Abstract
This manuscript details protocols for the ZnCl2 precipitation-assisted sample preparation (ZASP) for proteomic analysis. By inducing protein precipitation with ZASP precipitation buffer (ZPB, final concentration of ZnCl2 at 100 mM and 50% methanol), ZASP depletes harsh detergents and impurities, such as sodium dodecyl sulfate (SDS), Triton X-100, and urea, at high concentrations in solution from protein solutions prior to trypsin digestion. It is a practical, robust, and cost-effective approach for proteomic sample preparation. It has been observed that 90.2% of the proteins can be recovered from lysates by incubating with an equal volume of ZPB at room temperature for 10 min. In 1 h of data-dependent acquisition (DDA) analysis on an Exploris 480, 4,037 proteins and 25,626 peptides were quantified from 1 μg of mouse small intestine proteins, reaching a peak of 4,500 proteins and up to 30,000 peptides with 5 μg of input. Additionally, ZASP outperformed other common sample preparation methods such as sodium deoxycholate (SDC)-based in-solution digestion, acetone precipitation, filter-aided sample preparation (FASP), and single-pot, solid-phase-enhanced sample preparation (SP3). It demonstrated superior performance in protein (4,456 proteins) and peptide identification (29,871 peptides), lower missing cleavage rates (16.3%), and high reproducibility (Pearson correlation coefficients of 0.96 between replicates) with similar protein distributions and cellular localization patterns. Significantly, the cost of ZASP per sample with 100 μg of protein as input is lower than 30 RMB, including the expense of trypsin.
Key features
• ZASP is a user-friendly method that enables efficient, sensitive, and cost-effective proteomic analysis in a common biochemistry lab through simple incubation and centrifugation.
• ZASP enables 90% protein recovery from lysates with various trypsin and LC–MS-incompatible reagents by incubating at room temperature for 10 min.
• ZASP enables unbiased proteomic analysis of diverse biological samples, including cells, optimal cutting temperature (OCT)-embedded tissues, and formalin-fixed paraffin-embedded (FFPE) tissues.
Keywords: Sample preparation, LC–MS-incompatible reagents, ZnCl2 induced protein precipitation, Protein recovery, Proteomics.
Graphical overview

Background
Sample preparation is a critical step in proteomics analysis, including protein extraction, denaturation, reduction, alkylation, and digestion, followed by peptide purification. The quality of sample preparation directly impacts the reproducibility, accuracy, and sensitivity of LC–MS analysis [1,2]. Protein extraction is usually enhanced by adding chaotropes and detergents, such as SDS [3,4]. However, most of these additives impair subsequent protein enzymatic digestion and LC–MS analysis, thereby compromising the reliability and sensitivity of the LC–MS results [5,6]. Multiple methods, including affinity-based approaches [7], membrane filtration [8], and protein precipitation [9–11], have been developed to remove them. However, each method has its limitations, such as being labor-intensive or time-consuming, resulting in sample loss, high cost, or low throughput [8,10–14]. We aim to develop a cost-effective, simple, and widely applicable sample preparation technology to resolve these issues and enable high-quality proteomics analysis.
Zn2+-based precipitation methods have been commonly used for the initial fractionation of proteins based on their solubility, such as in the purification of antibodies from cell culture broth [15,16]. ZnSO4 and acetone aqueous solutions have been reported to rapidly recover peptides generated by trypsin and pepsin digestion prior to MS analysis [17]. Zn2+ may bind to histidine and cysteine residues on the surface of proteins via exposed imidazole and thiol groups to form relatively stable complexes [18]. Additionally, as a member of the Hofmeister series, Zn2+ can cause protein precipitation by altering the solubility of peptides and proteins in organic solvents [19–21]. Because of the stronger electrostatic interactions, Zn2+ is superior to other ions (e.g., Mg2+) in precipitating proteins from solutions [22].
Inspired by these studies, we recently developed a novel sample preparation method based on ZnCl2 and methanol-induced protein precipitation with in-solution digestion, termed ZnCl2 precipitation-assisted sample preparation (ZASP) [23]. ZASP achieves efficient recovery of proteins (recovery rate >90%) from lysates containing different trypsin- or LC–MS-incompatible detergents by incubating with an equal volume of ZASP precipitation buffer (ZPB) for 10 min at room temperature. We have demonstrated it as an efficient approach by comparing it with current popular sample preparation methods (e.g., FASP, SP3, SDC-based in-solution digestion) and profiling of different types of samples. The protocol for ZASP is described in this manuscript.
