJ002189) |
| Lama pacos (alpaca, Y19184) |
| Hippopotamus amphibius (hippopotamus, AJ010957) |
| Cetacea, whales, dolphins, porpoises |
| Balaenoptera physalus (fin whale, X61145) |
| Physeter macrocephalus (sperm whale, AJ277029) |
| Xenarthra, sloths, armadillos, anteaters |
| Dasypus novemcinctus (nine banded armadillo, Y11832) |
| Tamandua tetradactyla (lesser anteater, AJ421450) |
| Macroscelidea, elephant shrews |
| Macroscelides proboscideus (elephant shrew, AJ421452) |
| Proboscidea, elephants |
| Loxodonta africana (African elephant, AJ224821) |
| Sirenia, dugong, manatees |
| Dugong dugon (dugong, AJ421723) |
| Tubulidentata, aardvark |
| Orycteropus afer (aardvark, Y18475) |
| Tenrecomorpha, tenrecs, golden moles |
| Echinops telfairi (lesser hedgehog tenrec, AJ400734) |
| (Hyracoidea, hyraxes, not represented as yet) |
Accession nos. of new mtDNAs are shown in bold. The sequence of the hedgehog has been corrected compared to the original submission. The sequence of the brown rat is from a wild-caught animal. Erinaceomorpha, Tenrecomorpha, and Soricomorpha are traditionally included in the Lipotyphla.
Five eutherian orders are previously not represented by complete mtDNAs. To further complete the picture of eutherian mtg relationships we have added 11 complete mtDNAs to the eutherian data set, including four of these orders: Pholidota, Dermoptera, Sirenia, and Macroscelidea.
The phylogenetic position of Pholidota has been a matter of debate. A sister group relationship between Xenarthra and Pholidota in a basal position in the eutherian tree has been proposed (e.g., ref. 12). However, other authors (13) have challenged this proposal. Most morphological studies place Xenarthra at or close to the base of the eutherian tree and the term Epitheria has been coined for all eutherians except Xenarthra (14) or, alternatively, all eutherians except Xenarthra and Pholidota. Thus, the positions of Xenarthra and Pholidota are fundamental to the discussion of eutherian evolution. Xenarthra is currently represented by a single mtDNA, that of the armadillo. To examine the position of Xenarthra on the basis of more comprehensive sequence data we here add the mt genome of the lesser anteater to the mtg data set.
Also, the phylogenetic position of Macroscelidea has been contentious. For example, Simpson (15) joined Macroscelidea and Lipotyphla as sister groups in “Insectivora.” Other morphological proposals have joined Macroscelidea, Lagomorpha, and Rodentia on a common branch, a view endorsed by McKenna and Bell (16), who included this grouping in the Anagalida along with some extinct orders.
The morphological affinities between Proboscidea and Sirenia are well documented (e.g., ref. 17). The mt genome of the dugong allows firmer establishment of the position of Sirenia than was possible in a previous cyt b study (18).
The grouping of Primates, Dermoptera, Scandentia (tree shrews), and Chiroptera (bats) into the superordinal clade Archonta has been favored by morphologists (16). To examine the relationships between Primates and their presumed closest relatives we have added the flying lemur to the mtg sampling. The order Primates includes three basal lineages, Prosimii, Tarsioidea, and Anthropoidea. Anthropoidea is well represented by mtg data, but only one prosimian mt genome (Nycticebus coucang) has been described (19). The addition of the ring-tailed lemur to the data set splits the prosimian branch, allowing extended study of basal primate relationships in conjunction with the recent release of a tarsier sequence.
We add also the mt genomes of the brown hare (Lagomorpha), the tree shrew, the polar bear, the northern sea lion, and the walrus. Lagomorpha is currently represented by the mt genomes of the rabbit and the pika. The brown hare completes the sampling, providing additional data for analysis of the Glires hypothesis, which posits a sister group relationship between Rodentia and Lagomorpha. The phylogenetic position of the Scandentia has been studied (20) but the current taxon sampling has allowed further analysis of its position.
Pinniped relationships are of a particular interest because of the distinct difference between molecular results and recent morphological views (e.g., ref. 21) that posit a sister group relationship between Phocidae and Odobenidae to the exclusion of Otariidae. However, this proposal is inconsistent with chromosomal data (22) and analyses of cyt b and 12S rRNA sequences (23–25). The mtDNAs of the walrus, sea lion, and polar bear allow a firmer analysis of pinniped relationships than was previously possible.
Materials and Methods
Table 1 lists the 60 mammalian species studied. The mt genomes of the anteater, the flying lemur and the dugong were PCR-amplified with a series of pairs of conserved mt primers. mtDNA was prepared from frozen tissues from a single individual of the other species following described procedures (26). The brown hare mt genome was cloned as a single EcoRI fragment in pUC19. The new mt genomes conform to the organization of other eutherian mtDNAs. The control regions of all 11 species contain tandemly organized repeat motifs. The motifs of the new tree shrew mt genome differ somewhat from the described mtDNA (20), but only minor differences occur in the coding regions of the two mtDNAs.
