Abstract
In mammalian cells, DNA double-strand breaks (DSBs) cause rapid phosphorylation of the H2AX core histone variant (to form γ-H2AX) in megabase chromatin domains flanking sites of DNA damage. To investigate the role of H2AX in mammalian cells, we generated H2AX-deficient (H2AXΔ/Δ) mouse embryonic stem (ES) cells. H2AXΔ/Δ ES cells are viable. However, they are highly sensitive to ionizing radiation (IR) and exhibit elevated levels of spontaneous and IR-induced genomic instability. Notably, H2AX is not required for NHEJ per se because H2AXΔ/Δ ES cells support normal levels and fidelity of V(D)J recombination in transient assays and also support lymphocyte development in vivo. However, H2AXΔ/Δ ES cells exhibit altered IR-induced BRCA1 focus formation. Our findings indicate that H2AX function is essential for mammalian DNA repair and genomic stability.
The DNA in eukaryotic cells is packaged into chromatin, the fundamental unit of which is the nucleosome. The nucleosome consists of DNA wrapped around an octamer of the four core histones—H2A, H2B, H3, and H4 (1). The H2A histones consist of several subfamilies that contain distinct, conserved amino- and carboxyl-terminal amino acid sequences (2). The H2AX subfamily contains a conserved carboxyl-terminal Ser-Gln-Glu (SQE motif) amino acid sequence. This SQE motif represents the consensus in vitro phosphorylation site for members of the phosphoinositide 3-kinase related kinase (PIKK) family that includes the protein kinases DNA-dependent protein kinase catalytic subunit (DNA-PKcs), ataxia telangiectasia mutated (ATM), and ATM and Rad3-related (ATR) (3).
The repair of spontaneous and induced DNA double-strand breaks (DSBs) is critical for the maintenance of genomic integrity. In eukaryotic cells, the two major pathways of DSB repair are nonhomologous end-joining (NHEJ) and homologous recombination (HR; refs. 4 and 5). Covalent modifications of core histones via phosphorylation, acetylation, and methylation have been proposed to form a “histone code” that is read by cellular proteins to facilitate downstream molecular events (6). In response to DNA damage by agents that induce DNA double-strand breaks, Mec1, the Saccharomyces cerevisiae homologue of ATR, phosphorylates the SQE motif of H2A (7). This phosphorylation event is required for the efficient repair of chromosomal DSBs by NHEJ but does not appear to be as important for homologous recombination (7). In mammalian cells, H2AX is rapidly phosphorylated on the induction of DSBs by ionizing radiation (IR) and DNA damaging agents (8, 9), resulting in formation of γ-H2AX foci along megabase chromatin domains flanking DNA damage sites (9).
Foci of γ-H2AX also form at the immunoglobulin heavy chain locus during class switch recombination (CSR) in activated mature B cells (10). CSR occurs between large, highly repetitive S regions and also may be initiated by DSBs (10, 11) and completed by NHEJ factors (12–15). Notably, CSR is significantly impaired in the absence of H2AX (10). Earlier during lymphocyte development, exons that encode immunoglobulin and T cell receptor (TCR) variable regions are assembled by V(D)J recombination. Formation of γ-H2AX foci occurs at the TCRα locus during V(D)J recombination (16). V(D)J recombination is initiated by the recombination-activating gene 1 and 2 proteins (RAG1 and RAG2 or RAGs), which introduce DSBs between recombining V, D, or J segments and flanking recombination signal sequences (RSs) to generate blunt, 5′ phosphorylated RS ends and hairpinned coding ends (17). Joining of RS ends absolutely requires four NHEJ factors, including XRCC4, DNA ligase IV (Lig4), Ku70, and Ku80; whereas joining of coding ends requires these four factors plus DNA-PKcs and Artemis (17). Thus, completion of RAG-initiated V(D)J recombination in transient reporter substrates provides a strict assay for a direct function of known factors in mammalian NHEJ. In this context, a direct evaluation of the potential role of H2AX in V(D)J recombination has not been reported.
ATM, and possibly DNA-PKcs, phosphorylate H2AX after IR (18, 19). However, additional wortmannin-insensitive kinases also have been implicated (18). ATM and DNA-PKcs are both required for the repair of IR-induced DSBs because cells deficient for either of these factors are hypersensitive to IR and exhibit DNA repair defects. ATM-deficient cells also exhibit cell cycle checkpoint defects and dramatically increased genomic instability (20). In this context, DNA-PKcs is directly involved in the repair of DSBs (21) whereas ATM may have a more indirect role via phosphorylation of certain proteins involved in the DNA damage response (20). It has been argued that ATM and related kinases, including DNA-PKcs and ATR, may mediate some functions via phosphorylation of H2AX (18, 19). On IR, foci of the DNA repair proteins Mre11/RAD50/NBS1 (the MRN complex), RAD51, 53BP1, and BRCA1 colocalize with γ-H2AX foci (18, 22, 23). In this context, γ-H2AX may play a role in the recruitment of BRCA1, RAD51, and perhaps other DNA repair factors to the sites of DNA damage (18). Therefore, mammalian H2AX may be downstream of relevant phosphoinositide 3-kinase related kinases in the mediation of particular DNA damage responses and, in this context, theoretically could have a role in maintenance of genomic stability.