Materials and reagents
Biological materials
1. HEK 293T cells (ATCC, catalog number: CRL-3216)
2. Mouse tissues (C57BL/6) (obtained from The Beijing Proteome Research Center)
3. Yeast (ATCC, catalog number: 204508)
4. E. coli (ATCC, catalog number: 25922)
Reagents
1. Tris(hydroxymethyl)aminomethane (Tris) LC–MS grade (Pierce, catalog number: 17926) (keep in a dry, cool, and well-ventilated place)
2. Tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl) ≥98% (Sigma-Aldrich, catalog number: 646547) (store at 4 °C)
3. 2-Chloroacetamide (CAA) ≥99% (Sigma-Aldrich, catalog number: C0267) (keep in a dry, cool, and well-ventilated place)
4. Trypsin, MS grade (Promega, catalog number: 90057) (store at -20 °C)
5. Formic acid, ≥98% (Sigma-Aldrich, catalog number: 33015) (keep container tightly closed in a dry and well-ventilated place)
6. Acetonitrile (ACN) (Thermo Scientific, catalog number: A955-4) (keep container tightly closed in a dry and well-ventilated place)
7. Sodium dodecyl sulfate (SDS) (Solarbio, catalog number: S8010) (keep container tightly closed in a dry and well-ventilated place)
8. Zinc chloride (ZnCl2) ≥98% (Sigma-Aldrich, catalog number: 208086) (keep container tightly closed in a dry and well-ventilated place)
9. Methanol, LC–MS grade) (Sigma-Aldrich, catalog number: 1.06035) (keep container tightly closed in a dry and well-ventilated place)
10. Pierce BCA Protein Assay kit (Thermo Scientific, catalog number: 23227) (keep in a dry, cool, and well-ventilated place)
11. Sodium deoxycholate (SDC) ≥97% (Sigma-Aldrich, catalog number: D6750) (keep container tightly closed in a dry and well-ventilated place)
12. Dulbecco's modified Eagle's medium (DMEM) (Gibco, catalog number: C11995500BT) (store at 4 °C)
13. Fetal bovine serum (FBS) (NEWZERUM, catalog number: FBS-CS500) (store at -20 °C)
14. Penicillin-streptomycin (Gibco, catalog number: 15140122) (store at 4 °C)
15. Trypsin-EDTA solution (Gibco, catalog number: 25300054) (store at 4 °C)
16. Phosphate-buffered saline (PBS) (Gibco, catalog number: C10010500BT) (store at 4 °C)
17. Yeast extract peptone dextrose media (YEPD) (Sigma-Aldrich, catalog number: Y1500) (keep container tightly closed in a dry and well-ventilated place)
18. Luria-Bertani medium (LB) (Sigma-Aldrich, catalog number: L2542) (store at 4 °C and keep away from light)
Solutions
1. SDS lysis buffer (see Recipes)
2. ZASP precipitation buffer (see Recipes)
3. SDC lysis buffer (see Recipes)
4. Buffer A (see Recipes)
5. Buffer B (see Recipes)
Recipes
1. SDS lysis buffer
4% SDS (w/v) and 100 mM Tris-HCl in water at pH 8.5: mix 20 mL of 10% (w/v) SDS solution stock with 5 mL of 1 M Tris, followed by the addition of 25 mL of H2O.
2. ZASP precipitation buffer (ZPB)
Methanol consisting of 0.1% formic acid and 200 mM ZnCl2 (v/v): dissolve 1,363 mg of ZnCl2 and 50μL of formic acid in 50mL of methanol.
Note: ZnCl2 is hygroscopic and should be stored in a tightly sealed container under dry conditions. Formic acid is corrosive and should be handled with appropriate protective equipment (gloves, lab coat, and eye protection) in a fume hood.
3. SDC lysis buffer
1% SDC (w/v) and 100 mM Tris-HCl in water at pH 8.5: mix 30mL of 5% (w/v) SDC solution with 15mL of 1M Tris-HCl, then add 90mL of H2O.
4. Buffer A
0.1% formic acid aqueous solution: mix 10 μL of formic acid with 10mL of H2O.
5. Buffer B
60% acetonitrile solution containing 0.1% formic acid: mix 6 mL of acetonitrile with 4 mL of H2O, followed by the addition of 10 μL of formic acid.