The phylogenetic analyses were carried out on the concatenated amino acid and nucleotide (first plus second codon positions) sequences of the 12 heavy-strand encoded protein-coding genes. The light-strand encoded NADH6 gene was not included as it deviates markedly in nucleotide and amino acid composition from the other genes. The length of the alignment was 9,882 nucleotides, 3,294 amino acids, after removal of gaps and ambiguous sites adjacent to gaps. Analytical methods, as implemented in the tree-puzzle (27), phylip (28), molphy (29), paup (30), and pal/vanilla (31) packages were used to analyze phylogenetic relationships. The mtREV-24 model of amino acid sequence evolution (29) and the TN-93 model of nucleotide evolution (32) were used for distance and likelihood analyses. Parameter estimation was according to the software, using the nucleotide/amino acid frequencies of the data set. For nucleotide, the transition/transversion parameter was estimated to 1.7 and the pyrimidine/purine parameter to 1.4. The analyses were performed under the assumptions of both rate homogeneity and rate heterogeneity among sites, the latter with a Γ model plus four classes of variable sites (RH4, Table 3). The rate heterogeneity parameter alpha was 0.7 for amino acid and 0.16 for nucleotide sequences. SH test (33), likelihood values, SDs, and the number of substitutions and their SDs (34) were used for comparison of alternative trees relative to the best maximum likelihood (ML) tree. A χ2 test for compositional homogeneity as implemented in the tree-puzzle program was used to test for stationarity of nucleotide/amino acid composition.
Table 3.
Support for alternative topologies from amino acid sequences
| SHRH4 | ΔlnLRH4 | Steps | |
|---|---|---|---|
| (Derm,((Pro,Tars),Anth)) | 0.046 | −32.8 ± 17.7 | +38 ± 12 |
| (Anth,(Derm,(Pro,Tars))) | 0.045 | −32.1 ± 17.1 | +46 ± 12 |
| Pangolin on branch J | 0.231 | −13.4 ± 20.6 | +13 ± 12 |
| (sea lion,(seal,walrus)) | 0.004 | −56.9 ± 19.3 | +34 ± 9 |
| Cet sister group to Art | 0.004 | −65.4 ± 22.4 | +38 ± 10 |
| Glires on branch B | 0.010 | −63.6 ± 25.7 | +54 ± 15 |
| on branch D | 0.150 | −31.2 ± 29.8 | +52 ± 15 |
| Topology as in ref. 7 | 0.002 | −74.0 ± 34.0 | +92 ± 23 |
| Root at African clade | 0.110 | −35.5 ± 28.0 | +62 ± 15 |
The probability values according to the SH test for alternative relationships relative to the best likelihood tree based on amino acid sequences (Fig. 1) are shown next to the differences in log-likelihood values, the number of amino acid substitutions (steps), and their SDs. Anth, Anthropoidea; Art, Artiodactyla; Cet, Cetacea; Derm, Dermoptera; Pro, Prosimii; Tars, Tarsioidea.
Nomenclature.
(i) Dermosimii.
The joining of Dermoptera/Anthropoidea, Prosimii, and Tarsioidea on a common branch is the best phylogenetic alternative of the current study. We suggest that the ordinal name Primates be maintained as including these three basal lineages. Dermosimii acknowledges the contribution of both dermopterans and primates to the Dermoptera/Anthropoidea grouping (derma = Greek for skin; −simii = Latin, plural for apes or monkeys).
(ii) Erinaceomorpha.
The analyses united Erinaceus and Echinosorex on a common branch, separate from other lipotyphlans, thus forming the order Erinaceomorpha.
(iii) Soricomorpha.
A morphological relationship between Talpidae and Soricidae has been suggested (35).
(iv) Tenrecomorpha.
In accordance with the morphological grouping (36) of Tenrecidae and Chrysochloridae.
(v) Cetartiodactyla.
(37) = (Cetacea + Artiodactyla).
(vi) Cetancodonta.
(38) = (Cetacea + Hippopotamidae).
(vii) Ferae.
(16, 39) = (Carnivora + Pholidota).
(viii) Cetferungulata.
The grouping ((Artiodactyla/Cetacea)(Carnivora/Perissodactyla)) (40) has been given the name Cetferungulata (41). We suggest that this definition be extended to include Pholidota as well.
(ix) African clade.
The morphological similarities between some of these orders and their African distribution were discussed by Le Gros Clark and Sonntag (42).
Results and Discussion
The Positions of the New mtg Taxa.
The study includes four orders, Dermoptera, Sirenia, Macroscelidea, and Pholidota, which, hitherto, have not been represented by complete mtDNAs.
The ML analysis (both amino acid and nucleotide) grouped Dermoptera and Primates on a common branch with Dermoptera falling within Primates as the sister group of Anthropoidea, making Primates paraphyletic. The Dermoptera/Anthropoidea relationship is strongly supported (Table 2) and the alternative relationship, a sister group relationship between monophyletic Primates and Dermoptera, is statistically refuted (Table 3). There are morphological arguments for placing Primates and Dermoptera as sister groups. Beard (43) has proposed a close relationship between Dermoptera and the extinct families Micromomyidae, Plesiadapidae, and Paromomoyidae, which some other authors (e.g., ref. 16) have considered as the geologically oldest primates. Despite the anticipated close relationship between Dermoptera and Primates, the grouping of Dermoptera within the order Primates as the sister group of the Anthropoidea is highly unexpected. On the basis of the strong support for this relationship, we expect Dermoptera/Anthropoidea to constitute a natural group and propose a new name for it: Dermosimii.
Table 2.