Materials and Methods
Targeting Constructs and Probes.
The 5L/3N targeting vector was constructed in pLNTK (24). The 5′ homology arm is a 4.9-kb NotI/BstXI genomic fragment with an loxP site inserted into a BstXI site 5′ of the H2AX promoter. The 3′ homology arm is a 6.1-kb BstXI/BamHI genomic fragment. The 5′H2AX probe was generated via PCR with primers 5′-GGAGGGATCCTGTACTACGTCTACATGGGG-3′ and 5′-TCTCACCTTCCAGTTCCACCAGGTTG-3′. The 3′H2AX probe was generated via PCR with primers 5′-CTCTGGATCCCGTAGAGGGCAGAAGG-3′ and 5′-GCGCGGATCCTGATTTCAAACTGTATGCCAGGG-3′. The Neo probe was generated via PCR with primers 5′-GCAGCCAATATGGGATCGGC-3′ and 5′-GTTCGGCTGGCGCGAGCCCC-3′. The IntH2AX probe is a 300-bp AatII/BglII fragment.
Gene Targeting and Generation of Embryonic Stem (ES) Cells.
The 5L/3N targeting vector was electroporated into TC1 ES cells (25) as described (26). Targeted clones were identified by Southern blotting with the 3′H2AX probe on HindIII-digested DNA (7.2 kb H2AXNeo, 14.3 kb H2AXWT) and confirmed with the 5′H2AX probe on BamHI-digested DNA (8.8 kb H2AXNeo, 14.5 kb H2AXWT) and the IntH2AX probe on HindIII-digested DNA to detect integration of the 5′loxP site (5.0 kb). To remove the PGK-neor gene, 2.5 × 106 cells of independent H2AXWT/Neo clones were infected with recombinant AdenoCre. H2AXWT/Flox clones were identified via Southern blot analysis with the 5′H2AX probe on BamHI-digested DNA (8.4 kb). Independent H2AXWT/Flox clones were retransfected with 5L/3N. H2AXFlox/Neo ES cells were identified by Southern blotting with the 3′H2AX probe on HindIII-digested DNA (7.2 kb H2AXNeo, 9.3 kb H2AXFlox). H2AXFlox/Flox, H2AXFlox/Δ, and H2AXΔ/Δ ES cells were made from independent H2AXFlox/Neo ES clones. XRCC4−/− TC1 ES cells were obtained through sequential gene targeting to first generate ES cells in which exon 3 of XRCC4 was flanked by loxP sites (C.Y., unpublished data).
Preparation and Characterization of H2AX Antibodies.
CKATQASQEY and CKATQAS*QEY (the asterisk denotes phosphoserine) peptides were synthesized, coupled to keyhole limpet hemocyanin (Pierce), and used to generate high titer polyclonal antisera in rabbits. Affinity-purified antibody samples recognized a predominant ≈17-kDa band in histone preparations from human or wild-type mouse cells but not from mouse H2AXΔ/Δ ES cells. Only the Abs specific for S139-phosphorylated H2AX revealed a dose-dependent increase in immunoblot signal and characteristic nuclear foci by immunostaining after IR exposure of cells.
Histone Extraction and Western Blot Analysis.
Histone preparations were made as previously described (8). Western analysis was performed with anti-H2AX rabbit polyclonal antisera (0.1 μg/ml) and antibodies to histone H4 (Cell Signaling).
IR Sensitivity and Genomic Instability Assays.
ES cells passaged off feeder cells were plated onto gelatinized plates. For IR sensitivity assays, cells were irradiated 18 h later by the indicated doses of γ-rays, cultured for 7 days, then stained and counted. For genomic instability assays, cells were irradiated (150 rad) 24 h later, then cultured for 48 h. Metaphases were prepared and analyzed as described (27).
V(D)J Recombination Assays.
All ES cell assays were performed and analyzed as described (28).
FACS Staining of H2AXΔ/Δ Lymphocytes.
Lymphocytes isolated from H2AXΔ/Δ RAG chimeric mice (29) were stained with FITC-conjugated anti-CD8, anti-B220 as well as phycoerythrin-conjugated anti-CD4 and anti-IgM antibodies (PharMingen) and analyzed via a FACScan (Becton Dickinson).
Immunostaining and Confocal Microscopy.
ES cells were plated on gelatinized cover slips, and immunostaining was performed as described (30). Cells were stained with phospho-H2AX-specific polyclonal (1 μg/ml), mouse BRCA1-specific monoclonal (1:5 dilution), and RAD51 polyclonal (Oncogene Science; 1:5 dilution) antibodies, and rhodamine- and FITC-conjugated secondary antibodies (Jackson ImmunoResearch; 1:50 dilution). Nuclei were stained with To-pro-3 (Molecular Probes). Images were collected by confocal microscopy (Bio-Rad Radiance 2000) and processed by using Adobe photoshop (Adobe Systems, Mountain View, CA) software.