Laboratory supplies
1. C18 solid phase extraction (SPE) disks (3M Empore, catalog number: 98060402173)
2. C18 powder (Durashell, catalog number: DC930010-L)
3. 0.5 mL centrifuge tubes (Corning, catalog number: 3208)
4. 1.5 mL centrifuge tubes (Corning, catalog number: 3620)
5. 2.0 mL centrifuge tubes (Corning, catalog number: 3213)
6. 10 μL pipette tips (Axygen, catalog number: T-300-L)
7. 200 μL pipette tips (Axygen, catalog number: T-200-C-L)
8. 1,000 μL pipette tips (Axygen, catalog number: T-1,000-C)
9. Cell culture plates (Nunc EasyDish, catalog number: 150468)
Equipment
1. Sonicator Chromatin and DNA Shearing System (Qsonica, catalog number: Q800R3)
2. SpeedVac Integrated Vacuum Concentrator (Thermo Scientific, catalog number: SPD1030A-230)
3. Tissue homogenizer (OMNI, model: Bead Ruptor 24)
4. Centrifuge 5424R (Eppendorf, catalog number: 5404000090)
5. Research® Plus pipettes (0.1–2.5 μL) (Eppendorf, catalog number: 3123000217)
6. Research® Plus pipettes (2–20 μL) (Eppendorf, catalog number: 3123000292)
7. Research® Plus pipettes (20–200 μL) (Eppendorf, catalog number: 3123000250)
8. Research® Plus pipettes (100–1,000 μL) (Eppendorf, catalog number: 3123000268)
9. ThermoMixer® C (Eppendorf, catalog number: 5382 000015)
10. Freezer (-20 °C) (Haier, model: DW-25L262)
11. Freezer (-80 °C) (Thermo Scientific, catalog number: 907)
12. CO2 incubator (Thermo Scientific, catalog number: 3111)
13. Shaker (Jingda, model: DHZ-DA)
14. UV/Vis Spectrophotometer (Thermo Scientific)
15. Microplate Reader (BioTek, model: Synergy LX)
16. Orbitrap Exploris 480 Mass Spectrometer (Thermo Scientific, catalog number: BRE725533)
Software and datasets
1. DDA data analysis software (MaxQuant, v.2.0.1.0)
2. Fasta files were downloaded from the Swiss-Prot database, containing entries for human (20387), mouse (17098), Saccharomyces cerevisiae (strain ATCC 204508/S288c, Baker's yeast, 6729), and E. coli (strain K12, 4529) sequences, released in April 2021.
Procedure
A. Sample collection
1. HEK 293T cells
a. Culture HEK 293T cells in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin at 37 °C in an environment with 5% CO2 in a 10 cm cell culture dish.
b. Harvest the cells when they reach 80% confluence.
c. Discard the medium. Add 1 mL of 0.05% trypsin-EDTA solution and incubate for 1 min to detach cells.
d. Add 1 mL of DMEM supplemented with 10% FBS and 1% penicillin-streptomycin to terminate the digestion reaction. Centrifuge the cell suspension at 1,000× g for 5 min and remove the supernatant.
e. Wash the cells three times with PBS: add 5–10 mL of PBS to the cells, gently pipetting to create a suspension. Then, centrifuge the cell suspension at 1,000× g for 5 min and remove the supernatant. Repeat this a total of three times.
f. Store the cell pellets at -80 °C until further processing.
2. Yeast cells
a. Cultivate yeasts in YEPD media at 30 °C shaking at 200 rpm in a DHZ-DA shaker.
b. Collect the cells from a 5–10 mL culture by centrifugation at 4,000× g for 5 min when the optical density at 600 nm (OD600) reaches approximately 1.0. Use a UV/Vis spectrophotometer to measure optical density.
c. Wash the cells three times with PBS buffer: add 5–10 mL of PBS to the cells, gently pipetting to create a suspension. Then, centrifuge the cell suspension at 4,000× g for 5 min and remove the supernatant. Repeat this a total of three times.
d. Store the cell pellets at -80 °C until further processing.
3. E. coli
a. Grow E. coli cells in LB medium at 37 °C overnight with vigorous shaking in a DHZ-DA shaker.
b. Harvest the cells from a 5–10 mL culture during the stationary phase (OD600 between 0.6 and 1) by centrifugation at 4,000× g for 10 min.
c. Wash the cells twice with PBS buffer: add 5–10 mL of PBS to the cells, gently pipetting to create a suspension. Then, centrifuge the cell suspension at 4,000× g for 5 min and remove the supernatant. Repeat this step twice.
d. Store the cell pellets at -80 °C until further processing.