Bootstrap support values for internal branches
| Branch | LBP | NJ | FIT | MP | PUZ |
|---|---|---|---|---|---|
| A | 94 | 100 | 48 | 41 | — |
| B | 97 | 63 | 40 | 50 | — |
| C | 62 | 31 | 65 | 10 | — |
| D | 83 | 70 | 42 | 53 | — |
| E | 70 | 21 | 14 | — | — |
| F | 100 | 100 | 100 | 100 | 73 |
| G | 94 | 88 | 49 | 31 | 75 |
| H | 100 | 36 | 41 | 36 | — |
| I | 58 | — | — | — | — |
| J | 100 | 63 | 72 | 71 | 86 |
| K | 100 | 100 | 100 | 99 | 66 |
| L | 60 | 63 | — | — | — |
| M | 97 | 89 | 100 | 71 | 50 |
| N | 97 | 96 | 94 | 83 | — |
| O | 82 | — | — | — | — |
| P | 100 | 100 | 100 | 100 | 51 |
| Q | 99 | 99 | 100 | 99 | — |
| R | 100 | 76 | 87 | 99 | — |
| S | 100 | 97 | 99 | 100 | 86 |
A dash (—) indicates that this branch was not or differently resolved. LBP, local bootstrap probability; NJ, neighbor joining; FIT, fitch; MP, maximum parsimony; PUZ, tree-puzzle.
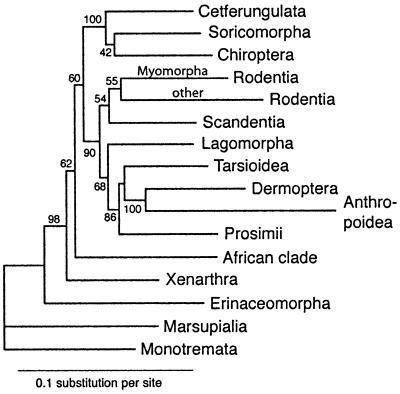
Compared with a recent primate study (19) the inclusion of the tarsier has allowed examination of the sister group relationship between Tarsioidea and Anthropoidea (Haplorrhini) to the exclusion of Strepsirrhini (Prosimii). The addition of the ring-tailed lemur to the data set is important for examining this particular question as it splits the prosimian branch. Analyses of short interspersed nuclear elements (44) support the Strepsirrhini/Haplorrhini hypothesis. This relationship was found in the nucleotide analysis (Fig. 2). It was not the preferred ML amino acid alternative, however, whether or not the flying lemur was included in the analysis.
Figure 2.
ML tree based on nucleotide analysis of the same 60 sequences as in Fig. 1. Major groups that were identified in both amino acid and nucleotide analyses have been joined on single branches for which average branch lengths were calculated. Support values indicate ML local bootstrap probability (29).
The phylogenetic position of Scandentia was addressed in a recent mtg study (20) that identified Scandentia and Lagomorpha as sister groups. The current results, which have a more comprehensive taxon representation, are consistent with the Scandentia study (20) in those parts of the tree where taxon sampling was comparable.
Both the dugong and the elephant shrew fell within the African clade, the dugong as the sister group of the elephant, and the elephant shrew as that of the tenrec. Morphological similarities between some orders belonging to this clade and their African location were pointed out by Le Gros Clark and Sonntag (42). Compared with a previous study (4), the addition of the dugong and the elephant shrew to the data set yielded increased support for the whole African clade as such. The lesser anteater grouped solidly with the armadillo. Interestingly, the depth of the divergence between the two taxa is similar or greater than that of ordinal divergences within the African clade.
Pholidota and Carnivora joined on a common branch, extending the sister group relationship between Carnivora and Perissodactyla to one between Carnivora/Pholidota (Ferae; refs. 16 and 39) and Perissodactyla. On this basis we suggest an extension of the designation Cetferungulata (sensu refs. 40 and 41) to include Pholidota as well. Even though the support for the Carnivora/Pholidota relationship is strong, a topology with Pholidota basal to the other cetferungulate taxa receives a similar likelihood value as the best tree and cannot be statistically rejected (Table 3).
The analysis identified monophyletic Pinnipedia with strong support. Also, the sister group relationship between Otariidae and Odobenidae was strongly supported.
The Eutherian mtg Tree.
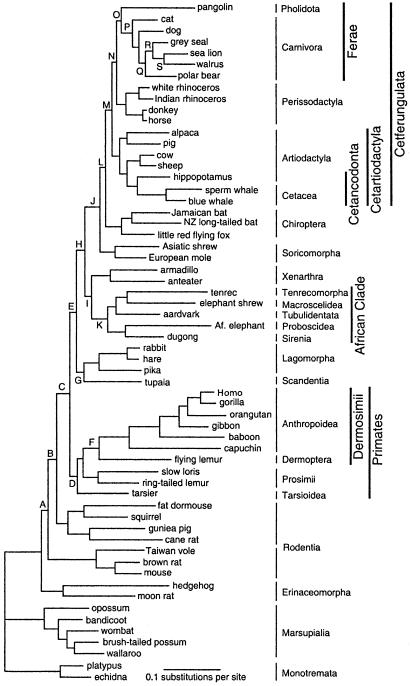
The current taxon sampling, taken together with the use of seven OG species for rooting the tree, allows a more comprehensive analysis of eutherian relationships than has been possible in previous mtg studies. Branches of particular relevance for the discussion have been labeled with capital letters (A-S) in Fig. 1, which is based on analysis of the amino acid data set. Support for branches is given in Table 2.