Results
Generation of H2AXΔ/Δ ES Cells.
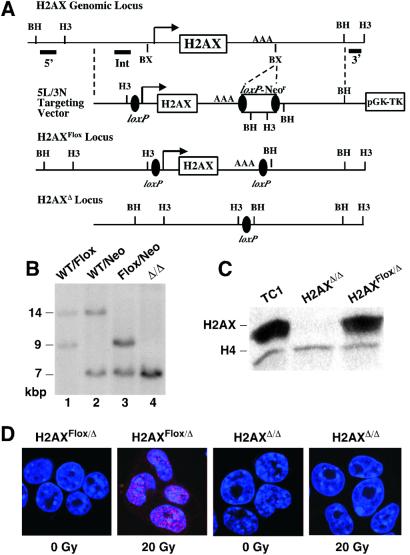
To directly investigate potential roles of H2AX in mammalian cells, we have generated and characterized H2AX-deficient mouse ES cells. We generated H2AX-deficient ES cells with a vector (5L/3N) designed to create conditional H2AX null alleles (Fig. 1A). Because expressed drug resistance markers can have negative effects on transcription of adjacent genes, we used a loxP-Neor cassette to delete the drug resistance gene after targeting. We first generated targeted TC1 ES clones with the H2AX gene flanked by a 5′loxP site and a 3′loxP-Neor cassette (H2AXWT/Neo; Fig. 1B, lane 2). Then, we used transient Cre expression to delete the Neor cassette to generate ES clones with a wild-type and a floxed H2AX allele (H2AXWT/Flox; Fig. 1B, lane 1). H2AXWT/Flox ES cells were retargeted with 5L/3N vector to generate clones with the H2AX gene floxed on one allele and flanked by a 5′loxP site and a 3′loxP-Neor cassette on the second allele (H2AXFlox/Neo; Fig. 1B, Lane 3). Transient Cre expression from a recombinant AdenoCre vector in H2AXFlox/Neo clones resulted in the generation of ES cells with deletion of the H2AX gene on both alleles (H2AXΔ/Δ; Fig. 1B, Lane 4). This same AdenoCre infection also generated H2AXFlox/Flox and H2AXFlox/Δ ES cells that serve as controls for potential effects of Cre expression. Western blotting with an antibody specific for nonphosphorylated H2AX, the major pool of H2AX in cells (8), confirmed that H2AXΔ/Δ ES cells lack detectable H2AX protein (Fig. 1C). Thus, H2AX is not required for ES cell viability; however, potential growth defects remain to be examined. Consistent with complete deletion of H2AX, H2AXΔ/Δ ES cells lacked IR-induced γ-H2AX foci (Fig. 1D).
Figure 1.
Generation and characterization of H2AX-deficient ES Cells. (A) Schematic diagram of the mouse H2AX genomic locus, 5L/3N targeting vector, and H2AXFlox and H2AXΔ alleles. The H2AX promoter, exon, and polyadenylation site are shown, respectively, as an arrow, open box, and AAA. Filled bars show the relative locations of the 5′H2AX, IntH2AX, and 3′H2AX probes. Restriction site designations: BX, BstXI; BH, BamHI; H3, HindIII. (B) Southern blot analysis of H2AXWT/Neo (lane 2), H2AXWT/Flox (lane 1), H2AXFlox/Neo (lane 3), and H2AXΔ/Δ (lane 4) HindIII-digested DNA probed with 3′H2AX. The sizes of the bands are indicated. (C) Western blot analysis of TC1, H2AXFlox/Δ (no. 40), and H2AXΔ/Δ (no. 45) ES cells with anti-H2AX and anti-H4 antibodies (loading control). (D) Unirradiated and irradiated (15 min post 20 Gy) H2AXFlox/Δ and H2AXΔ/Δ cells immunostained for γ-H2AX foci (red).
H2AX-Deficient ES Cells Are Hypersensitive to Ionizing Radiation.