B. Protein extraction
1. Add SDS lysis buffer to the cell pellets or tissues and boil at 95 °C for 10 min to denature the proteins. The volume ratio of cell pellets to lysis buffer is 1:10.
2. For tissues, yeast cells, and E. coli cells, homogenize samples using an OMNI Bead Ruptor 24 Elite instrument (6.6 m/s, 3 cycles, 30 s on, 15 s off) after being cooled in ice water.
3. Sonicate the lysates or HEK 293T cell suspension for 10 min using a Qsonica Q800R3 sonicator (85% power, 10 s on, 10 s off).
4. Quantify protein concentrations by measuring absorbance at 562 nm using a BioTek Synergy LX Multimode Reader, employing the Pierce BCA Protein Assay kit following the manufacturer’s instructions.
5. Aliquot the lysates to 100 μg of protein and store at -80 °C until further use.
C. Protein precipitation
1. Sample volumes are adjusted based on the amount of input protein using SDS lysis buffer: 50 μL for 1–10 μg, 100 μL for 20–100 μg, 200 μL for 500 μg, and 500 μL for 1,000 μg of input protein.
2. Add an equal volume of ZPB to each sample, mixing gently by pipetting. Let the mixture incubate at room temperature for 10 min. For the precipitation step, ensure that the final solution has a concentration of 50% methanol and 100 mM ZnCl2.
Note: Cloudiness occurred during incubation for protein inputs exceeding 5 μg.
3. Collect protein pellets by centrifugation at 19,000× g for 5 min at 4 °C. Protein particles are not visible for inputs under 5 μg.
D. Washing protein pellets
1. Wash the protein pellets twice with the same volume of freshly prepared methanol containing 0.1% formic acid as the ZPB, assisted by sonication using a Qsonica Q800R3 sonicator (85% power, 10 s on, 10 s off, 1 min).
2. Evaporate in the air for at least 10 min at room temperature to ensure complete removal of residual formic acid.
E. Resuspension proteins for in-solution digestion
1. Add an appropriate volume of SDC lysis buffer (e.g., 50μL for 1–10μg, 100μL for 20–100μg, 200μL for 500μg, or 500μL for 1,000μg of input protein) to resuspend the resulting clean protein pellet and sonicate for 1 min (85% power, 10 s on, 10 s off) to aid solubilization.
2. Optional: To calculate the recovery rate of proteins, protein concentrations can be measured using a BCA assay to determine the amount of resuspended protein.
3. Add TCEP stock (pH 7) to a final concentration of 10 mM and CAA stock to a final concentration of 40 mM and incubate for 10 min at 65 °C for reduction and alkylation.
F. In-solution digestion
1. Add trypsin at an enzyme-to-protein ratio of 1:30 and digest at 37 °C for 14 h.
2. After digestion, add formic acid to the sample until the pH reaches 2–3 to terminate the digestion reaction.
G. Preparation of C18 StageTips
1. For inputs under 20 μg, fill the C18 StageTip with four layers of C18 solid phase extraction membrane with a punch.
2. For inputs between 20 and 100 μg, fill the C18 StageTip with a C18 solid phase extraction membrane and C18 powder. Weigh the C18 powder (C18 powder is a hydrophobic, reverse-phase silica-based material with a particle size of 3μm and a pore size of 150Å). Add 200 μL of 100% acetonitrile to reconstitute the C18 suspension. Next, transfer the C18 suspension into 200 μL tips containing one layer of the C18 disk. The volume of C18 suspension is calculated based on the amount of protein input corresponding to the mass of C18. For reference, 3 mg of C18 powder is suitable for 100 μg of protein. After centrifuging to remove the acetonitrile, store the tips in a tip box until use. The details of making C18 StageTips are described in Video 1.
Video 1. Step-by-step guide for the manufacturing of C18 StageTips.

Note: The C18 StageTip can be kept in a tip box at room temperature for several months. Although we recommend preparing it one day before use to avoid any issues during storage, this is not strictly necessary. In addition to the homemade C18 StageTip, commercial C18 columns (e.g., PierceTM C18 Tips for 80 μg of total peptide, catalog number: 87784) can be used for desalting.