Figure 1.
mtg ML amino acid tree for 53 eutherian plus 7 OG taxa. Branches discussed in the text are labeled with capital letters (A–S). For nomenclature see Materials and Methods. It is probable that branch E (alternatively H) corresponds to the zhelestids (refs. 64–66 and text).
The traditional order Lipotyphla splits into three lineages: Erinaceomorpha, Tenrecomorpha, and Soricomorpha. On the basis of their separate phylogenetic positions and the depths of their origin, each of these lineages is recognized as having ordinal status. The ML amino acid analysis placed Erinaceomorpha as the basal eutherian lineage, separated from the remaining eutherian taxa (branch A). The nucleotide and amino acid composition of the hedgehog mtDNA (45) deviates from that of the other eutherians (41), a circumstance that might affect its position in the tree. However, when the same χ2 statistics are applied to the current taxon sampling, the amino acid composition of the hedgehog is no longer significantly different. The moon rat mtDNA shows no such deviation and, irrespective of whether the hedgehog or the moon rat was used alone or in combination, the same basal position of the Erinaceomorpha was reconstructed.
Rodents were identified as the most basal group among the remaining eutherians. Consistent with other mtg studies (10, 11), the ML amino acid analysis did not favor a monophyletic Rodentia but rather split the order into two groups, one with myomorph rodents and the Taiwan vole, the other consisting of the two hystricognaths and the dormouse and the squirrel.
The remaining taxa (branch C) split into two groups, one (branch D) including Primates and Dermoptera, the other (branch E) comprising the morphologically heterogeneous group of all of the remaining eutherians. Within branch E the ML analysis identified a sister group relationship between Lagomorpha/Scandentia (branch G) and the remaining orders (branch H). The latter branch (H) splits into two major groups, branch I, which includes Xenarthra and the African clade, and branch J, with Soricomorpha, Chiroptera, and extended Cetferungulata. Branch I, which joins Xenarthra and African clade, is short and the support for this sister group relationship is limited. The support for branch K (African clade) is conclusive but the relationships between the aardvark, the elephant shrew, and the tenrec remained largely unresolved.
Branch J is consistently recovered by various methods and data sets. The best tree had Soricomorpha as the sister group of Chiroptera and Cetferungulata, whereas the second best alternative joined Soricomorpha and Chiroptera. Branch M includes five morphologically distinct orders (Carnivora, Pholidota, Perissodactyla, Artiodactyla, and Cetacea). Previous mtg analyses (40) had identified the Artiodactyla/Cetacea/Perissodactyla/Carnivora grouping, with sister group relationships between Artiodactyla and Cetacea and between Perissodactyla and Carnivora. The relationship between Carnivora and Perissodactyla (40) was morphologically unexpected, but this relationship has been generally supported in subsequent molecular studies.
Branch P (Cetartiodactyla) and the sister group relationship between Hippopotamidae and Cetacea (Cetancodonta; branch Q) were strongly supported. Sarich (46) discussed the hippopotamid/cetacean affinity, but did not give experimental details. The sister group relationship between Hippopotamidae and Cetacea was demonstrated in analyses of complete cyt b genes (18, 47), and statistical evidence for the Cetancodonta clade was provided in mtg analyses, which also suggested that this divergence had taken place ≈55 million years (MY) B.P. (38). These findings have been supported by different nuclear data (e.g., ref. 48). The analysis joined the pig (Suina) and the alpaca (Tylopoda) on a common branch, but the support for this arrangement was not conclusive. The second best alternative placed Suina as the sister group of all other cetartiodactyls (see also refs. 48–50).
The relationship between artiodactyls and cetaceans was recently addressed in two morphological/paleontological studies (51, 52). The conclusions of these studies were inconsistent. Gingerich et al. (51), in agreement with molecular results, concluded that cetaceans have their origin within Artiodactyla, whereas Thewissen et al. (52) inferred that Artiodactyla and Cetacea were sister groups. Test of the latter phylogeny relative to the best mtg tree found the latter relationship as highly improbable (Table 3). Although the morphological conclusions may initially seem incongruent, both might be correct if Archaeoceti is paraphyletic. Archaeocete paraphyly has been suggested (53–56) and it is possible that the study by Thewissen et al. focused on taxa that do not form a monophyletic group together with extant cetaceans.
The amino acid analyses reconstructed trees that were largely congruent irrespective of the method used. In comparison, the nucleotide trees differed considerably depending on the analytical approach used. ML analyses of both amino acid and nucleotide data sets reconstructed trees that were similar with respect to most intraordinal relationships, whereas the relationships between some basal lineages differed. Erinaceomorpha remained a basal eutherian taxon in both data sets, whereas in the nucleotide analyses, Xenarthra (followed by the African clade) became a basal taxon among remaining eutherians. As a result of this shift, Rodentia became a monophyletic group that joined Scandentia, Lagomorpha, and Primates/Dermoptera. The ML nucleotide tree of ordinal and clade relationships is shown in Fig. 2.