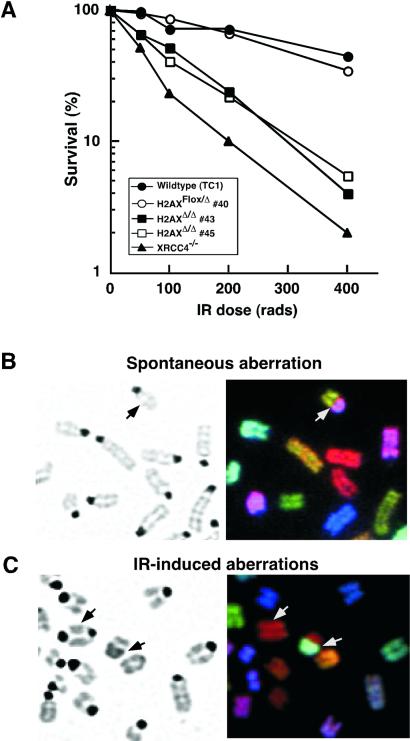
To evaluate the role of H2AX in DNA DSB repair, we used a colony formation assay to assess the IR sensitivity of H2AX mutant vs. normal ES cells. Two independent H2AXΔ/Δ ES cell subclones were highly sensitive to IR as compared with wild-type TC1 ES cells, but apparently less sensitive than XRCC4−/− ES cells (Fig. 2A). However, H2AXFlox/Δ ES cells did not exhibit increased IR sensitivity (Fig. 2A). Two additional H2AXΔ/Δ ES subclones from a second independently targeted ES cell, but neither H2AXFlox/Δ nor H2AXFlox/Flox ES cells from the same clone, were also highly IR sensitive (data not shown). H2AXΔ/Δ ES cells also showed increased sensitivity to the radiometric drug bleomycin (data not shown). However, like S. cerevisiae with H2A lacking the SQE motif, H2AXΔ/Δ ES cells were not obviously hypersensitive to UV irradiation (data not shown). Therefore, H2AX function contributes to the survival of mouse ES cells in the presence of ionizing radiation and other DNA damaging agents that induce chromosomal DSBs.
Figure 2.
Increased ionizing radiation sensitivity and genomic instability of H2AX-deficient ES cells. (A) IR sensitivity of wild-type (TC1), XRCC4−/−, H2AXFlox/Δ (no. 40), and two independent H2AXΔ/Δ (nos. 43 and 45) ES cell lines. Data are plotted as the percentage of colonies that grew out at a given γ-ray dose over unirradiated cells. The plotted numbers were obtained from triplicate data points of a representative experiment. (B and C) Metaphase spreads of unirradiated (B) and γ-irradiated (C) H2AXΔ/Δ ES cells. (Left) 4′,6-diamidino-2-phenylindole (DAPI)-stained chromosomes. (Right) SKY analysis. Arrow points to chromosomal translocations between two different chromosomes (B and C) and a dicentric chromosome (C).
H2AX-Deficient ES Cells Exhibit Genomic Instability.
The potential role of H2AX in suppressing genomic instability was examined by using spectral karyotyping (SKY) to examine levels and types of chromosomal aberrations in early passage H2AXΔ/Δ ES cells in comparison with H2AXFlox/Δ cells cultured for the same time. SKY chromosomal painting allows unambiguous identification of every mouse chromosome (31). We observed that 17–29% of metaphases from three independently derived H2AXΔ/Δ cells lines (nos. 45, 85, and 91) exhibited chromosomal abnormalities, as opposed to 3–8% of metaphases from H2AXFlox/Δ control cell lines (nos. 40 and 79; Table 1). The types of chromosomal aberrations observed comprised fragments, detached centromeres, fusions, and translocations (Fig. 2B). Stable, high level Cre expression in mouse embryonic fibroblasts can also lead to genomic instability (32, 33). However, in our studies, Cre expression was transient; moreover, increased instability was observed in H2AXΔ/Δ vs. H2AXFlox/Δ lines that were generated from the exact same AdenoCre infections of H2AXFlox/Neo parental cells. AdenoCre-infected H2AXFlox/Flox control cells and other infected or uninfected ES cell lines show baseline instability that, at the resolution of our current assays, was not clearly different from that of H2AXFlox/Δ cells (ref. 34; D.O.F. and J.C.F., unpublished observations). We conclude that early passage H2AXΔ/Δ ES cells exhibited significantly increased spontaneous chromosomal instability compared with controls. However, without additional studies, we cannot rule out a minor effect of the retained loxP sites or of H2AX haploinsufficiency on maintenance of genomic stability.
Table 1.
Increased spontaneous genomic instability in H2AXΔ/Δ ES cells
| Genotype | H2AXFlox/Δ no. 40 | H2AXFlox/Δ no. 79 | H2AXΔ/Δ no. 45 | H2AXΔ/Δ no. 85 | H2AXΔ/Δ no. 91 |
|---|---|---|---|---|---|
| Fragmented chromosomes | 2 | 1 | 0 | 4 | 1 |
| Detached centromeres | 0 | 0 | 2 | 3 | 1 |
| Translocations | 0 | 0 | 4 | 4* | 2 |
| Fusions | 0 | 0 | 0 | 0 | 2 |
| Unusual structures | 0 | 1 | 0 | 0 | 0 |
| Total anomalies (%)† | 2 (7%) | 2 (8%) | 6 (20%) | 11 (46%) | 6 (25%) |
| Total metaphases | 29 | 26 | 30 | 24 | 24 |
| Percentage with abnormalities‡ | 3% | 8% | 17% | 29% | 21% |
One translocation was reciprocal.
The percentage represents events per 100 metaphases.
Percentage of metaphases with at least one aberration.