3. For higher protein input, Sep-Pak C18 cartridges are a good choice.
H. Desalting
1. For StageTips packed with C18 powder, centrifuge tips at 1,000× g for 45 s at 4 °C before use.
2. Wetting StageTips: Transfer 100% acetonitrile to C18 StageTips and centrifuge at 1,000× g for 2 min to remove acetonitrile. Add buffer B to StageTips and centrifuge at 1,000× g for 2 min at 4 °C.
3. Pre-equilibrium StageTips: Add buffer A to StageTips and centrifuge at 1,500× g for 2 min at 4 °C. Repeat this step 2 times.
4. Load the tryptic peptides: Transfer the digest to StageTips and centrifuge at 1,000× g for around 1.5 min at 4 °C. Collect the flowthrough, transfer it to the StageTips, and then centrifuge at 1,000× g for around 1.5 min at 4 °C.
5. Washing: Transfer buffer A to the StageTips and centrifuge at 1,500× g for 2 min. Repeat this step once.
6. Elution: Transfer buffer B to StageTips. Centrifuge the tips at 1,000× g until they are fully dried (approximately 2 min) at 4 °C.
7. Dry down the elutes and store them at -20 °C before LC–MS analysis. This desalting procedure was performed following established protocols as previously described [24,25]. The detailed volume of buffers added at each step for desalting using different C18 StageTips and cartridges is listed in Table 1.
Table 1. Volume of buffers added during each step of the desalting process.
| Wetting with acetonitrile | Wetting with buffer B | Pre-equilibrium with buffer A | Loading samples | Flowthrough | Washing with buffer A | Eluting with buffer B | |
|---|---|---|---|---|---|---|---|
| Homemade C18 StageTip | 1×250 μL | 1×250 μL | 2×250 μL | 1× | 1× | 2×250 μL | 1×100 μL |
| Sep-Pak C18 cartridge | 3×1 mL | 3×1 mL | 1× | 1× | 5×1 mL | 1×0.3 mL |
Notes:
1. Load samples slowly on the StageTip column.
2. Check how long each step of centrifugation will take if it is your first time doing it.
3. Do not let the C18 become fully dried (paper white) before elution.
LC–MS/MS analysis
We analyzed the peptides using a 60-min data-dependent acquisition method with an UltiMateTM 3000 RSLC nanoliquid chromatography system (Thermo Fisher Scientific), coupled to an Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific). Further details can be found in our original work, published in Molecular & Cellular Proteomics [23]. The samples can also be measured using other types of mass spectrometers and methods. Our work primarily focuses on sample preparation.
Data analysis
The raw files were processed via MaxQuant software (version 2.0.1.0). Protein sequences retrieved from the SwissProt database (released in April 2021), encompassing entries for humans (20,387), mouse (17,098), S. cerevisiae (strain ATCC 204508/S288c, Baker's yeast. 6729), and E. coli (strain K12, 4529) sequences, were utilized. Trypsin/P was specified as the proteolytic enzyme with a maximum of two missed cleavage sites. Cysteine carboxamidomethylation was set as a fixed modification, while methionine oxidation and protein N-terminal acetylation were considered variable modifications.
Protein recovery rate was calculated using the following formula: Protein recovery rate (%) = (precipitated protein amount)/(protein amount in lysate) × 100. The precipitated protein amount = protein concentration × volume of resuspension buffer. Protein concentrations were measured using a Pierce BCA Protein Assay kit (Thermo Scientific). To evaluate digestion efficiency, missed cleavages were assessed. The number of missed enzymatic cleavages is reported in the "Evidence.txt" file generated by MaxQuant. The missed cleavage rate was calculated as follows: Missed cleavage rate (%) = (number of peptides with missed cleavages) / (total number of identified peptides) × 100. The quality of the LC–MS data was assessed based on the shape of the total ion current (TIC) and base peak, the number of MS1 and MS2 scans, and the signals from these scans, along with the results of the database search. During method development, optimization, and evaluation phases, three technical replicates were performed in parallel. The comparison between ZASP and conventional methods focused on the number of identified peptides and proteins, the missed cleavage rate, the correlation between replicates, and the characteristics of identified proteins. Statistical analyses were conducted using two-sided t-tests, with the FDR calculated via the Benjamini-Hochberg method (significance cutoff: 5%).