The mtg results challenge several traditional or semitraditional morphological hypotheses for eutherian relationships. The distinctly separate positions of Xenarthra and Pholidota in the tree are incongruent with systematic schemes that suggest a close relationship between these two orders. The lipotyphlan polyphyly reconstructed in the mtg studies is of particular interest as it might aid in reevaluation of the morphological characteristics used to unite this “order.” It is, nevertheless, of interest that when the current lipotyphlan relationships are considered in isolation, the results are consistent with morphological studies (35) that place Erinaceidae as a basal sister group to remaining lipotyphlans. The separate positions of Rodentia, Lagomorpha, and Macroscelidea in the tree are inconsistent with the Anagalida hypothesis. Also, the Archonta hypothesis (Primates/Dermoptera/Scandentia/Chiroptera) remained unsupported as shown in previous mtg studies (20, 57, 58). The position of Dermoptera within Primates as the sister group of Anthropoidea is highly unexpected, even though there are morphological arguments in favor of a close relationship between the two orders (44). The Glires hypothesis (Rodentia + Lagomorpha) was not supported in the amino acid analyses (Table 3). Finally, the pinniped results lend no support to the morphological hypothesis of a sister group relationship between Odobenidae and Phocidae to the exclusion of Otariidae (21), underlining the problems associated with basing phylogenetic conclusions on anatomical features that may have strong adaptive values.
Temporal Aspects on Eutherian Divergences.
Previous mtg datings using inter- or intraordinal eutherian calibration points (19, 47, 49, 58, 59) have suggested that only one eutherian order, Cetacea (≈55 MY B.P.; ref. 39), has originated after the K/T boundary (65 MY B.P.). The inclusion of the orders Dermoptera, Sirenia, Macroscelidea, and Pholidota does not change this picture, as their origin is considerably deeper than that of the Cetacea. Thus, the analyses underline the Cretaceous origin of virtually all eutherian orders, even though their diversification has largely taken place after the Cretaceous/Tertiary (K/T) boundary (50), as also is shown in analyses of nuclear sequences (60–62).
The suggestions of a Cretaceous origin of eutherian orders have been met with suspicion by many paleontologists. It has been argued that the molecular estimates suggesting early origin of eutherian orders are artifactual and caused by accelerated molecular evolution at the Cretaceous/Tertiary (K/T) boundary or immediately thereafter (63). However, a post-K/T acceleration of this kind would actually have an opposite effect, as calibration points such as A/C-60 (the split between ruminant artiodactyls and cetancodonts 60 MY B.P.) (58), placed within the postulated window of accelerated evolution, would tend to shrink the estimated datings of earlier divergences. Furthermore, comparisons between estimates based on eutherian calibration points of different ages do not suggest any effects of this kind (49).
It has been tentatively proposed (19) that the heterogenous eutherian group (branch E in Fig. 1) might be closely connected to the zhelestids (64–66). The oldest zhelestid fossils are ≈85–90 MY old, ≈10 MY younger than the molecular dating (19) of the split between Primates and the heterogenous group (branch E in Fig. 1). These fossils show that some zhelestids had already ≈85 MY B.P. begun to differentiate morphologically toward herbivory. If the proposed relationship between the zhelestids and the heterogenous eutherian group is phylogenetically correct, the zhelestid fossils support mtg analyses that place primate origin and divergences much deeper than commonly conceived (2, 19, 49, 58, 59).
Comparison with Non-mtg Studies.
The mtg results allow comparison with recent phylogenetic studies based on both real (5–7) and real plus constructed data sets (67). The trees reconstructed in the real data studies and the mtg analyses show pronounced similarities when they are viewed as unrooted. The relationships within the order-rich African clade and Cetferungulata are common to all studies. Similarly, the model tree of the real plus constructed data sets (67) is consistent with the mtg amino acid findings.
The real data studies do not permit direct comparison of the performance of mt and nuclear data sets as such as these studies (5–7) all used a combination of mt (rRNA genes) and nuclear data. The capacity of mt rRNA genes for resolving ordinal mammalian relationships may be more limited than generally believed (e.g., ref. 68). Only the stem regions of the mt rRNA genes seem to carry a useful (albeit marginal) phylogenetic signal and inclusion of the other parts of these sequences may increase the background noise and promote the selection of the wrong signal (20). Also the mt/nuclear analyses (6, 7) included third codon positions. These positions may show a high degree of randomization in studies of deep divergences (e.g., ref. 69). In addition, the constraints that were put on several relationships in these analyses (7) complicate a direct comparison with the current results.
Rosenberg and Kumar (67) use a different way for examining phylogenetic findings. An initial analysis of the effect of taxon sampling and sequence length on the phylogenetic outcome showed that sequence length is highly critical to the reliability of the results (see also ref. 70). These conclusions were substantiated by two sets of computer simulations. The first involved 50 hypothetical genes, the evolution of which conformed with the data set used by Murphy et al. (6), whereas the second simulation was based on evolutionary parameters estimated from 1,167 real genes. Analysis of sequences fitting these parameters reliably reconstructed the tree OG(Rodentia(Primates,(Lagomorpha(Artiodactyla(Carnivora,Perissodactyla))))). This relationship is the same as in mtg amino acid studies (e.g., Fig. 1) but at variance with the recent mt/nuclear results (5–7).
The Root of the mtg Eutherian Tree.
One of the main differences between the mtg results and the recent mt/nuclear studies (7) is related to the position of Erinaceomorpha, which in mtg analyses (both amino acid and nucleotide) has a basal position in the eutherian tree. This finding is inconsistent with nucleotide analyses of nuclear sequences, which preferably place Erinaceomorpha with Soricomorpha in a less basal position.