We also examined karyotypic consequences of exposure to IR in H2AXΔ/Δ, XRCC4−/−, and wild-type ES cells. Early passage ES cells were exposed to 150 rad of IR and allowed to recover for 48 h. SKY analyses revealed a significant increase in the percentage of metaphases with chromosomal abnormalities in the H2AXΔ/Δ and XRCC4−/− cells (70% and 55% of metaphases with at least 1 aberration, respectively) compared with wild-type controls (15% with abnormalities; Table 2). Furthermore, the total number of abnormalities observed in irradiated H2AX-deficient and wild-type ES cells differed by ≈8-fold (1.7 per metaphase vs. 0.2 per metaphase, respectively). XRCC4-deficient ES cells exhibited a similar, albeit lower (≈1.0 per metaphase), level of IR-induced chromosomal aberrations than H2AX-deficient cells (Table 2). After IR treatment, dramatic chromosomal fragmentation, detached centromeres, fusions, and translocations were observed in the H2AX-deficient cells (Fig. 2C). Furthermore, irradiated H2AX-deficient cells exhibited a significant increase in the percentage of metaphases with aneuploidy (83%) in comparison with XRCC4−/− (23%) and wild-type controls (20%) (Fig. 2C).
Table 2.
Increased IR-induced genomic instability in H2AXΔ/Δ ES cells
| Genotype | Wild-type TC1 | XRCC4−/− | H2AXΔ/Δ no. 45 |
|---|---|---|---|
| Fragmented chromosomes | 1 | 7 | 13 |
| Detached centromeres | 1 | 2 | 5 |
| Translocations | 1 | 5 | 10* |
| Fusions | 0 | 3 | 6 |
| Unusual structures | 1 | 4 | 6 |
| Aneuploid (%)† | 4 (20%) | 5 (23%) | 19 (83%) |
| Total anomalies (%)† | 4 (20%) | 21 (95%) | 40 (174%) |
| Total metaphases | 20 | 22 | 23 |
| Percentage with abnormalities‡ | 15% | 55% | 70% |
Forty-eight hours post 150 rad.
One translocation was reciprocal.
The percentage represents events per 100 metaphases.
Percentage of metaphases with at least 1 aberration.
DNA damage results in the activation of cell cycle checkpoints, and cells deficient in checkpoint proteins often exhibit increased chromosomal instability (35). Similar to wild-type ES cells (36, 37), H2AXΔ/Δ ES cells, as well as XRCC4−/− ES cells, arrest at G2 on exposure to 1,000 rad of IR (Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org). Therefore, the H2AX gene and, by inference, the formation of γ-H2AX foci, is not required for the normal DNA damage checkpoint of ES cells. Consequently, the increased genomic instability in H2AX deficient ES cells is most likely due to a DNA repair defect.
H2AX Is Not Required for V(D)J Joining in a Transient Assay.
Deficiencies in all known NHEJ proteins have major effects on the joining of RAG-liberated coding and/or RS ends in the context of transient V(D)J recombination substrates. Thus, to further examine potential roles of H2AX, we examined the ability of H2AX-deficient ES cell lines to support coding and RS joining within extrachromosomal V(D)J recombination substrates by using an established transient assay (28, 38). Two independently derived H2AXΔ/Δ ES cell lines (nos. 45 and 85), as well as two independent H2AXFlox/Δ (nos. 40 and 79), were assayed. V(D)J recombination percentages observed in wild-type, H2AXFlox/Δ, and H2AXΔ/Δ cells were quite similar (Table 3), thus demonstrating that the efficiency of coding and RS joining within plasmid substrates is not significantly impaired in H2AX-deficient ES cells. Structures of coding joins from plasmids recovered from H2AXΔ/Δ ES cells also were indistinguishable from those of wild-type cells (Fig. 5, which is published as supporting information on the PNAS web site). RS joins from H2AX-deficient cells, like those of wild-type ES cells, were precise (Table 3). Thus, H2AX is not required for normal processing and ligating of either blunt or hairpinned, RAG-mediated DSBs in extrachromosomal V(D)J substrates.
Table 3.