Validation of protocol
During protocol optimization, the combination of 100 mM ZnCl2 and 50% methanol (final concentration) was determined to be optimal, achieving the highest protein recovery rate (90.2%) and yielding the most protein and peptide identifications (Figure 1). To assess analytical performance, ZASP was further evaluated using a range of protein input amounts (1–1,000 μg). Remarkably, over 4,000 proteins could be reliably identified from as little as 1 μg of input, with low peptide-level variation (median CV < 20%) and strong reproducibility across replicates and input levels (Figure 2). In addition, the TIC, MS1, and MS2 signal intensities obtained from ZASP were comparable to those from the conventional in-solution digestion, indicating equivalent chromatographic performance and ionization efficiency (Figure 3). Finally, comparative analysis demonstrated that ZASP outperformed conventional sample preparation methods—including acetone precipitation, FASP, and SP3—in both the depth of protein and peptide identifications and lower missed cleavage rates (Figure 4) [23].
Figure 1. Development and optimization of ZnCl2 precipitation-assisted sample preparation (ZASP).
(A) Protein recovery associated with various ZnCl2 and methanol mixtures. (B) Enumeration of proteins and peptides identified in HEK 293T cells treated with diverse ZnCl2 and methanol combinations. For comparative analyses of protein and peptide identifications, only those without missing values were considered. AC and MT denote acetone and methanol, respectively. For each condition, three technical replicates were conducted. The bar plots show the average protein recovery rates and the average number of identified proteins, with error bars indicating the standard deviation.
Figure 2. Performance evaluation of ZnCl2 precipitation-assisted sample preparation (ZASP).
(A) The number of proteins identified in mouse small intestinal tissue with protein inputs ranging from 1 μg to 1,000 μg. For each condition, three technical replicates were performed to determine the average number of identified proteins. The error bars represent the standard deviation of these identifications. (B) Ranked plots illustrating protein abundance in samples with 1 μg and 1,000 μg of protein input. (C) Coefficients of variation for peptides across samples with varying protein inputs. (D) Pearson correlation coefficients for protein identification in samples with 1 μg and 1000 μg of protein input.
Figure 3. Comparison of total ion current (TIC), base peak (MS1), and MS2 profiles between in-solution digestion (ISD) and ZnCl2 precipitation-assisted sample preparation (ZASP).
All the figures above are reprinted/adapted from Shao et al. [23]. © The Authors. Published by Elsevier Inc. on behalf of the American Society for Biochemistry and Molecular Biology. Distributed under a Creative Commons Attribution License 4.0 (CC BY) http://creativecommons.org/licenses/by/4.0/.
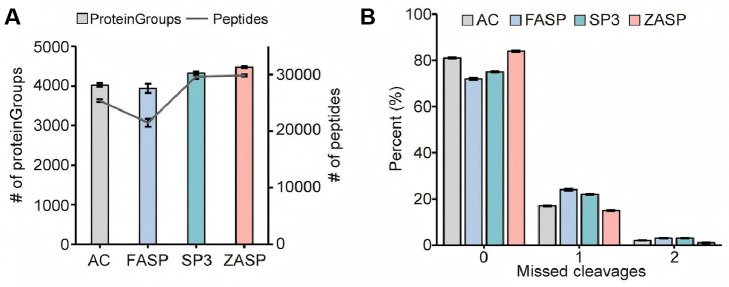
Figure 4. Comparative analysis of ZnCl2 precipitation-assisted sample preparation (ZASP) and other detergent-depletion sample preparation techniques.
(A) Number of proteins and peptides identified using acetone precipitation (AC), filter-aided sample preparation (FASP), solid-phase-enhanced sample preparation (SP3), and ZASP methods. (B) Missed cleavage rates of peptides identified by each of the four methods. For each method, three technical replicates were performed. The bar plots show the average number of identified proteins and the average missed cleavages in three replicates, with error bars indicating the standard deviation.
The protocol was published and validated in [23].
General notes and troubleshooting
1. Adjust the solvent volumes based on the protein input; use smaller volumes for lower protein amounts and larger volumes for higher input.
2. After precipitation, proteins can be resolubilized in buffers like 1% SDC lysis buffer or other suitable buffers, depending on the analysis requirements.
3. For the reduction and alkylation of proteins, it is recommended to use a TCEP solution with a pH of 7. The TCEP solution, when prepared by dissolving TCEP powder in water, tends to be strongly acidic. This acidity can negatively impact the resolubilization of protein pellets in SDC buffer.