Shifts in the relative positions of basal taxa in any phylogenetic tree may have a pronounced effect on the general topology of the tree. For this reason we have examined whether the exclusion of some basal taxa in Fig. 1 would affect the positions of other groupings.
Exclusion of Erinaceomorpha did not affect other relationships. Next to the Erinaceomorpha, rodents have a basal position in the mtg amino acid tree. The effects of excluding rodents from the data set were investigated in two steps, first by removing myomorph rodents and the Taiwan vole, second by removing all rodents. Contrary to the exclusion of Erinaceomorpha, the exclusion of myomorph rodents and the vole had a pronounced effect on eutherian relationships. Thus, Xenarthra and the African clade moved into basal positions with a disrupted sister group relationship, whereas caviomorph/sciurignath rodents became the sister group of Primates, and Lagomorpha became the sister group of Primates. Removal of all rodents had the same effect, except for a restored relationship between Scandentia and Lagomorpha. When both Erinaceomorpha and Rodentia were excluded, Xenarthra maintained its basal position, whereas the African clade remained as the sister group of Soricomorpha/Chiroptera/Cetferungulata.
As is evident from the branch lengths in Fig. 1, the mt evolutionary rate of the anthropoids is fast compared with other eutherian lineages. The influence of the anthropoids on the mtg topology is therefore of a particular interest. The tree reconstructed after excluding the anthropoids placed Erinaceomorpha in a basal position followed by the African clade. In addition, Rodentia became the sister group of remaining primates/Dermoptera, whereas Xenarthra became the sister group of an assembly that included Primates/Dermoptera/Rodentia and Lagomorpha/Scandentia.
The nucleotide data set was also sensitive to exclusion of basal taxa. In this case removal of Erinaceomorpha as well as Erinaceomorpha/Rodentia promoted a basal position of the African clade followed by Xenarthra. Coincident with this, Primates moved into a less basal position, grouping with Dermoptera, Lagomorpha, Scandentia, and monophyletic Rodentia.
Superficially, the trees shown in Figs. 1 and 2 may appear strikingly different. However, the differences are essentially related to the rooting point of the tree above the position of Erinaceomorpha. Independent of analytical method, all amino acid analyses placed this point on the branch leading to myomorph rodents and the vole, whereas ML nucleotide analysis placed it on the short Xenarthra branch, alternatively on the branch leading to the African clade. However, as a result of the short branches separating basal eutherian divergences, an mtg amino acid tree with the root placed on the African clade branch is not statistically refuted under a rate heterogeneity model when Erinaceomorpha is excluded from the data set (Table 3).
The number of described marsupial and monotreme mtDNAs has gradually increased during the time of eutherian mtg phylogenetics. It is therefore of interest that the same general eutherian mtg tree has been reconstructed irrespective of whether a single taxon (2) or several OG taxa have been used to root the tree. Similarly, the choice of methods and weighting approaches used in the past appear to have had little or no effect on the topology of the mtg tree (20, 71, 72).
The position of the root of any phylogenetic tree defines the direction of evolution in the tree and provides information as to the order of divergences within the same tree. The current mtg analyses and the recent mt/nuclear studies identify different rooting points in the eutherian tree. It is as yet not clear whether this inconsistency is related to the OG or to the eutherian data sets, or both. Whatever the reasons for this discrepancy, it is evident that the establishment of the rooting point of the eutherian tree is of paramount importance to the discussion of eutherian evolution, both molecular and morphological.
Acknowledgments
We thank François Catzeflis, Wilfried W. de Jong, the late Francis H. Fay, Eberhart Fuchs, Carsten Grøndahl, and Lars Olsson for samples. We also thank Kerryn Slack for valuable help with the manuscript. This study was supported by the Swedish Research Council, European Commission Grant ERB-FMRX-CT98-0221, the Crawford Foundation, and by the Nilsson–Ehle Foundation.
Abbreviations
- ML
maximum likelihood
- mt
mitochondrial
- mtg
mitogenomic
- MY
million years
- OG
outgroup
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the EMBL database (see Table 1 for accession nos.).