Analysis of signal and coding joining in H2AXΔ/Δ ES cells
| Cell lines | (Ampr + Camr)/Ampr | % | Relative level | Fidelity (%) |
|---|---|---|---|---|
| pJH290 (coding joining) | ||||
| Exp. I | ||||
| TC1 | 702/234,500 | 0.3 | 1.0 | |
| H2AXFlox/Δ (no. 40) | 668/188,000 | 0.4 | 1.3 | |
| H2AXFlox/Δ (no. 79) | 760/197,500 | 0.4 | 1.3 | |
| H2AXΔ/Δ (no. 45) | 456/94,000 | 0.5 | 1.7 | |
| H2AXΔ/Δ (no. 85) | 533/109,000 | 0.5 | 1.7 | |
| XRCC4−/− | 0/124,000 | <0.0008 | <0.002 | |
| Exp. II | ||||
| H2AXFlox/Δ (no. 40) | 205/45,200 | 0.5 | 1.0 | |
| H2AXΔ/Δ (no. 43) | 1,268/182,200 | 0.7 | 1.4 | |
| H2AXΔ/Δ (no. 45) | 170/32,200 | 0.6 | 1.2 | |
| XRCC4−/− | 5/85,400 | 0.006 | 0.01 | |
| pJH200 (signal joining) | ||||
| Exp. I | ||||
| TC1 | 1,168/511,000 | 0.2 | 1.0 | ND |
| H2AXFlox/Δ (no. 40) | 1,096/727,000 | 0.2 | 1.0 | ND |
| H2AXFlox/Δ (no. 79) | 1,436/550,000 | 0.3 | 1.5 | ND |
| H2AXΔ/Δ (no. 45) | 633/278,500 | 0.2 | 1.0 | 15/15 (100%) |
| H2AXΔ/Δ (no. 85) | 506/285,000 | 0.2 | 1.0 | 14/14 (100%) |
| XRCC4−/− | 5/578,000 | 0.0009 | 0.0004 | ND |
| Exp. II | ||||
| H2AXFlox/Δ (no. 40) | 210/115,000 | 0.2 | 1.0 | |
| H2AXΔ/Δ (no. 43) | 1,332/425,000 | 0.3 | 1.5 | |
| H2AXΔ/Δ (no. 45) | 184/94,733 | 0.2 | 1.0 | |
| XRCC4−/− | 7/64,533 | 0.01 | 0.05 |
H2AX Is Not Required for Chromosomal V(D)J Recombination.
Mice carrying homozygous inactivating mutations of the NHEJ factors are severely impaired in lymphocyte development because of inability of progenitor lymphocytes to complete V(D)J joining (17). To further examine potential roles of H2AX in V(D)J recombination and lymphocyte development, we examined effects of H2AX deficiency on lymphocyte development via RAG-2-deficient blastocyst complementation (RDBC; ref. 29). Analysis of RAG-deficient chimeras generated from H2AXΔ/Δ ES cells revealed the accumulation of significant numbers of mature (CD4+, CD8+ and B220+, IgM+) peripheral T and B lymphocytes (Fig. 6A, which is published as supporting information on the PNAS web site), although the absolute numbers were significantly reduced compared with those of wild-type mice. However, we cannot make quantitative conclusions regarding the role of H2AX in lymphocyte development because of inherent limitations of RDBC. Thus, H2AX, in marked contrast to all known NHEJ factors, is not required for chromosomal V(D)J recombination or lymphocyte development. We also PCR-amplified and sequenced TCRβ coding joins from an H2AXΔ/Δ thymus and found that they were indistinguishable from those of wild-type (Fig. 6B). Thus, H2AX is not required absolutely for the normal ligation of RAG-liberated DSBs in developing lymphocytes.
Altered IR-Induced BRCA1 and RAD51 Focus Formation in H2AXΔ/Δ ES Cells.
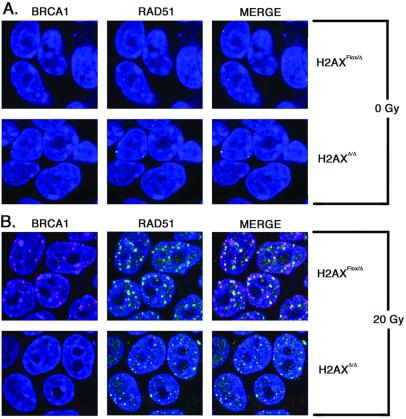
The formation of γ-H2AX foci is a very early event after induction of DSBs and has been proposed to function in recruiting downstream DNA repair factors, including homologous recombination factors BRCA1 and RAD51, to DNA damage sites (18). To assess BRCA1 and RAD51 focus formation in H2AXΔ/Δ ES cells, we examined IR-induced foci by immunofluorescence. In the absence of irradiation, both H2AXFlox/Δ and H2AXΔ/Δ cells exhibited on average one or two BRCA1 and RAD51 foci (Fig. 3A). Within 6 h postirradiation, nearly all wild-type and H2AXFlox/Δ cells had at least five foci per cell (Fig. 3B). Notably, in H2AXΔ/Δ cells, a much more modest induction of BRCA1 foci was observed on irradiation, although, when viewed at higher gain, some focal staining was clearly detectable (Fig. 3B). In contrast, similar numbers of RAD51 foci were observed 6 h postirradiation in H2AXΔ/Δ and H2AXFlox/Δ cells. However, these RAD51 foci appeared smaller in H2AXΔ/Δ cells than H2AXFlox/Δ cells (Fig. 3B). The differences seen in the pattern of RAD51 foci could reflect induction of different subpopulations of RAD51 foci in H2AXΔ/Δ vs. H2AXFlox/Δ cells. Alternatively, the total numbers of RAD51 molecules recruited to individual DNA damage sites may be reduced in H2AXΔ/Δ cells. Regardless, our data indicate that H2AX contributes to BRCA1 and, apparently, RAD51 focus formation responses to IR.
Figure 3.