4. Choosing and preparing C18 StageTips depend on the input of proteins. If protein quantity is lower than 20 μg, it is recommended to use C18 StageTips packed with a C18 membrane. For protein amounts between 20 and 100 μg, homemade StageTips filled with C18 powder are a cost-effective option. For higher protein input, Sep-Pak C18 cartridges are a good choice.
5. Commercial tips and columns can also be used instead of homemade C18 StageTips.
6. While standard plasticware was used in this protocol, users working with sub-microgram protein amounts or, particularly, adsorption-sensitive samples, may consider using certified low-binding tubes and tips to further reduce potential sample loss.
7. To ensure reliable performance, all key instruments—particularly high-speed centrifuges and sonicators—should be regularly calibrated and maintained according to the manufacturer's specifications.
Acknowledgments
We would like to thank all the members of the He laboratory at the National Center for Protein Science-Beijing and our colleagues at the π-Hub Infrastructure. This work was supported by the China National Key R&D Program (Grant No.2021YFA1301601), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2019-I2M-5-063), and the Major Project of Guangzhou National Laboratory (Grant No. GZNL2023A01004). This protocol was used in our original study [23].
Competing interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. Scheerlinck E., Dhaenens M., Van Soom A., Peelman L., De Sutter P., Van Steendam K. and Deforce D.(2015). Minimizing technical variation during sample preparation prior to label-free quantitative mass spectrometry. Anal Biochem. 490: 14 19 19. 10.1016/j.ab .2015.08.018 [DOI] [PubMed] [Google Scholar]
- 2. Mottaz-Brewer H. M., Norbeck A. D., Adkins J. N., Manes N. P., Ansong C., Shi L., Rikihisa Y., Kikuchi T., Wong S. W., Estep R. D., et al.(2008). Optimization of proteomic sample preparation procedures for comprehensive protein characterization of pathogenic systems. J Biomol Tech. 19(5): 285 295 295 . https://pubmed.ncbi.nlm.nih.gov/19183792/ [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou J. Y., Dann G. P., Shi T., Wang L., Gao X., Su D., Nicora C. D., Shukla A. K., Moore R. J., Liu T., et al.(2012). Simple Sodium Dodecyl Sulfate-Assisted Sample Preparation Method for LC-MS-Based Proteomics Applications. Anal Chem. 84(6): 2862 2867 2867. 10.1021/ac203394r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doellinger J., Schneider A., Hoeller M. and Lasch P.(2020). Sample Preparation by Easy Extraction and Digestion(SPEED)- A Universal, Rapid, and Detergent-free Protocol for Proteomics Based on Acid Extraction. Mol Cell Proteomics. 19(1): 209 222 222. 10.1074/mcp.tir119 .001616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ma H., Zou T., Li H. and Cheng H.(2020). The interaction of sodium dodecyl sulfate with trypsin: Multi-spectroscopic analysis, molecular docking, and molecular dynamics simulation. Int J Biol Macromol. 162: 1546 1554 1554. 10.1016/j.ijbiomac .2020.08.020 [DOI] [PubMed] [Google Scholar]
- 6. Rundlett K. L. and Armstrong D. W.(1996). Mechanism of Signal Suppression by Anionic Surfactants in Capillary Electrophoresis−Electrospray Ionization Mass Spectrometry. Anal Chem. 68(19): 3493 3497 3497. 10.1021/ac960472p [DOI] [PubMed] [Google Scholar]
- 7. Gan G., Xu X., Chen X., Zhang X. F., Wang J. and Zhong C. Q.(2021). SCASP: A Simple and Robust SDS-Aided Sample Preparation Method for Proteomic Research. Mol Cell Proteomics. 20: 100051 10.1016/j.mcpro .2021.100051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wiśniewski J. R., Zougman A., Nagaraj N. and Mann M.(2009). Universal sample preparation method for proteome analysis. Nat Methods. 6(5): 359 362 362. 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- 9. Hughes C. S., Moggridge S., Müller T., Sorensen P. H., Morin G. B. and Krijgsveld J.(2018). Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat Protoc. 14(1): 68 85 85. 10.1038/s41596-018-0082-x [DOI] [PubMed] [Google Scholar]