References
- 1.Arnason U, Johnsson E. J Mol Evol. 1992;34:493–505. doi: 10.1007/BF00160463. [DOI] [PubMed] [Google Scholar]
- 2.Janke A, Feldmaier-Fuchs G, Thomas W K, von Haeseler A, Pääbo S. Genetics. 1994;137:243–256. doi: 10.1093/genetics/137.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mouchaty S K, Gullberg A, Janke A, Arnason U. Mol Biol Evol. 2000;17:60–67. doi: 10.1093/oxfordjournals.molbev.a026238. [DOI] [PubMed] [Google Scholar]
- 4.Mouchaty S K, Gullberg A, Janke A, Arnason U. Zool Scr. 2000;29:307–317. [Google Scholar]
- 5.Madsen O, Scally M, Douady C J, Kao D J, DeBry R W, Adkins R, Amrine H M, Stanhope M J, de Jong W W, Springer M S. Nature (London) 2001;409:610–614. doi: 10.1038/35054544. [DOI] [PubMed] [Google Scholar]
- 6.Murphy W J, Eizirik E, Johnson W E, Zhang Y P, Ryder O A, O'Brien S J. Nature (London) 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 7.Murphy W J, Eizirik E, O'Brien S J, Madsen O, Scally M, Douady C J, Teeling E, Ryder O A, Stanhope M J, de Jong W W, Springer M S. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 8.Graur D, Hide A H, Li W H. Nature (London) 1991;351:649–652. doi: 10.1038/351649a0. [DOI] [PubMed] [Google Scholar]
- 9.D'Erchia A M, Gissi C, Pesole G, Saccone C, Arnason U. Nature (London) 1996;381:597–600. doi: 10.1038/381597a0. [DOI] [PubMed] [Google Scholar]
- 10.Reyes A, Pesole G, Saccone C. Gene. 2000;259:177–187. doi: 10.1016/s0378-1119(00)00438-8. [DOI] [PubMed] [Google Scholar]
- 11.Mouchaty S K, Catzeflis F, Janke A, Arnason U. Mol Phylogenet Evol. 2001;18:127–135. doi: 10.1006/mpev.2000.0870. [DOI] [PubMed] [Google Scholar]
- 12.Novacek M J. Nature (London) 1992;356:121–125. doi: 10.1038/356121a0. [DOI] [PubMed] [Google Scholar]
- 13.Rose K D, Emry R J. In: Mammal Phylogeny 2, Placentals. Szalay F S, Novacek M J, McKenna M C, editors. New York: Springer; 1993. pp. 81–102. [Google Scholar]
- 14.McKenna M C. In: Phylogeny of the Primates. Luckett W P, Szalay F S, editors. New York: Plenum; 1975. pp. 21–46. [Google Scholar]
- 15.Simpson G G. Bull Am Mus Nat Hist. 1945;85:1–350. [Google Scholar]
- 16.McKenna M C, Bell S K. Classification of Mammals Above the Species Level. New York: Columbia Univ. Press; 1997. [Google Scholar]
- 17.Gaeth A P, Short R V, Renfree M B. Proc Natl Acad Sci USA. 1999;96:5555–5558. doi: 10.1073/pnas.96.10.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irwin D M, Arnason U. J Mamm Evol. 1994;2:37–55. [Google Scholar]
- 19.Arnason U, Gullberg A, Schweizer Burguete A, Janke A. Hereditas. 2000;133:217–228. doi: 10.1111/j.1601-5223.2000.00217.x. [DOI] [PubMed] [Google Scholar]
- 20.Schmitz J, Ohme M, Zischler H. Mol Biol Evol. 2000;17:1334–1343. doi: 10.1093/oxfordjournals.molbev.a026417. [DOI] [PubMed] [Google Scholar]
- 21.Wyss A R, Flynn J. In: Mammal Phylogeny 2, Placentals. Szalay F S, Novacek M J, McKenna M C, editors. New York: Springer; 1993. pp. 32–52. [Google Scholar]
- 22.Arnason U. Hereditas. 1977;87:227–242. doi: 10.1111/j.1601-5223.1978.tb01266.x. [DOI] [PubMed] [Google Scholar]
- 23.Arnason U, Bodin K, Gullberg A, Ledje C, Mouchaty S. J Mol Evol. 1995;40:78–85. doi: 10.1007/BF00166598. [DOI] [PubMed] [Google Scholar]
- 24.Ledje C, Arnason U. J Mol Evol. 1996;43:641–649. doi: 10.1007/BF02202112. [DOI] [PubMed] [Google Scholar]
- 25.Ledje C, Arnason U. J Mol Evol. 1996;42:135–144. doi: 10.1007/BF02198839. [DOI] [PubMed] [Google Scholar]
- 26.Arnason U, Gullberg A, Widegren B. J Mol Evol. 1991;33:556–568. doi: 10.1007/BF02102808. [DOI] [PubMed] [Google Scholar]
- 27.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 28.Felsenstein J. PHYLIP. Seattle: Univ. of Washington; 1993. [Google Scholar]
- 29.Adachi J, Hasegawa M. Comput Sci Monogr. 1996;28:1–150. [Google Scholar]
- 30.Swofford D L. PAUP. Sunderland, MA: Sinauer; 2000. [Google Scholar]
- 31.Drummond A, Strimmer K. Bioinformatics. 2001;17:662–663. doi: 10.1093/bioinformatics/17.7.662. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Nei M. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 33.Shimodaira H, Hasegawa M. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 34.Templeton A R. Evolution (Lawrence, Kans) 1983;37:221–224. [Google Scholar]
- 35.Butler P M. In: The Phylogeny and Classification of the Tetrapods, Mammals. Benton M J, editor. Vol. 2. Oxford: Clarendon; 1988. pp. 117–141. [Google Scholar]
- 36.Eisenberg J F. The Mammalian Radiations. Chicago: Univ. Chicago Press; 1981. p. xx. +1–610. [Google Scholar]