Formation of IR-induced BRCA1 and RAD51 foci in H2AXΔ/Δ vs. H2AXFlox/Δ ES cells. Immunofluorescence of H2AXFlox/Δ and H2AXΔ/Δ cells unirradiated (A) or 6 h postirradiation with 20 Gy (B) to visualize foci of BRCA1 (rhodamine, red) and RAD51 (FITC, green). Nuclei appear blue (Topro-3). In the merged images, overlapping foci appear yellow.
Discussion
H2AX-Deficient ES Cells Exhibit Genomic Instability.
We have demonstrated a critical role for the H2AX core histone in the cellular response to chromosomal DSBs. H2AX-deficient ES cells have increased IR sensitivity and elevated levels of both spontaneous and IR-induced genomic instability. Because H2AX is not required for the normal IR-induced DNA damage checkpoint, increased genomic instability in H2AX-deficient ES cells is likely due to a DNA repair defect. In S. cerevisiae, the H2A core histone also functions in repair of DSBs (7). In addition, a parallel gene-targeted mutation study reached similar conclusions regarding mammalian H2AX function (39). Therefore, the essential function of a core histone in the cellular response to chromosomal DSBs is conserved throughout evolution, from yeast to mammals. Given the role of H2AX in suppression of genomic instability and its role in recruitment of BRCA1 to DNA damage sites, H2AX also may be predicted to function as a tumor suppressor.
H2AX Is Not Required for Catalysis of V(D)J Recombination.
Absence of H2AX had no effect on V(D)J recombination (either efficiency or quality) within extrachromosomal V(D)J recombination substrates in ES cells and allowed generation of normal V(D)J coding joins in H2AX-deficient thymocytes. Because deficiencies in known NHEJ factors severely impair V(D)J recombination in both contexts, our findings indicate that H2AX does not directly participate in catalysis of this reaction and, by extension, in catalysis of classical NHEJ. Given that H2AX is not directly involved in V(D)J recombination, localization of RAG-dependent γ-H2AX foci to the TCRα locus may function more generally to effect a DNA damage response to RAG-initiated DSBs (16), possibly in the context of ATM-mediated surveillance (40). However, H2AX still may be required to effect efficient NHEJ-mediated repair of other chromosomal DSBs perhaps via accessibility/recruitment functions. In this context, the observations that G1-specific γ-H2AX and NBS foci are linked to the IgH locus during CSR and that CSR is substantially impaired in H2AX−/− lymphocytes (10) clearly implicate H2AX in CSR, a recombination process likely completed by NHEJ.
Function of H2AX in DNA Repair.
We find that H2AX deficiency causes a dramatic decrease in BRCA1 focus formation after IR, and a more subtle decrease in the size of IR-induced RAD51 foci. These effects could reflect inefficient propagation of specialized chromatin tracts around a break, a reduction in the density of chromatin-associated BRCA1 and RAD51 complexes, or both. Therefore, by virtue of its molecular associations, the H2AX protein likely has a role in modulating DNA repair via homologous recombination as well as NHEJ. Consistent with this notion, H2AX-deficient ES cells appear hypersensitive to the cross-linking agent mitomycin C (ref. 39; C.H.B. and H.S., unpublished data), a phenotype shared with ES cells deficient in the homologous recombination factor RAD54 (41), but not NHEJ-deficient ES cells (J.S., unpublished data). Phosphorylation of H2A (in yeast) and, presumably, H2AX (in mammals) causes an alteration in chromatin structure that may facilitate DNA repair (7). Therefore, H2AX may function to promote “accessible” chromatin and thus facilitate the kinetics through which DNA repair factors associate with and repair DSBs. Finally, the IR-dependent physical association between γ-H2AX and 53BP1 (23) suggests that γ-H2AX may also function as a docking site for factors involved in the cellular response to DNA damage. Overall, H2AXΔ/Δ ES cells should provide an important model system for the molecular analysis of the precise nature of H2AX function in the repair of chromosomal DSBs.
Supplementary Material
Acknowledgments
We thank Barbara Woodman and Laurie Davidson for technical assistance, and Drs. Jianzhu Chen, Ronald DePinho, and Andre Nussenzweig for critical review of this manuscript. C.H.B. is a Research Associate of the Howard Hughes Medical Institute. J.S. is a Special Fellow of the Leukemia and Lymphoma Society. K.F.C. is a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research. F.W.A. is an Investigator of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grant AI35714 (F.W.A.), National Cancer Institute (NCI) Grant CA92625 (F.W.A.), and a NCI Howard Temin Award (R.S.).