- 10. Méchin V., Damerval C. and Zivy M.(2007). Total protein extraction with TCA-acetone. Methods Mol Biol. 355: 1 8 8. 10.1385/1-59745-227-0 :1 [DOI] [PubMed] [Google Scholar]
- 11. Hughes C. S., Foehr S., Garfield D. A., Furlong E. E., Steinmetz L. M. and Krijgsveld J.(2014). Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. 10(10): e20145625. 10.15252/msb.20145625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duong V. A. and Lee H.(2023). Bottom-Up Proteomics: Advancements in Sample Preparation. Int J Mol Sci. 24(6): 5350 10.3390/ijms24065350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosenfeld J., Capdevielle J., Guillemot J. C. and Ferrara P.(1992). In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 203(1): 173 179 179. 10.1016/0003-2697 (92)90061-b [DOI] [PubMed] [Google Scholar]
- 14. Zougman A., Selby P. J. and Banks R. E.(2014). Suspension trapping(STrap) sample preparation method for bottom‐up proteomics analysis. Proteomics. 14(9): 1006 1000 1000. 10.1002/pmic.201300553 [DOI] [PubMed] [Google Scholar]
- 15. Li Z., Gu Q., Coffman J. L., Przybycien T. and Zydney A. L.(2019). Continuous precipitation for monoclonal antibody capture using countercurrent washing by microfiltration. Biotechnol Progr. 35(6): e2886. https://doi.org/ 10.1002/btpr.2886 [DOI] [PubMed] [Google Scholar]
- 16. Gu Q., Li Z., Coffman J. L., Przybycien T. M. and Zydney A. L.(2020). High throughput solubility and redissolution screening for antibody purification via combinedPEGand zinc chloride precipitation. Biotechnol Progr. 36(6): e3041. https://doi.org/ 10.1002/btpr.3041 [DOI] [PubMed] [Google Scholar]
- 17. Baghalabadi V., Razmi H. and Doucette A.(2021). Salt-Mediated Organic Solvent Precipitation for Enhanced Recovery of Peptides Generated by Pepsin Digestion. Proteomes. 9(4): 44 10.3390/proteomes9040044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang T., Cleland J. L., Lam X., Meyer J. D., Jones L. S., Randolph T. W., Manning M. C. and Carpenter J. F.(2000). Effect of zinc binding and precipitation on structures of recombinant human growth hormone and nerve growth factor. J Pharm Sci. 89(11): 1480 1485 1485. 10.1002/1520-6017 (200011)89:11<1480::aid-jps10>3.0.co;2-m [DOI] [PubMed] [Google Scholar]
- 19. Melander W. and Horváth C.(1977). Salt effects on hydrophobic interactions in precipitation and chromatography of proteins: An interpretation of the lyotropic series. Arch Biochem Biophys. 183(1): 200 215 215. 10.1016/0003-9861 (77)90434-9 [DOI] [PubMed] [Google Scholar]
- 20. Baghalabadi V. and Doucette A. A.(2020). Mass spectrometry profiling of low molecular weight proteins and peptides isolated by acetone precipitation. Anal Chim Acta. 1138: 38 48 48. 10.1016/j.aca .2020.08.057 [DOI] [PubMed] [Google Scholar]
- 21. Schwierz N., Horinek D., Sivan U. and Netz R. R.(2016). Reversed Hofmeister series—The rule rather than the exception. Curr Opin Colloid Interf Sci. 23: 10 18 18. 10.1016/j.cocis .2016.04.003 [DOI] [Google Scholar]
- 22. Flores S. C., Kherb J., Konelick N., Chen X. and Cremer P. S.(2012). The Effects of Hofmeister Cations at Negatively Charged Hydrophilic Surfaces. J Phys Chem. 116(9): 5730 5734 5734. 10.1021/jp210791j [DOI] [Google Scholar]
- 23. Shao X., Huang Y., Xu R., He Q., Zhang M., He F. and Wang D.(2024). ZASP: A Highly Compatible and Sensitive ZnCl2 Precipitation-Assisted Sample Preparation Method for Proteomic Analysis. Mol Cell Proteomics. 23(10): 100837 10.1016/j.mcpro .2024.100837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kulak N. A., Pichler G., Paron I., Nagaraj N. and Mann M.(2014). Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 11(3): 319 324 324. 10.1038/nmeth.2834 [DOI] [PubMed] [Google Scholar]
- 25. Ruprecht B., Zecha J., Zolg D. P. and Kuster B.(2017). High pH Reversed-Phase Micro-Columns for Simple, Sensitive, and Efficient Fractionation of Proteome and(TMT labeled) Phosphoproteome Digests. Methods Mol Biol. 1550: 83 98 98. 10.1007/978-1-4939-6747-6_8 [DOI] [PubMed] [Google Scholar]