- 37.Montgelard C, Catzeflis F M, Douzery E. Mol Biol Evol. 1997;14:550–559. doi: 10.1093/oxfordjournals.molbev.a025792. [DOI] [PubMed] [Google Scholar]
- 38.Ursing B M, Arnason U. Proc R Soc London Ser B. 1998;265:2251–2255. doi: 10.1098/rspb.1998.0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shoshani J, McKenna M C. Mol Phylogenet Evol. 1998;9:572–584. doi: 10.1006/mpev.1998.0520. [DOI] [PubMed] [Google Scholar]
- 40.Xu X, Janke A, Arnason U. Mol Biol Evol. 1996;13:1167–1173. doi: 10.1093/oxfordjournals.molbev.a025681. [DOI] [PubMed] [Google Scholar]
- 41.Arnason U, Gullberg A, Janke A. Proc R Soc London B Biol Sci. 1999;266:339–345. doi: 10.1098/rspb.1999.0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Gros Clark W E, Sonntag C F. Proc Zool Soc (London) 1926;30:445–485. [Google Scholar]
- 43.Beard K C. In: Mammal Phylogeny 2, Placentals. Szalay F S, Novacek M J, McKenna M C, editors. New York: Springer; 1993. pp. 129–150. [Google Scholar]
- 44.Schmitz J, Ohme M, Zischler H. Genetics. 2001;157:777–784. doi: 10.1093/genetics/157.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krettek A, Gullberg A, Arnason U. J Mol Evol. 1995;41:952–957. doi: 10.1007/BF00173175. [DOI] [PubMed] [Google Scholar]
- 46.Sarich V M. In: Mammal Phylogeny 2, Placentals. Szalay F S, Novacek M J, McKenna M C, editors. New York: Springer; 1993. pp. 103–113. [Google Scholar]
- 47.Arnason U, Gullberg A. Mol Biol Evol. 1996;13:407–417. doi: 10.1093/oxfordjournals.molbev.a025599. [DOI] [PubMed] [Google Scholar]
- 48.Gatesy J, O'Leary M A. Trends Ecol Evol. 2001;16:562–570. [Google Scholar]
- 49.Arnason U, Gullberg A, Gretarsdottir S, Ursing B, Janke A. J Mol Evol. 2000;50:569–578. doi: 10.1007/s002390010060. [DOI] [PubMed] [Google Scholar]
- 50.Ursing B M, Slack K, Arnason U. Zool Scr. 2000;29:83–88. [Google Scholar]
- 51.Gingerich P D, ul Haq M, Zalmout I S, Kahn I H, Malkani M S. Science. 2001;293:2239–2242. doi: 10.1126/science.1063902. [DOI] [PubMed] [Google Scholar]
- 52.Thewissen J G M, Williams E M, Roe L J, Hussain S T. Nature (London) 2001;413:277–281. doi: 10.1038/35095005. [DOI] [PubMed] [Google Scholar]
- 53.Kellogg A R. Carnegie Inst Wash Publ. 1936;482:1–366. [Google Scholar]
- 54.Slijper E I. Capita Zool. 1936;7:1–590. [Google Scholar]
- 55.Yablokov A V, Belkovich V M, Borisov V I. Whales and Dolphins. Nauka Moscow; 1972. pp. 1–472. (Russian). [Google Scholar]
- 56.Fordyce R E, Barnes L G. Annu Rev Earth Planet Sci. 1994;22:419–455. [Google Scholar]
- 57.Pumo D E, Finamore P S, Franek W R, Carleton C J, Tarzami S, Balzarano D. J Mol Evol. 1998;47:709–717. doi: 10.1007/pl00006430. [DOI] [PubMed] [Google Scholar]
- 58.Arnason U, Gullberg A, Janke A, Xu X. J Mol Evol. 1996;43:650–661. doi: 10.1007/BF02202113. [DOI] [PubMed] [Google Scholar]
- 59.Arnason U, Gullberg A, Janke A. J Mol Evol. 1998;47:718–727. doi: 10.1007/pl00006431. [DOI] [PubMed] [Google Scholar]
- 60.Hedges S B, Parker P H, Sibley C G, Kumar S. Nature (London) 1996;381:226–229. doi: 10.1038/381226a0. [DOI] [PubMed] [Google Scholar]
- 61.Kumar S, Hedges S B. Nature (London) 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 62.Easteal S. BioEssays. 1999;21:1052–1058. doi: 10.1002/(SICI)1521-1878(199912)22:1<1052::AID-BIES9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 63.Benton M J. BioEssays. 1999;21:1043–1051. doi: 10.1002/(SICI)1521-1878(199912)22:1<1043::AID-BIES8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 64. Nessov, L. A. (1985) Vestn. Leningr. Univ. Ser. 7 17, 8–18.
- 65.Archibald J D. Science. 1996;272:1150–1153. doi: 10.1126/science.272.5265.1150. [DOI] [PubMed] [Google Scholar]
- 66.Archibald J D, Averianov A O, Ekdale E G. Nature (London) 2001;414:62–65. doi: 10.1038/35102048. [DOI] [PubMed] [Google Scholar]
- 67.Rosenberg M S, Kumar S. Proc Natl Acad Sci USA. 2001;98:10751–10756. doi: 10.1073/pnas.191248498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McNiff B E, Allard M W. Am J Phys Anthropol. 1998;107:225–241. doi: 10.1002/(SICI)1096-8644(199811)107:3<225::AID-AJPA1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 69.DeBry R W, Sagel R M. Mol Phylogenet Evol. 2001;19:290–301. doi: 10.1006/mpev.2001.0945. [DOI] [PubMed] [Google Scholar]
- 70.Poe S, Swofford D L. Nature (London) 1999;398:299–300. doi: 10.1038/18592. [DOI] [PubMed] [Google Scholar]
- 71.Penny D, Hasegawa M, Waddell P J, Hendy M D. Syst Biol. 1999;48:76–93. doi: 10.1080/106351599260454. [DOI] [PubMed] [Google Scholar]
- 72.Waddell P J, Cao Y, Hauf J, Hasegawa M. Syst Biol. 1999;48:31–53. doi: 10.1080/106351599260427. [DOI] [PubMed] [Google Scholar]