Abbreviations
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- ATM
ataxia telangiectasia mutated
- ATR
ATM and Rad3-related
- DSB
double-strand break
- NHEJ
nonhomologous end-joining
- HR
homologous recombination
- IR
ionizing radiation
- CSR
class switch recombination
- TCR
T cell receptor
- RAG
recombination-activating gene protein
- RS
recombination signal sequence
- ES
embryonic stem
- SKY
spectral karyotyping
References
- 1.Wolffe A P. Chromatin Structure and Function. New York: Academic; 1998. [Google Scholar]
- 2.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Curr Opin Genet Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 3.Abraham R T. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 4.Hoeijmakers J H. Nature (London) 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 5.Cromie G A, Connelly J C, Leach D R. Mol Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 6.Strahl B D, Allis C D. Nature (London) 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Downs J A, Lowndes N F, Jackson S P. Nature (London) 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 8.Rogakou E P, Pilch D R, Orr A H, Ivanova V S, Bonner W M. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 9.Rogakou E P, Boon C, Redon C, Bonner W M. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen S, Casellas R, Reina-San-Martin B, Chen H T, Difilippantonio M J, Wilson P C, Hanitsch L, Celeste A, Muramatsu M, Pilch D R, et al. Nature (London) 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenter A, Wuerffel R. Ann N Y Acad Sci. 1999;870:206–217. doi: 10.1111/j.1749-6632.1999.tb08880.x. [DOI] [PubMed] [Google Scholar]
- 12.Manis J P, Gu Y, Lansford R, Sonoda E, Ferrini R, Davidson L, Rajewsky K, Alt F W. J Exp Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casellas R, Nussenzweig A, Wuerffel R, Pelanda R, Reichlin A, Suh H, Qin X F, Besmer E, Kenter A, Rajewsky K, Nussenzweig M C. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rolink A, Melchers F, Andersson J. Immunity. 1996;5:319–330. doi: 10.1016/s1074-7613(00)80258-7. [DOI] [PubMed] [Google Scholar]
- 15.Manis J P, Dudley D, Kaylor L, Alt F W. Immunity. 2002;16:607–617. doi: 10.1016/s1074-7613(02)00306-0. [DOI] [PubMed] [Google Scholar]
- 16.Chen H T, Bhandoola A, Difilippantonio M J, Zhu J, Brown M J, Tai X, Rogakou E P, Brotz T M, Bonner W M, Ried T, Nussenzweig A. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassing C H, Swat W, Alt F W. Cell. 2002;109:S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 18.Paull T T, Rogakou E P, Yamazaki V, Kirchgessner C U, Gellert M, Bonner W M. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 19.Burma S, Chen B P, Murphy M, Kurimasa A, Chen D J. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 20.Shiloh Y. Biochem Soc Trans. 2001;29:661–666. doi: 10.1042/0300-5127:0290661. [DOI] [PubMed] [Google Scholar]
- 21.Jeggo P A. Radiat Res. 1998;150:S80–91. [PubMed] [Google Scholar]
- 22.Schultz L B, Chehab N H, Malikzay A, Halazonetis T D. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rappold I, Iwabuchi K, Date T, Chen J. J Cell Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorman J R, van der Stoep N, Monroe R, Cogne M, Davidson L, Alt F W. Immunity. 1996;5:241–252. doi: 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- 25.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 26.Sleckman B P, Bardon C G, Ferrini R, Davidson L, Alt F W. Immunity. 1997;7:505–515. doi: 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- 27.Ferguson D O, Sekiguchi J M, Chang S, Frank K M, Gao Y, DePinho R A, Alt F W. Proc Natl Acad Sci USA. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu Y, Jin S, Gao Y, Weaver D T, Alt F W. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Lansford R, Stewart V, Young F, Alt F W. Proc Natl Acad Sci USA. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scully R, Chen J, Ochs R L, Keegan K, Hoekstra M, Feunteun J, Livingston D M. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 31.Liyanage M, Coleman A, du Manoir S, Veldman T, McCormack S, Dickson R B, Barlow C, Wynshaw-Boris A, Janz S, Wienberg J, et al. Nat Genet. 1996;14:312–315. doi: 10.1038/ng1196-312. [DOI] [PubMed] [Google Scholar]
- 32.Silver D P, Livingston D M. Mol Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 33.Loonstra A, Vooijs M, Beverloo H B, Allak B A, van Drunen E, Kanaar R, Berns A, Jonkers J. Proc Natl Acad Sci USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.d'Adda di Fagagna F, Hande M P, Tong W M, Roth D, Lansdorp P M, Wang Z Q, Jackson S P. Curr Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 35.Dasika G K, Lin S C, Zhao S, Sung P, Tomkinson A, Lee E Y. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt-Kastner P K, Jardine K, Cormier M, McBurney M W. Oncogene. 1998;16:3003–3011. doi: 10.1038/sj.onc.1201835. [DOI] [PubMed] [Google Scholar]
- 37.Hirao A, Kong Y Y, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge S J, Mak T W. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 38.Hesse J E, Lieber M R, Gellert M, Mizuuchi K. Cell. 1987;49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- 39.Celeste A, Petersen S, Romanienko P J, Fernandez-Capetillo O, Chen H T, Sedelnikova O, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio M J, et al. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perkins E J, Nair A, Cowley D O, Van Dyke T, Chang Y, Ramsden D A. Genes Dev. 2002;16:159–164. doi: 10.1101/gad.956902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Essers J, Hendriks R W, Swagemakers S M, Troelstra C, de Wit J, Bootsma D, Hoeijmakers J H, Kanaar R. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.